Using Selective Enzymes to Measure Noncanonical DNA Building Blocks: dUTP, 5-Methyl-dCTP, and 5-Hydroxymethyl-dCTP
Abstract
:1. Introduction
2. Methods
2.1. Mycobacterial Strains and Growth Conditions
2.2. dUTPase Expression and Purification
2.3. Dcd:dut Expression and Purification
2.4. Measuring dCTP Deaminase Activity
2.5. dNTP Pool Extraction
2.6. dNTP Quantification
2.7. Statistical Analysis
3. Results
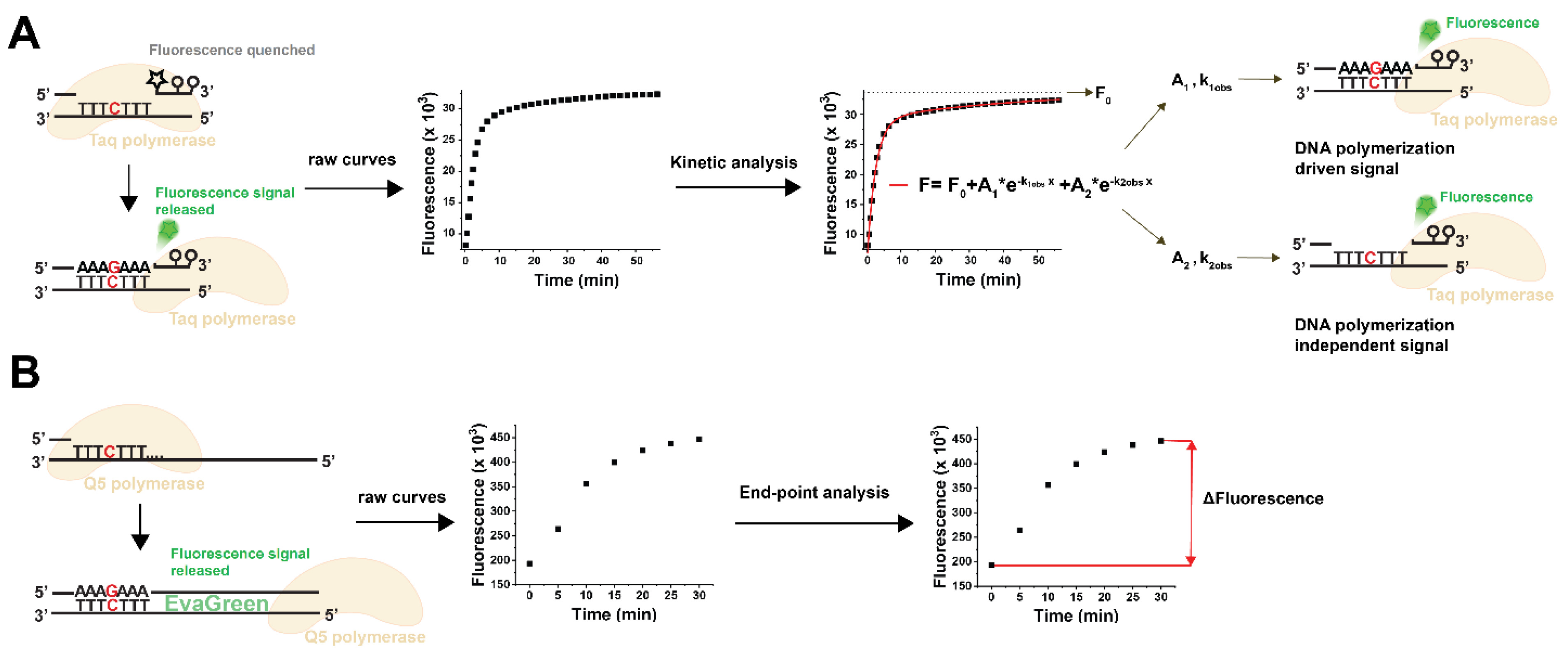
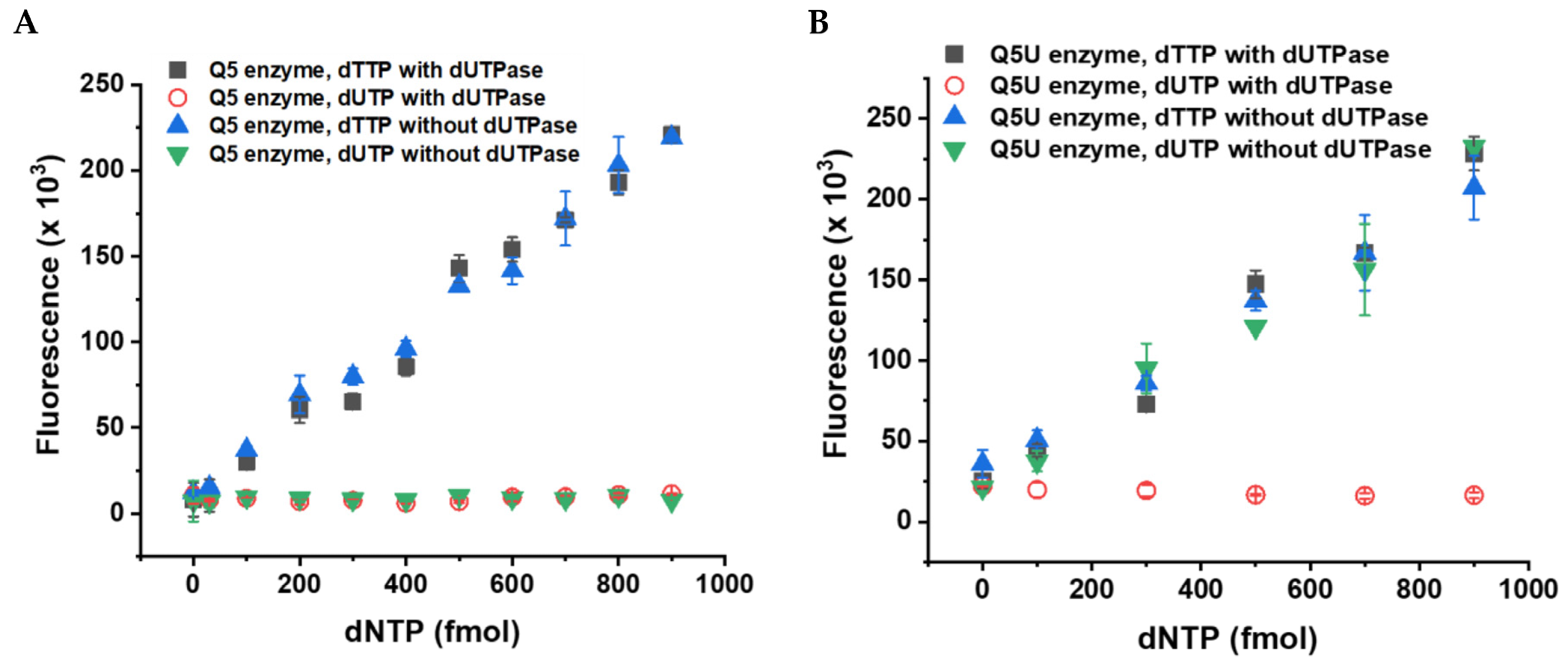
3.1. Choosing the Suitable Fluorescent Method for Non-Canonical dNTP Determination and Establishment of the dUTP Measurement Technique
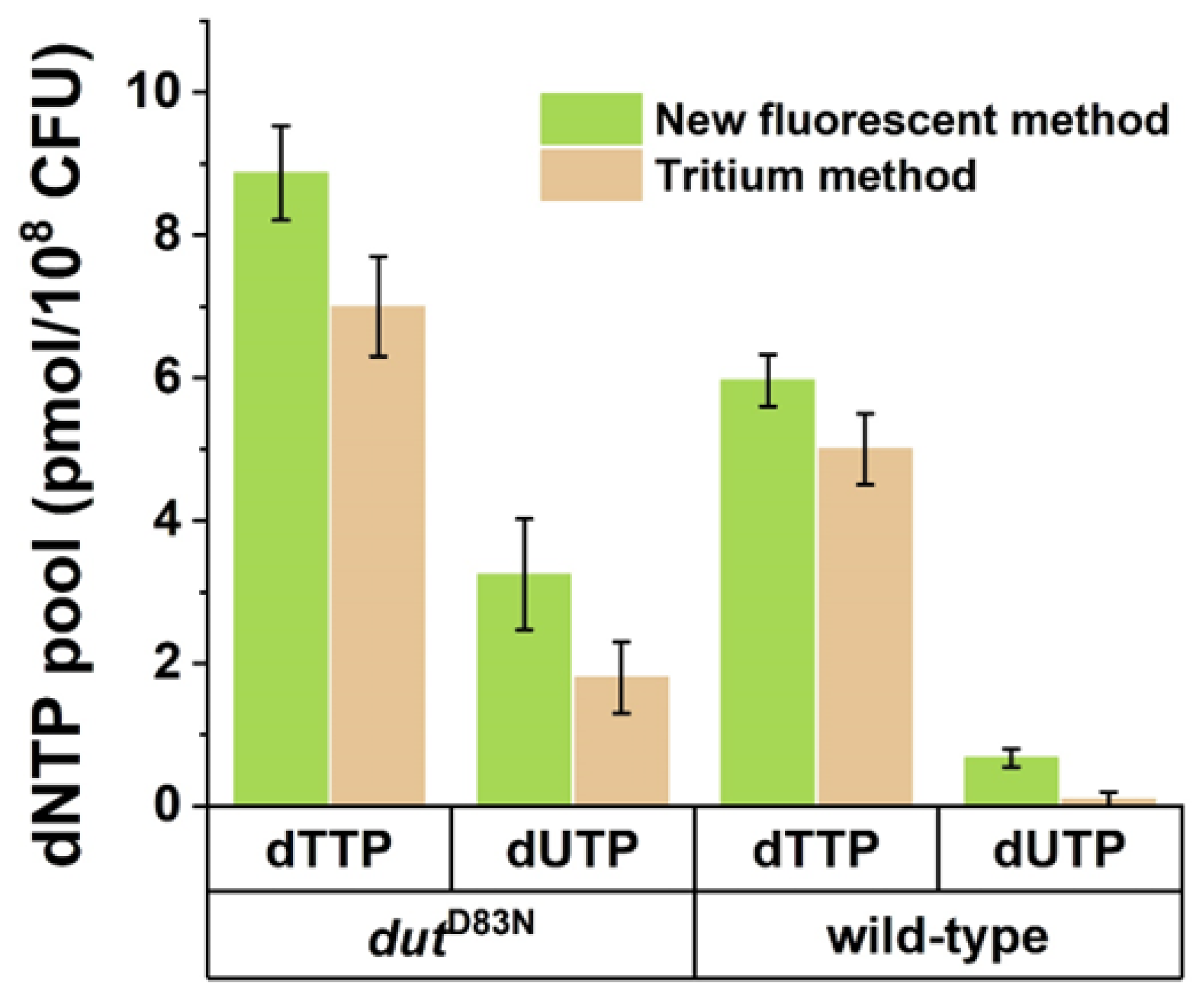
3.2. Validation of the Fluorescent dUTP Measurement Technique
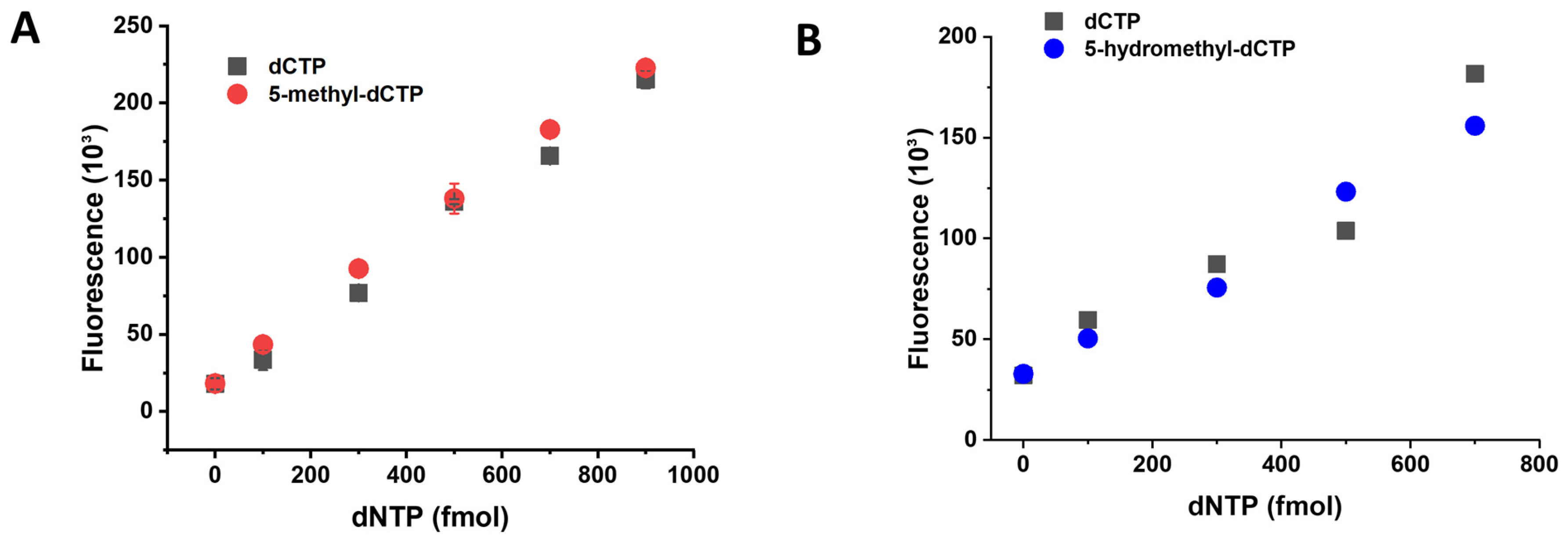
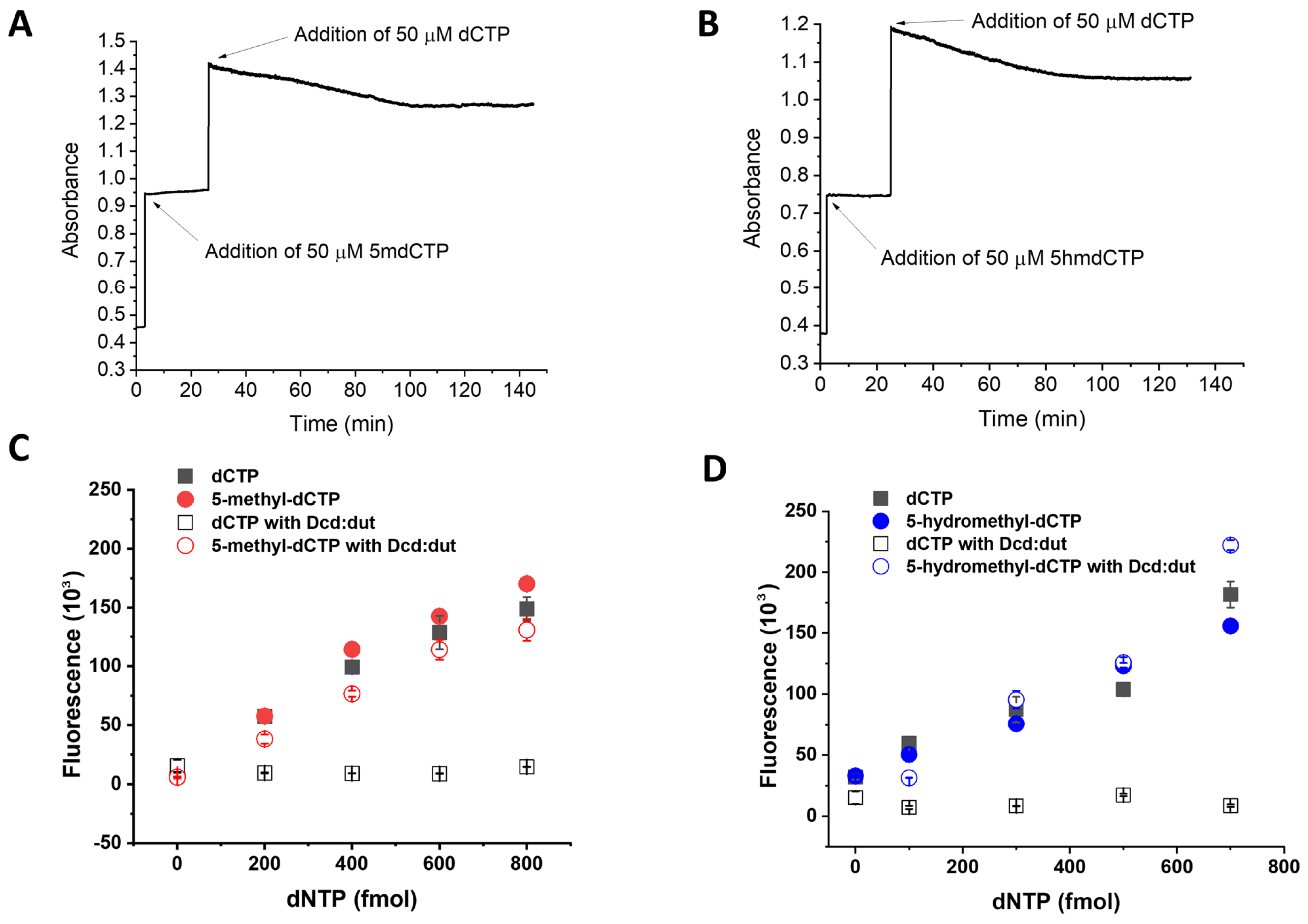
3.3. Establishment of the 5-Methyl-dCTP and the 5-Hydroxymethyl-dCTP Measurement Technique
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Szabó, J.E.; Surányi, É.V.; Mébold, B.S.; Trombitás, T.; Cserepes, M.; Tóth, J. A User-friendly, high-throughput tool for the precise quantitation of deoxyribonucleoside triphosphates from biological samples. Nucleic Acids Res. 2020, 48, e45. [Google Scholar] [CrossRef] [PubMed]
- Purhonen, J.; Banerjee, R.; McDonald, A.E.; Fellman, V.; Kallijärvi, J. A sensitive assay for dNTPs based on long synthetic oligonucleotides, EvaGreen dye and inhibitor-resistant high-fidelity DNA polymerase. Nucleic Acids Res. 2020, 48, e87. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Hsin, P.; Huang, C.-Y.; Chang, Z.-F. A Convenient and Sensitive Method for Deoxynucleoside Triphosphate Quantification by the Combination of Rolling Circle Amplification and Quantitative Polymerase Chain Reaction. Anal. Chem. 2021, 93, 14247–14255. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Yagüe-Capilla, M.; González-Pacanowska, D.; Chang, Z.F. Quantitation of deoxynucleoside triphosphates by click reactions. Sci. Rep. 2020, 10, 611. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, P.; Franzolin, E.; Pontarin, G.; Reichard, P.; Bianchi, V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010, 38, e85. [Google Scholar] [CrossRef] [PubMed]
- Straube, H.; Niehaus, M.; Zwittian, S.; Witte, C.-P.; Herde, M. Enhanced nucleotide analysis enables the quantification of deoxynucleotides in plants and algae revealing connections between nucleoside and deoxynucleoside metabolism. Plant Cell 2021, 33, 270–289. [Google Scholar] [CrossRef]
- Dong, J.; Wu, T.; Xiao, Y.; Xu, L.; Fang, S.; Zhao, M. A fuel-limited isothermal DNA machine for the sensitive detection of cellular deoxyribonucleoside triphosphates. Chem. Commun. 2016, 52, 11923–11926. [Google Scholar] [CrossRef]
- Kong, Z.; Jia, S.; Chabes, A.L.; Appelblad, P.; Lundmark, R.; Moritz, T.; Chabes, A. Simultaneous determination of ribonucleoside and deoxyribonucleoside triphosphates in biological samples by hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry. Nucleic Acids Res. 2018, 46, e66. [Google Scholar] [CrossRef]
- Pai, C.-C.; Kearsey, S.E. A Critical Balance: dNTPs and the Maintenance of Genome Stability. Genes 2017, 8, 57. [Google Scholar] [CrossRef]
- Ke, P.Y.; Kuo, Y.Y.; Hu, C.M.; Chang, Z.F. Control of dTTP pool size by anaphase promoting complex/cyclosome is essential for the maintenance of genetic stability. Genes Dev. 2005, 19, 1920–1933. [Google Scholar] [CrossRef]
- Hu, C.M.; Chang, Z.F. Mitotic control of dTTP pool: A necessity or coincidence? J. Biomed. Sci. 2007, 14, 491–497. [Google Scholar] [CrossRef]
- Chen, S.; Bonifati, S.; Qin, Z.; St Gelais, C.; Wu, L. SAMHD1 Suppression of Antiviral Immune Responses. Trends Microbiol. 2019, 27, 254–267. [Google Scholar] [CrossRef]
- Thientosapol, E.S.; Bosnjak, D.; Durack, T.; Stevanovski, I.; van Geldermalsen, M.; Holst, J.; Jahan, Z.; Shepard, C.; Weninger, W.; Kim, B.; et al. SAMHD1 enhances immunoglobulin hypermutation by promoting transversion mutation. Proc. Natl. Acad. Sci. USA 2018, 115, 4921–4926. [Google Scholar] [CrossRef]
- Szabo, J.E.; Németh, V.; Papp-Kádár, V.; Nyíri, K.; Leveles, I.; Bendes, A.A.; Zagyva, I.; Rona, G.; Pálinkás, H.L.; Besztercei, B.; et al. Highly potent dUTPase inhibition by a bacterial repressor protein reveals a novel mechanism for gene expression control. Nucleic Acids Res. 2014, 42, 11912–11920. [Google Scholar] [CrossRef]
- Requena, C.E.; Pérez-Moreno, G.; Ruiz-Pérez, L.M.; Vidal, A.E.; González-Pacanowska, D. The NTP pyrophosphatase DCTPP1 contributes to the homoeostasis and cleansing of the dNTP pool in human cells. Biochem. J. 2014, 459, 171–180. [Google Scholar] [CrossRef]
- Mathews, C.K. DNA precursor metabolism and genomic stability. FASEB J. 2006, 20, 1300–1314. [Google Scholar] [CrossRef] [PubMed]
- Vértessy, B.G.; Tóth, J. Keeping uracil out of DNA: Physiological role, structure and catalytic mechanism of dUTPases. Acc. Chem. Res. 2009, 42, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Maki, H.; Sekiguchi, M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 1992, 355, 273–275. [Google Scholar] [CrossRef]
- Sang, P.B.; Varshney, U. Biochemical properties of MutT2 proteins from Mycobacterium tuberculosis and M. smegmatis and their contrasting antimutator roles in Escherichia coli. J. Bacteriol. 2013, 195, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Moroz, O.V.; Wilson, K.S.; Murzin, A.G. House cleaning, a part of good housekeeping. Mol. Microbiol. 2006, 59, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Nagy, G.N.; Leveles, I.; Vértessy, B.G. Preventive DNA repair by sanitizing the cellular (deoxy)nucleoside triphosphate pool. FEBS J. 2014, 281, 4207–4223. [Google Scholar] [CrossRef]
- Pancsa, R.; Fichó, E.; Molnár, D.; Surányi, V.; Trombitás, T.; Füzesi, D.; Lóczi, H.; Szijjártó, P.; Hirmondó, R.; E Szabó, J.; et al. dNTPpoolDB: A manually curated database of experimentally determined dNTP pools and pool changes in biological samples. Nucleic Acids Res. 2022, 50, D1508–D1514. [Google Scholar] [CrossRef]
- Sherman, P.A.; Fyfe, J.A. Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal. Biochem. 1989, 180, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Diamond, T.L.; Roshal, M.; Jamburuthugoda, V.K.; Reynolds, H.M.; Merriam, A.R.; Lee, K.Y.; Balakrishnan, M.; Bambara, R.A.; Planelles, V.; Dewhurst, S.; et al. Macrophage tropism of HIV-1 depends upon efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 2004, 279, 51545–51553. [Google Scholar] [CrossRef] [PubMed]
- Hirmondo, R.; Lopata, A.; Suranyi, E.V.; Vertessy, B.G.; Toth, J. Differential control of dNTP biosynthesis and genome integrity maintenance by the dUTPase superfamily enzymes. Sci. Rep. 2017, 7, 6043. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.M.; LaBonte, M.J.; Russell, J.; Louie, S.; Ghobrial, A.A.; Ladner, R.D. A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic Acids Res. 2011, 39, e112. [Google Scholar] [CrossRef] [PubMed]
- Kovári, J.; Barabás, O.; Varga, B.; Békési, A.; Tölgyesi, F.; Fidy, J.; Nagy, J.; Vértessy, B.G. Methylene substitution at the α-β bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site. Proteins Struct. Funct. Genet. 2008, 71, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Takács, E.; Grolmusz, V.K.; Vértessy, B.G. A tradeoff between protein stability and conformational mobility in homotrimeric dUTPases. FEBS Lett. 2004, 566, 48–54. [Google Scholar] [CrossRef]
- Goude, R.; Parish, T. Mycobacteria Protocols; Humana Press: Totowa, TJ, USA, 2009; Volume 465, pp. 13–22. [Google Scholar]
- Varga, B.; Migliardo, F.; Takacs, E.; Vertessy, B.; Magazù, S.; Telling, M.T.F. Study of solvent-protein coupling effects by neutron scattering. J. Biol. Phys. 2010, 36, 207–220. [Google Scholar] [CrossRef]
- Helt, S.S.; Thymark, M.; Harris, P.; Aagaard, C.; Dietrich, J.; Larsen, S.; Willemoes, M. Mechanism of dTTP inhibition of the bifunctional dCTP deaminase:dUTPase encoded by Mycobacterium tuberculosis. J. Mol. Biol. 2008, 376, 554–569. [Google Scholar] [CrossRef]
- Di Noia, J.M.; Neuberger, M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007, 76, 1–22. [Google Scholar] [CrossRef]
- Pálinkás, H.L.; Rácz, G.A.; Gál, Z.; Hoffmann, O.I.; Tihanyi, G.; Róna, G.; Gócza, E.; Hiripi, L.; Vértessy, B.G. Crispr/cas9-mediated knock-out of dutpase in mice leads to early embryonic lethality. Biomolecules 2019, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Muha, V.; Horváth, A.; Békési, A.; Pukáncsik, M.; Hodoscsek, B.; Merényi, G.; Róna, G.; Batki, J.; Kiss, I.; Jankovics, F.; et al. Uracil-containing DNA in Drosophila: Stability, stage-specific accumulation, and developmental involvement. PLoS Genet. 2012, 8, e1002738. [Google Scholar]
- Leveles, I.; Németh, V.; Szabó, J.E.; Harmat, V.; Nyíri, K.; Bendes, A.A.; Papp-Kádár, V.; Zagyva, I.; Róna, G.; Ozohanics, O.; et al. Structure and enzymatic mechanism of a moonlighting dUTPase. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 2298–2308. [Google Scholar] [CrossRef] [PubMed]
- Róna, G.; Marfori, M.; Borsos, M.; Scheer, I.; Takács, E.; Tóth, J.; Babos, F.; Magyar, A.; Erdei, A.; Bozóky, Z.; et al. Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: Structural and mechanistic insights. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, Z.; Cedar, H. DNA methylation: A molecular lock. Curr. Biol. 1997, 7, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Bessman, M.J.; Lehman, I.R.; Adler, J.; Zimmerman, S.B.; Simms, E.S.; Kornberg, A. Enzymatic synthesis of deoxyribonucleic acid. III. The incorporation of pyrimidine and purine analogues into deoxyribonucleic acid. Proc. Natl. Acad. Sci. USA 1958, 44, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.-H.; Huang, L.-H.; Ehrlich, M. A bacteriophage-induced 5-methyldeoxycytidine 5′-monophosphate kinase. Biochim. Biophys. Acta-Gene Struct. Expr. 1982, 696, 31–36. [Google Scholar] [CrossRef]
- Holliday, R.; Ho, T. Evidence for gene silencing by endogenous DNA methylation. Proc. Natl. Acad. Sci. USA 1998, 95, 8727–8732. [Google Scholar] [CrossRef]
- Mathews, C.K.; Brown, F.; Cohen, S.S. Virus-induced acquisition of metabolic function. VII. Biosynthesis de novo of deoxycytidylate hydroxymethylase. J. Biol. Chem. 1964, 239, 2957–2963. [Google Scholar] [CrossRef]
- O’Handley, S.F.; Dunn, C.A.; Bessman, M.J. Orf135 from Escherichia coli Is a Nudix Hydrolase Specific for CTP, dCTP, and 5-Methyl-dCTP. J. Biol. Chem. 2001, 276, 5421–5426. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, M.; Tsuchimoto, D.; Sakumi, K.; Nakabeppu, Y. Mouse RS21-C6 is a mammalian 2′-deoxycytidine 5′-triphosphate pyrophosphohydrolase that prefers 5-iodocytosine. FEBS J. 2009, 276, 1654–1666. [Google Scholar] [CrossRef] [PubMed]
| Tested Range (pmol) | R2 | LOD (pmol) | LOQ (pmol) | Accuracy Low (%) | Accuracy High (%) | Inter-Assay CV% | Intra-Assay CV% | |
|---|---|---|---|---|---|---|---|---|
| dUTP | 30–1000 | 0.95 ± 0.01 | 66 ± 35 | 110 ± 59 | 108 ± 13 | 102 ± 4 | 9.65 ± 5.33 | 14.96 ± 1.17 |
| hmdCTP/5m-dCTP | 30–1000 | 0.96 ± 0.04 | 37 ± 1 | 61 ± 1 | 99 ± 11 | 101 ± 1 | 8.82 ± 12.11 | 13.46 ± 12.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Surányi, É.V.; Perey-Simon, V.; Hirmondó, R.; Trombitás, T.; Kazzazy, L.; Varga, M.; Vértessy, B.G.; Tóth, J. Using Selective Enzymes to Measure Noncanonical DNA Building Blocks: dUTP, 5-Methyl-dCTP, and 5-Hydroxymethyl-dCTP. Biomolecules 2023, 13, 1801. https://doi.org/10.3390/biom13121801
Surányi ÉV, Perey-Simon V, Hirmondó R, Trombitás T, Kazzazy L, Varga M, Vértessy BG, Tóth J. Using Selective Enzymes to Measure Noncanonical DNA Building Blocks: dUTP, 5-Methyl-dCTP, and 5-Hydroxymethyl-dCTP. Biomolecules. 2023; 13(12):1801. https://doi.org/10.3390/biom13121801
Chicago/Turabian StyleSurányi, Éva Viola, Viktória Perey-Simon, Rita Hirmondó, Tamás Trombitás, Latifa Kazzazy, Máté Varga, Beáta G. Vértessy, and Judit Tóth. 2023. "Using Selective Enzymes to Measure Noncanonical DNA Building Blocks: dUTP, 5-Methyl-dCTP, and 5-Hydroxymethyl-dCTP" Biomolecules 13, no. 12: 1801. https://doi.org/10.3390/biom13121801
APA StyleSurányi, É. V., Perey-Simon, V., Hirmondó, R., Trombitás, T., Kazzazy, L., Varga, M., Vértessy, B. G., & Tóth, J. (2023). Using Selective Enzymes to Measure Noncanonical DNA Building Blocks: dUTP, 5-Methyl-dCTP, and 5-Hydroxymethyl-dCTP. Biomolecules, 13(12), 1801. https://doi.org/10.3390/biom13121801









