Expression Mapping and Functional Analysis of Orphan G-Protein-Coupled Receptor GPR158 in the Adult Mouse Brain Using a GPR158 Transgenic Mouse
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Gpr158-3×Flag-TeV-HA-T2A-tdTomato and Gpr158−/− Mice
2.2. Animals and Behavioral Tests
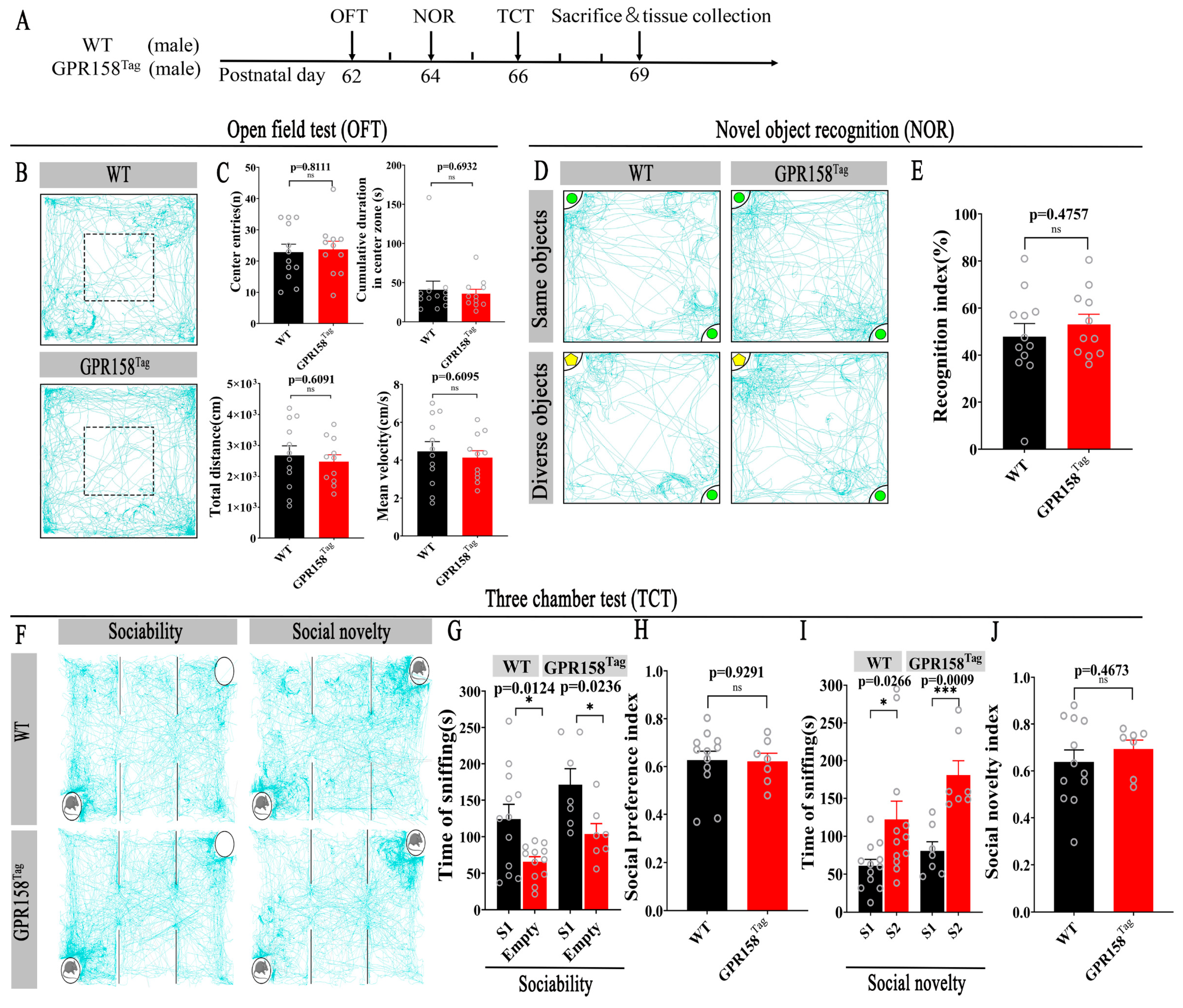
2.3. Open Field Test (OFT)
2.4. Three-Chamber Social Interaction Test
2.5. Novel Object Recognition Test
2.6. Immunofluorescence Staining
2.7. Fluorescent Images Quantification
2.7.1. Localization and Quantification of tdTomato+ Neurons in Various Brain Regions in the Sagittal Plane of the Brain of Gpr158Tag Mice Brains
2.7.2. Co-Localization of Subcells and Synapses in 3D tdTomato+ Cells
2.8. Co-Immunoprecipitation and Protein Mass Spectrometry Identification
2.9. RNA Sequencing (RNA-seq)
2.10. Statistical Analysis
3. Results
3.1. Generation of GPR158Tag Mice
3.2. GPR158Tag Mice Displayed No Deficits in Motor and Motor and Cognitive Behavior or Social Behavior and Social Memory, and No Anxiety-like Behavior
3.3. Expression Pattern of tdTomato-Labeled GPR158+ Cells in Mouse Brain
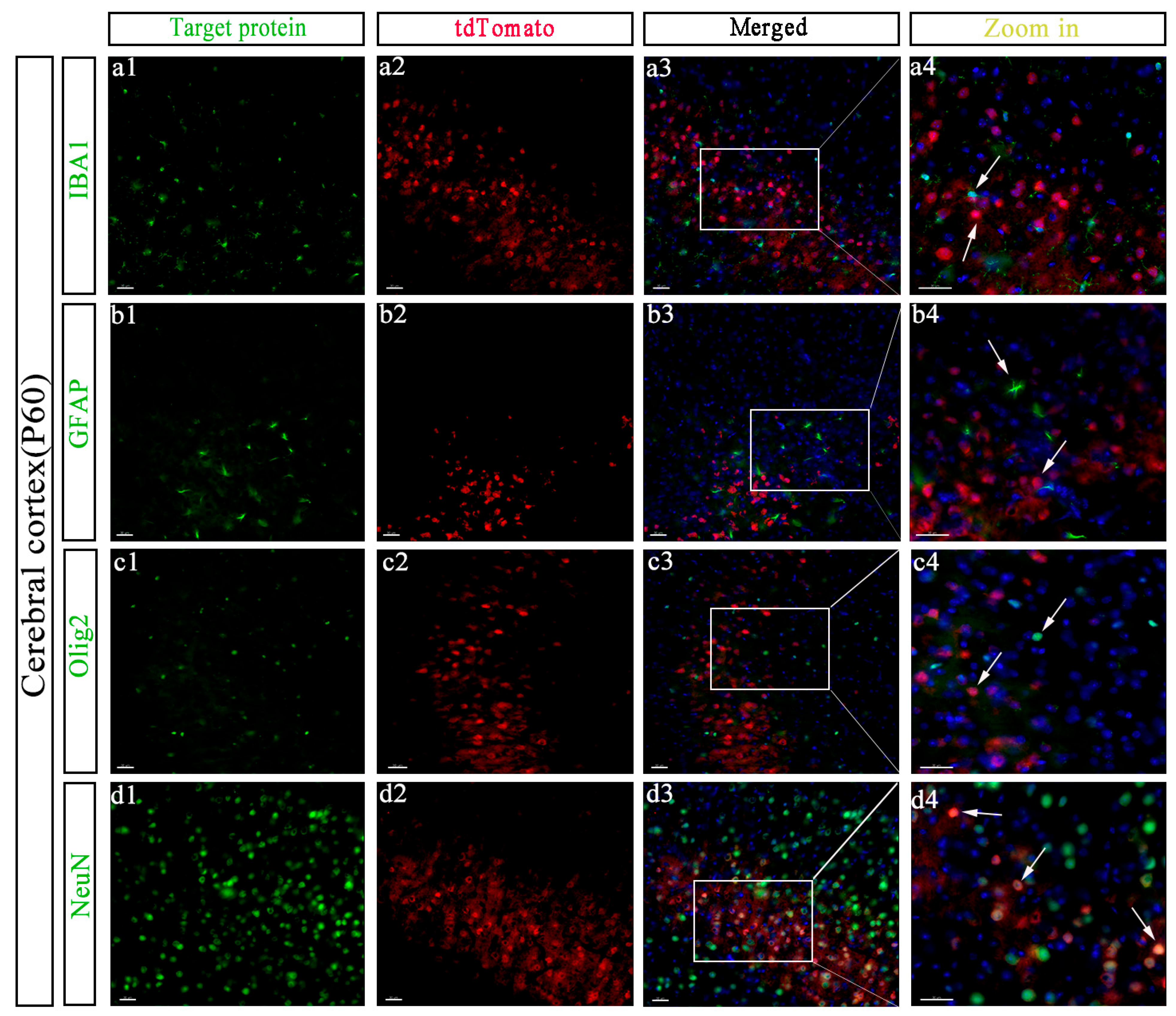
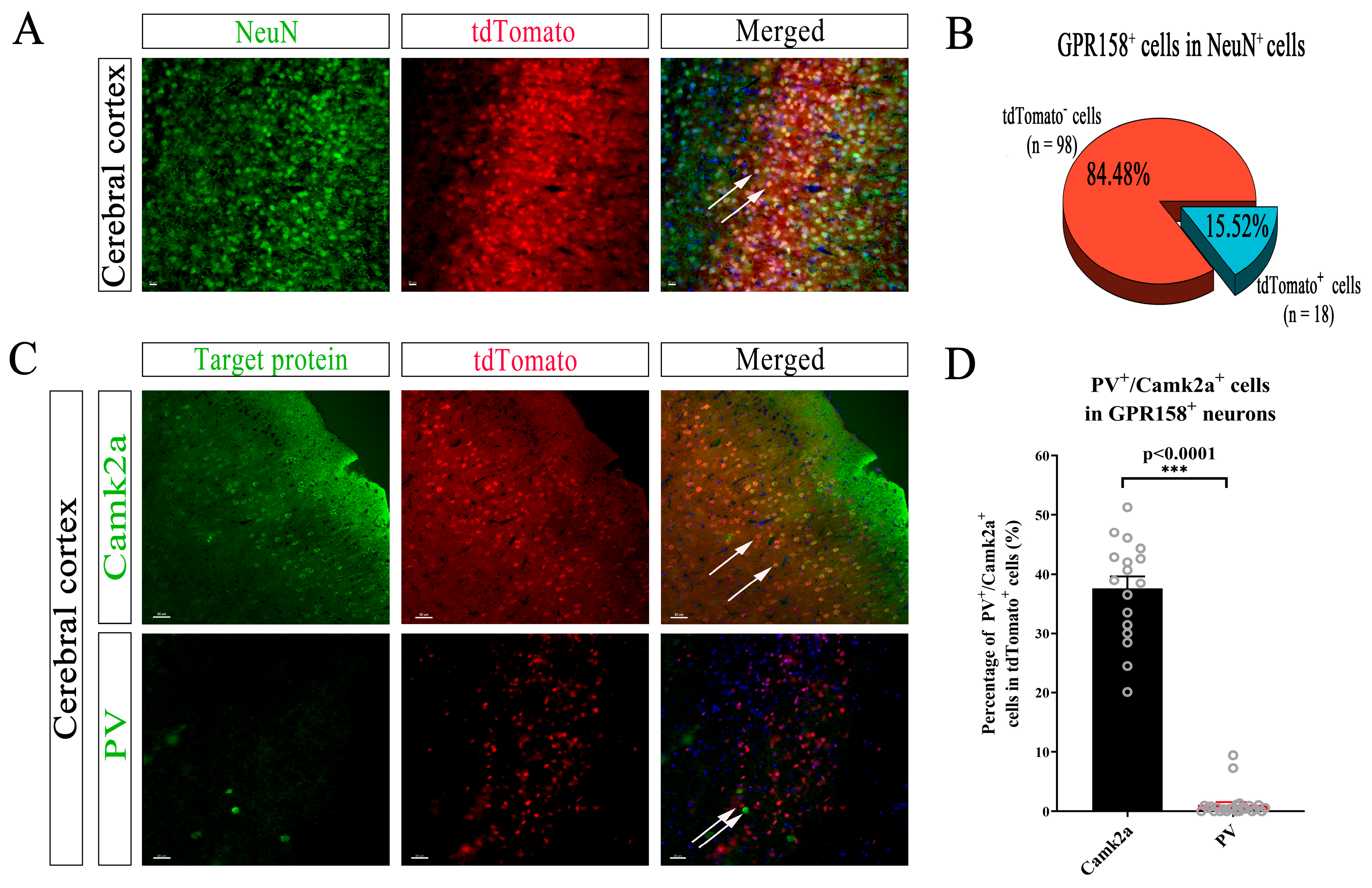
3.4. Specific Cell Types Expressing GPR158 in the Cerebral Cortex
3.5. Specific Neuron Types with GPR158 Expression in the Cerebral Cortex of GPR158Tag Mice
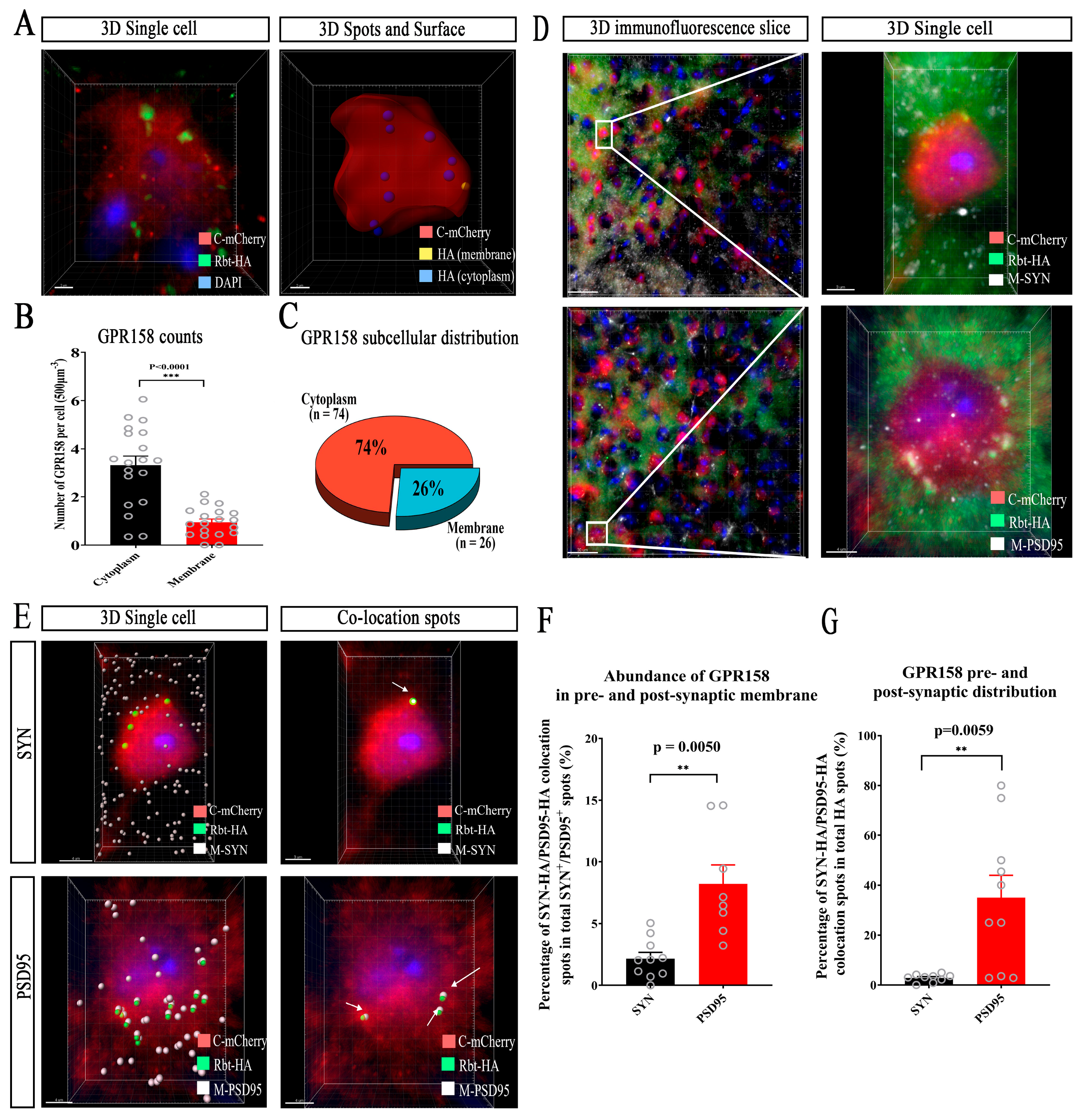
3.6. Subcellular Localization of GPR158 in Cortical Layer 2/3 Neurons in GPR158Tag Mice
3.7. Identification and Analysis of GPR158 Interacting Proteins via Mass Spectrometry and RNA Sequencing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sriram, K.; Insel, P.A. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol. Pharmacol. 2018, 93, 251–258. [Google Scholar] [CrossRef]
- Watkins, L.R.; Orlandi, C. In vitro profiling of orphan G protein coupled receptor (GPCR) constitutive activity. Br. J. Pharmacol. 2021, 178, 2963–2975. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Watkins, L.R.; Orlandi, C. Orphan G Protein Coupled Receptors in Affective Disorders. Genes 2020, 11, 694. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.I.; Kobilka, B.K. The Molecular Basis of G Protein-Coupled Receptor Activation. Annu. Rev. Biochem. 2018, 87, 897–919. [Google Scholar] [CrossRef] [PubMed]
- Maudsley, S.; Martin, B.; Luttrell, L.M. The origins of diversity and specificity in g protein-coupled receptor signaling. J. Pharmacol. Exp. Ther. 2005, 314, 485–494. [Google Scholar] [CrossRef]
- Magalhaes, A.C.; Dunn, H.; Ferguson, S.S. Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br. J. Pharmacol. 2012, 165, 1717–1736. [Google Scholar] [CrossRef]
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef]
- Alexander, S.P.H.; Christopoulos, A.; Davenport, A.P.; Kelly, S.; Mathie, A.; Peters, J.A.; Veale, E.L.; Armstrong, J.F.; Faccenda, E.; Harding, S.D.; et al. The Concise Guide to Pharmacology 2019/20: G protein-coupled receptors. Br. J. Pharmacol. 2019, 176 (Suppl. 1), S21–S141. [Google Scholar] [CrossRef]
- Fu, X.; Wei, S.; Wang, T.; Fan, H.; Zhang, Y.; Costa, C.D.; Brandnere, Y.; Yang, G.; Pan, Y.; He, Y.; et al. Research Status of the Orphan G Protein Coupled Receptor 158 and Future Perspectives. Cells 2022, 11, 1334. [Google Scholar] [CrossRef]
- Komolov, K.E.; Du, Y.; Duc, N.M.; Betz, R.M.; Rodrigues, J.; Leib, R.D.; Patra, D.; Skiniotis, G.; Adams, C.M.; Dror, R.O.; et al. Structural and Functional Analysis of a beta2-Adrenergic Receptor Complex with GRK5. Cell 2017, 169, 407–421.e6. [Google Scholar] [CrossRef] [PubMed]
- Itakura, T.; Webster, A.; Chintala, S.K.; Wang, Y.; Gonzalez, J.M., Jr.; Tan, J.C.; Vranka, J.A.; Acott, T.; Craft, C.M.; Sibug Saber, M.E.; et al. GPR158 in the Visual System: Homeostatic Role in Regulation of Intraocular Pressure. J. Ocul. Pharmacol. Ther. 2019, 35, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Itakura, T.; Jeong, S.; Liao, C.P.; Roy-Burman, P.; Zandi, E.; Groshen, S.; Pinski, J.; Coetzee, G.A.; Gross, M.E.; et al. Expression and functional role of orphan receptor GPR158 in prostate cancer growth and progression. PLoS ONE 2015, 10, e0117758. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, C.; Sutton, L.P.; Muntean, B.S.; Song, C.; Martemyanov, K.A. Homeostatic cAMP regulation by the RGS7 complex controls depression-related behaviors. Neuropsychopharmacology 2019, 44, 642–653. [Google Scholar] [CrossRef]
- Orlandi, C.; Xie, K.; Masuho, I.; Fajardo-Serrano, A.; Lujan, R.; Martemyanov, K.A. Orphan Receptor GPR158 Is an Allosteric Modulator of RGS7 Catalytic Activity with an Essential Role in Dictating Its Expression and Localization in the Brain. J. Biol. Chem. 2015, 290, 13622–13639. [Google Scholar] [CrossRef]
- Mallah, K.; Quanico, J.; Raffo-Romero, A.; Cardon, T.; Aboulouard, S.; Devos, D.; Kobeissy, F.; Zibara, K.; Salzet, M.; Fournier, I. Mapping Spatiotemporal Microproteomics Landscape in Experimental Model of Traumatic Brain Injury Unveils a link to Parkinson’s Disease. Mol. Cell. Proteom. 2019, 18, 1669–1682. [Google Scholar] [CrossRef]
- Zhu, M.; Jia, L.; Li, F.; Jia, J. Identification of KIAA0513 and Other Hub Genes Associated With Alzheimer Disease Using Weighted Gene Coexpression Network Analysis. Front. Genet. 2020, 11, 981. [Google Scholar] [CrossRef]
- Song, C.; Orlandi, C.; Sutton, L.P.; Martemyanov, K.A. The signaling proteins GPR158 and RGS7 modulate excitability of L2/3 pyramidal neurons and control A-type potassium channel in the prelimbic cortex. J. Biol. Chem. 2019, 294, 13145–13157. [Google Scholar] [CrossRef]
- Sutton, L.P.; Orlandi, C.; Song, C.; Oh, W.C.; Muntean, B.S.; Xie, K.; Filippini, A.; Xie, X.; Satterfield, R.; Yaeger, J.D.W.; et al. Orphan receptor GPR158 controls stress-induced depression. eLife 2018, 7, e33273. [Google Scholar] [CrossRef]
- Koroknai, V.; Szasz, I.; Hernandez-Vargas, H.; Fernandez-Jimenez, N.; Cuenin, C.; Herceg, Z.; Vizkeleti, L.; Adany, R.; Ecsedi, S.; Balazs, M. DNA hypermethylation is associated with invasive phenotype of malignant melanoma. Exp. Dermatol. 2020, 29, 39–50. [Google Scholar] [CrossRef]
- Ilijazi, D.; Pulverer, W.; Ertl, I.E.; Lemberger, U.; Kimura, S.; Abufaraj, M.; D’Andrea, D.; Pradere, B.; Bruchbacher, A.; Graf, A.; et al. Discovery of Molecular DNA Methylation-Based Biomarkers through Genome-Wide Analysis of Response Patterns to BCG for Bladder Cancer. Cells 2020, 9, 1839. [Google Scholar] [CrossRef]
- Engqvist, H.; Parris, T.Z.; Kovacs, A.; Nemes, S.; Werner Ronnerman, E.; De Lara, S.; Biermann, J.; Sundfeldt, K.; Karlsson, P.; Helou, K. Immunohistochemical validation of COL3A1, GPR158 and PITHD1 as prognostic biomarkers in early-stage ovarian carcinomas. BMC Cancer 2019, 19, 928. [Google Scholar] [CrossRef] [PubMed]
- Engqvist, H.; Parris, T.Z.; Kovacs, A.; Ronnerman, E.W.; Sundfeldt, K.; Karlsson, P.; Helou, K. Validation of Novel Prognostic Biomarkers for Early-Stage Clear-Cell, Endometrioid and Mucinous Ovarian Carcinomas Using Immunohistochemistry. Front. Oncol. 2020, 10, 162. [Google Scholar] [CrossRef] [PubMed]
- Oka, D.; Yamashita, S.; Tomioka, T.; Nakanishi, Y.; Kato, H.; Kaminishi, M.; Ushijima, T. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: A target for risk diagnosis and prevention of esophageal cancers. Cancer 2009, 115, 3412–3426. [Google Scholar] [CrossRef] [PubMed]
- Fenner, A. Prostate cancer: Orphan receptor GPR158 finds a home in prostate cancer growth and progression. Nat. Rev. Urol. 2015, 12, 182. [Google Scholar] [CrossRef]
- Li, N.; Zhang, Y.; Sidlauskas, K.; Ellis, M.; Evans, I.; Frankel, P.; Lau, J.; El-Hassan, T.; Guglielmi, L.; Broni, J.; et al. Inhibition of GPR158 by microRNA-449a suppresses neural lineage of glioma stem/progenitor cells and correlates with higher glioma grades. Oncogene 2018, 37, 4313–4333. [Google Scholar] [CrossRef]
- Fini, M.E.; Schwartz, S.G.; Gao, X.; Jeong, S.; Patel, N.; Itakura, T.; Price, M.O.; Price, F.W., Jr.; Varma, R.; Stamer, W.D. Steroid-induced ocular hypertension/glaucoma: Focus on pharmacogenomics and implications for precision medicine. Prog. Retin. Eye Res. 2017, 56, 58–83. [Google Scholar] [CrossRef]
- Patel, N.; Itakura, T.; Gonzalez, J.M., Jr.; Schwartz, S.G.; Fini, M.E. GPR158, an orphan member of G protein-coupled receptor Family C: Glucocorticoid-stimulated expression and novel nuclear role. PLoS ONE 2013, 8, e57843. [Google Scholar] [CrossRef]
- Khrimian, L.; Obri, A.; Ramos-Brossier, M.; Rousseaud, A.; Moriceau, S.; Nicot, A.S.; Mera, P.; Kosmidis, S.; Karnavas, T.; Saudou, F.; et al. Gpr158 mediates osteocalcin’s regulation of cognition. J. Exp. Med. 2017, 214, 2859–2873. [Google Scholar] [CrossRef]
- Condomitti, G.; Wierda, K.D.; Schroeder, A.; Rubio, S.E.; Vennekens, K.M.; Orlandi, C.; Martemyanov, K.A.; Gounko, N.V.; Savas, J.N.; de Wit, J. An Input-Specific Orphan Receptor GPR158-HSPG Interaction Organizes Hippocampal Mossy Fiber-CA3 Synapses. Neuron 2018, 100, 201–215.e9. [Google Scholar] [CrossRef]
- Orlandi, C.; Posokhova, E.; Masuho, I.; Ray, T.A.; Hasan, N.; Gregg, R.G.; Martemyanov, K.A. GPR158/179 regulate G protein signaling by controlling localization and activity of the RGS7 complexes. J. Cell Biol. 2012, 197, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Kosmidis, S.; Polyzos, A.; Harvey, L.; Youssef, M.; Denny, C.A.; Dranovsky, A.; Kandel, E.R. RbAp48 Protein Is a Critical Component of GPR158/OCN Signaling and Ameliorates Age-Related Memory Loss. Cell Rep. 2018, 25, 959–973.e6. [Google Scholar] [CrossRef] [PubMed]
- Kapust, R.B.; Tozser, J.; Fox, J.D.; Anderson, D.E.; Cherry, S.; Copeland, T.D.; Waugh, D.S. Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001, 14, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.L.L.; Hughes, L.E.; Luke, G.; Mendoza, H.; Ten Dam, E.; Gani, D.; Ryan, M.D. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 2001, 82 Pt 5, 1027–1041. [Google Scholar] [CrossRef]
- Kraeuter, A.K.; Guest, P.C.; Sarnyai, Z. The Open Field Test for Measuring Locomotor Activity and Anxiety-Like Behavior. Methods Mol. Biol. 2019, 1916, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Rein, B.; Ma, K.; Yan, Z. A standardized social preference protocol for measuring social deficits in mouse models of autism. Nat. Protoc. 2020, 15, 3464–3477. [Google Scholar] [CrossRef]
- Chin, E.W.M.; Goh, E.L.K. Behavioral Characterization of MeCP2 Dysfunction-Associated Rett Syndrome and Neuropsychiatric Disorders. Methods Mol. Biol. 2019, 2011, 593–605. [Google Scholar] [CrossRef]
- Micheau, J.; Vimeney, A.; Normand, E.; Mulle, C.; Riedel, G. Impaired hippocampus-dependent spatial flexibility and sociability represent autism-like phenotypes in GluK2 mice. Hippocampus 2014, 24, 1059–1069. [Google Scholar] [CrossRef]
- Kaidanovich-Beilin, O.; Lipina, T.; Vukobradovic, I.; Roder, J.; Woodgett, J.R. Assessment of social interaction behaviors. J. Vis. Exp. 2011, 48, e2473. [Google Scholar] [CrossRef]
- Stoppel, L.J.; Kazdoba, T.M.; Schaffler, M.D.; Preza, A.R.; Heynen, A.; Crawley, J.N.; Bear, M.F. R-Baclofen Reverses Cognitive Deficits and Improves Social Interactions in Two Lines of 16p11.2 Deletion Mice. Neuropsychopharmacology 2018, 43, 513–524. [Google Scholar] [CrossRef]
- Denninger, J.K.; Smith, B.M.; Kirby, E.D. Novel Object Recognition and Object Location Behavioral Testing in Mice on a Budget. J. Vis. Exp. 2018, 141, e58593. [Google Scholar] [CrossRef]
- Leger, M.; Quiedeville, A.; Bouet, V.; Haelewyn, B.; Boulouard, M.; Schumann-Bard, P.; Freret, T. Object recognition test in mice. Nat. Protoc. 2013, 8, 2531–2537. [Google Scholar] [CrossRef] [PubMed]
- Buffington, S.A.; Di Prisco, G.V.; Auchtung, T.A.; Ajami, N.J.; Petrosino, J.F.; Costa-Mattioli, M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 2016, 165, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Franklin, K.B.J. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates, 4th ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Boston, MA, USA, 2013. [Google Scholar]
- Yokoo, H.; Nobusawa, S.; Takebayashi, H.; Ikenaka, K.; Isoda, K.; Kamiya, M.; Sasaki, A.; Hirato, J.; Nakazato, Y. Anti-human Olig2 antibody as a useful immunohistochemical marker of normal oligodendrocytes and gliomas. Am. J. Pathol. 2004, 164, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.J.; Greer, C.A.; Firestein, S. Expression pattern of alpha CaMKII in the mouse main olfactory bulb. J. Comp. Neurol. 2002, 443, 226–236. [Google Scholar] [CrossRef] [PubMed]
- McDonald, A.J.; Muller, J.F.; Mascagni, F. GABAergic innervation of alpha type II calcium/calmodulin-dependent protein kinase immunoreactive pyramidal neurons in the rat basolateral amygdala. J. Comp. Neurol. 2002, 446, 199–218. [Google Scholar] [CrossRef]
- Jones, E.G.; Huntley, G.W.; Benson, D.L. Alpha calcium/calmodulin-dependent protein kinase II selectively expressed in a subpopulation of excitatory neurons in monkey sensory-motor cortex: Comparison with GAD-67 expression. J. Neurosci. 1994, 14, 611–629. [Google Scholar] [CrossRef]
- Benson, D.L.; Isackson, P.J.; Hendry, S.H.; Jones, E.G. Differential gene expression for glutamic acid decarboxylase and type II calcium-calmodulin-dependent protein kinase in basal ganglia, thalamus, and hypothalamus of the monkey. J. Neurosci. 1991, 11, 1540–1564. [Google Scholar] [CrossRef]
- Di Cristo, G.; Wu, C.; Chattopadhyaya, B.; Ango, F.; Knott, G.; Welker, E.; Svoboda, K.; Huang, Z.J. Subcellular domain-restricted GABAergic innervation in primary visual cortex in the absence of sensory and thalamic inputs. Nat. Neurosci. 2004, 7, 1184–1186. [Google Scholar] [CrossRef]
- Meng, F.T.; Ni, R.J.; Zhang, Z.; Zhao, J.; Liu, Y.J.; Zhou, J.N. Inhibition of oestrogen biosynthesis induces mild anxiety in C57BL/6J ovariectomized female mice. Neurosci. Bull. 2011, 27, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Cetereisi, D.; Kramvis, I.; Gebuis, T.; van der Loo, R.J.; Gouwenberg, Y.; Mansvelder, H.D.; Li, K.W.; Smit, A.B.; Spijker, S. Gpr158 Deficiency Impacts Hippocampal CA1 Neuronal Excitability, Dendritic Architecture, and Affects Spatial Learning. Front. Cell. Neurosci. 2019, 13, 465. [Google Scholar] [CrossRef] [PubMed]
- Coley, A.A.; Gao, W.J. PSD95: A synaptic protein implicated in schizophrenia or autism? Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 82, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Hunt, C.A.; Schenker, L.J.; Kennedy, M.B. PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J. Neurosci. 1996, 16, 1380–1388. [Google Scholar] [CrossRef]
- El-Husseini, A.E.; Schnell, E.; Chetkovich, D.M.; Nicoll, R.A.; Bredt, D.S. PSD-95 involvement in maturation of excitatory synapses. Science 2000, 290, 1364–1368. [Google Scholar] [CrossRef]
- Wiedenmann, B.; Franke, W.W.; Kuhn, C.; Moll, R.; Gould, V.E. Synaptophysin: A marker protein for neuroendocrine cells and neoplasms. Proc. Natl. Acad. Sci. USA 1986, 83, 3500–3504. [Google Scholar] [CrossRef]
- Ferguson, S.S. Evolving concepts in G protein-coupled receptor endocytosis: The role in receptor desensitization and signaling. Pharmacol. Rev. 2001, 53, 1–24. [Google Scholar]
- Tayou, J.; Wang, Q.; Jang, G.F.; Pronin, A.N.; Orlandi, C.; Martemyanov, K.A.; Crabb, J.W.; Slepak, V.Z. Regulator of G Protein Signaling 7 (RGS7) Can Exist in a Homo-oligomeric Form That Is Regulated by Galphao and R7-binding Protein. J. Biol. Chem. 2016, 291, 9133–9147. [Google Scholar] [CrossRef]
- Benson, D.L.; Isackson, P.J.; Gall, C.M.; Jones, E.G. Contrasting patterns in the localization of glutamic acid decarboxylase and Ca2+/calmodulin protein kinase gene expression in the rat central nervous system. Neuroscience 1992, 46, 825–849. [Google Scholar] [CrossRef]
- Sik, A.; Hajos, N.; Gulacsi, A.; Mody, I.; Freund, T.F. The absence of a major Ca2+ signaling pathway in GABAergic neurons of the hippocampus. Proc. Natl. Acad. Sci. USA 1998, 95, 3245–3250. [Google Scholar] [CrossRef]
- Achterberg, K.G.; Buitendijk, G.H.; Kool, M.J.; Goorden, S.M.; Post, L.; Slump, D.E.; Silva, A.J.; van Woerden, G.M.; Kushner, S.A.; Elgersma, Y. Temporal and region-specific requirements of alphaCaMKII in spatial and contextual learning. J. Neurosci. 2014, 34, 11180–11187. [Google Scholar] [CrossRef] [PubMed]
- Sakimoto, Y.; Oo, P.M.; Goshima, M.; Kanehisa, I.; Tsukada, Y.; Mitsushima, D. Significance of GABAA Receptor for Cognitive Function and Hippocampal Pathology. Int. J. Mol. Sci. 2021, 22, 12456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, D.; Sun, T.; Xu, J.; Chiang, H.C.; Shin, W.; Wu, L.G. The SNARE proteins SNAP25 and synaptobrevin are involved in endocytosis at hippocampal synapses. J. Neurosci. 2013, 33, 9169–9175. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.G.; Hamid, E.; Shin, W.; Chiang, H.C. Exocytosis and endocytosis: Modes, functions, and coupling mechanisms. Annu. Rev. Physiol. 2014, 76, 301–331. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, J.; Song, Z.; Wei, S.; Zhou, Y.; Ju, J.; Yao, P.; Jiang, Y.; Jin, H.; Chi, X.; Li, N. Expression Mapping and Functional Analysis of Orphan G-Protein-Coupled Receptor GPR158 in the Adult Mouse Brain Using a GPR158 Transgenic Mouse. Biomolecules 2023, 13, 479. https://doi.org/10.3390/biom13030479
Chang J, Song Z, Wei S, Zhou Y, Ju J, Yao P, Jiang Y, Jin H, Chi X, Li N. Expression Mapping and Functional Analysis of Orphan G-Protein-Coupled Receptor GPR158 in the Adult Mouse Brain Using a GPR158 Transgenic Mouse. Biomolecules. 2023; 13(3):479. https://doi.org/10.3390/biom13030479
Chicago/Turabian StyleChang, Jinlong, Ze Song, Shoupeng Wei, Yunxia Zhou, Jun Ju, Peijia Yao, Youheng Jiang, Hui Jin, Xinjin Chi, and Ningning Li. 2023. "Expression Mapping and Functional Analysis of Orphan G-Protein-Coupled Receptor GPR158 in the Adult Mouse Brain Using a GPR158 Transgenic Mouse" Biomolecules 13, no. 3: 479. https://doi.org/10.3390/biom13030479
APA StyleChang, J., Song, Z., Wei, S., Zhou, Y., Ju, J., Yao, P., Jiang, Y., Jin, H., Chi, X., & Li, N. (2023). Expression Mapping and Functional Analysis of Orphan G-Protein-Coupled Receptor GPR158 in the Adult Mouse Brain Using a GPR158 Transgenic Mouse. Biomolecules, 13(3), 479. https://doi.org/10.3390/biom13030479





