Improvement in Clinical Features of L-NAME-Induced Preeclampsia-like Rats through Reduced SERPINA5 Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal
2.2. Animal Experimental Procedures
- (1)
- The NC group (n = 6) was given 200 μL/day saline solution from day 9 to day 19 of gestation.
- (2)
- (3)
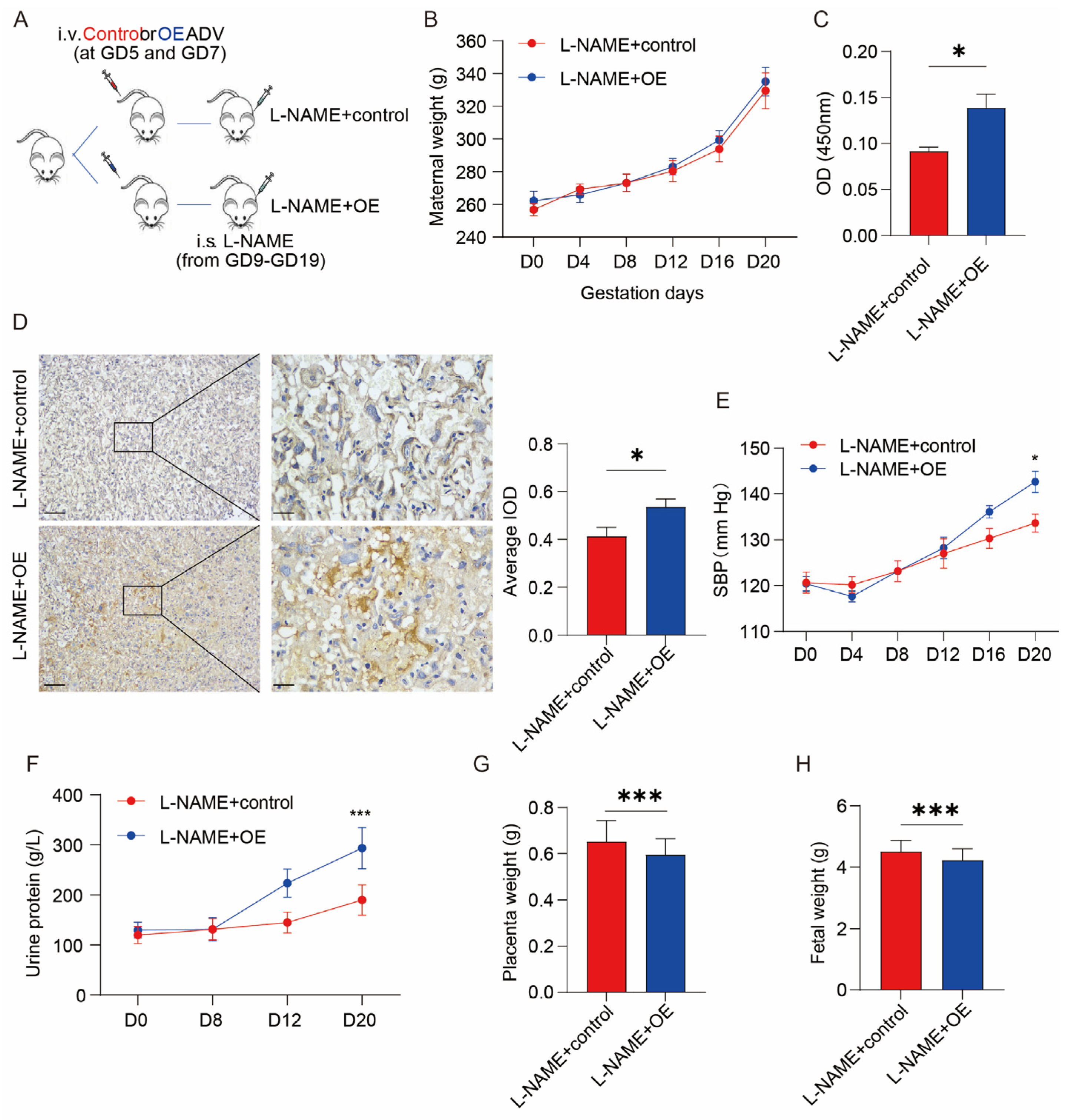
- L-NAME + OE group (n = 7): Recombinant ADV with the SERPINA5 gene (ADV-pCMV-SERPINA5) was used to overexpressed SERPINA5 in vivo. ADV-pCMV-SERPINA5 (200 μL, 1 × 109 PFU/rat) was administered to rats in GD5 and GD7 by tail injection [23]. At the same time, this group of rats received L-NAME treatment at a dose of 75 mg/kg/day, injected subcutaneously from day 9 to day 19 of gestation. One of the rats died after two times of ADV injections, so only 7 rats were included in the results. ADV was made by GeneChem Ltd. (Shanghai, China).
- (4)
- L-NAME + OE-control (n = 6): ADV-pCMV-empty was used as a control for SERPINA5 in vivo expression. This group of rats also received L-NAME treatment, as described above.
- (5)
- L-NAME + KD (n = 6): In vivo gene knockdown was performed using an ADV vector. A shRNA fragment targeting the rat SERPINA5 gene (NM_001244739) was cloned into the PLko-U6-MCS-CAG-EGFP vector. Knockdown vectors (200 μL, 1 × 109 PFU/rat) were given via tail injection on GD5 and GD7. As described above, these rats also received L-NAME at 75 mg/kg/day subcutaneously.
- (6)
- L-NAME + KD-control (n = 6): An empty PLko-U6-MCS-CAG-EGFP vector was used as a control for in vivo gene knockdown. The control vector (200 μL, 1 × 109 PFU/rat) was given via tail injection on GD5 and GD7. As described above, these rats also received L-NAME at 75 mg/kg/day subcutaneously.
2.3. Blood Pressure Measurement
2.4. Sample Collection
2.5. Semi-Quantification of Serum SERPINA5 Levels
2.6. Urine Protein Measurement
2.7. RNA Sequencing
2.8. Histology
2.9. Statistical Analysis
3. Results
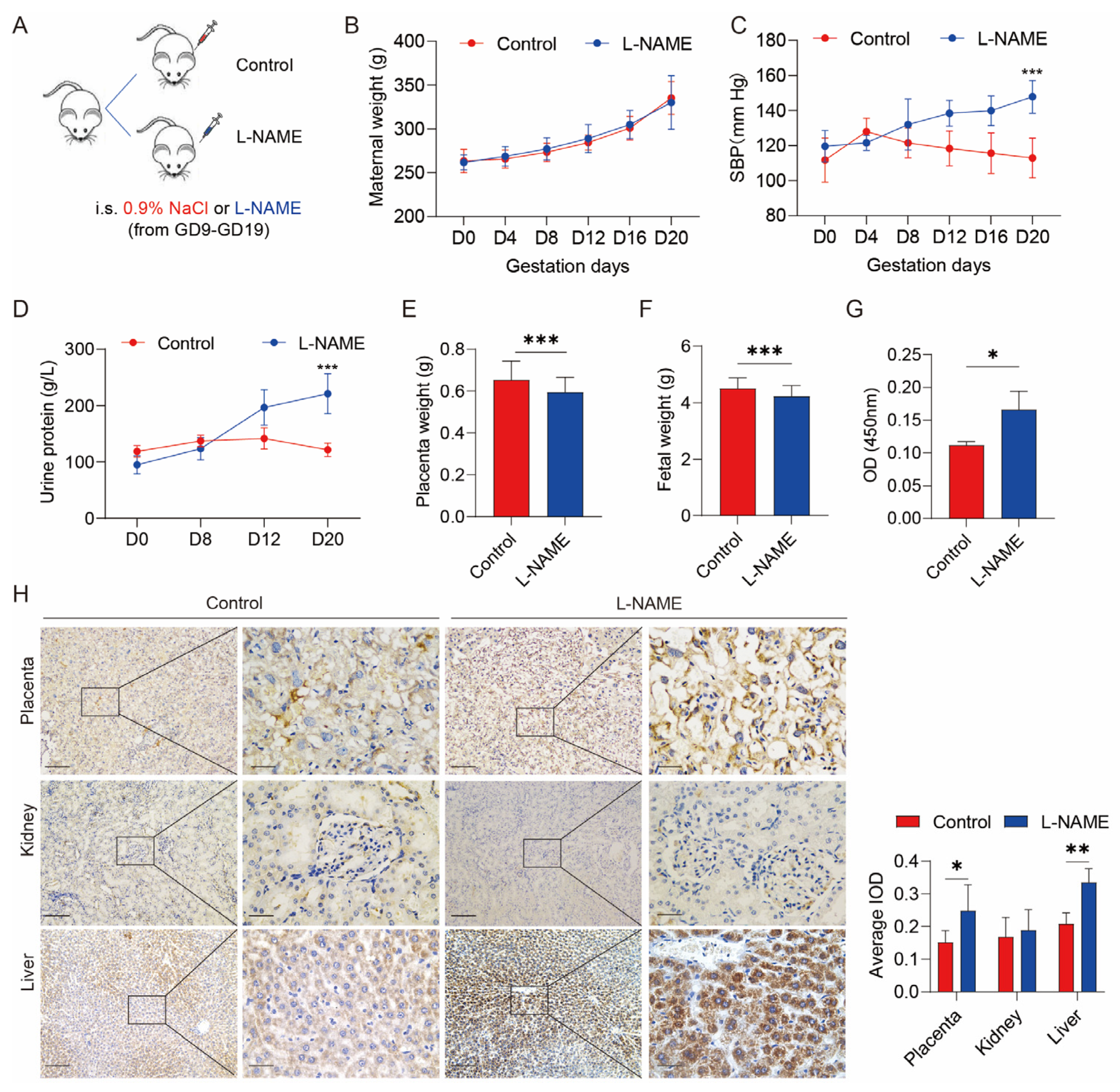
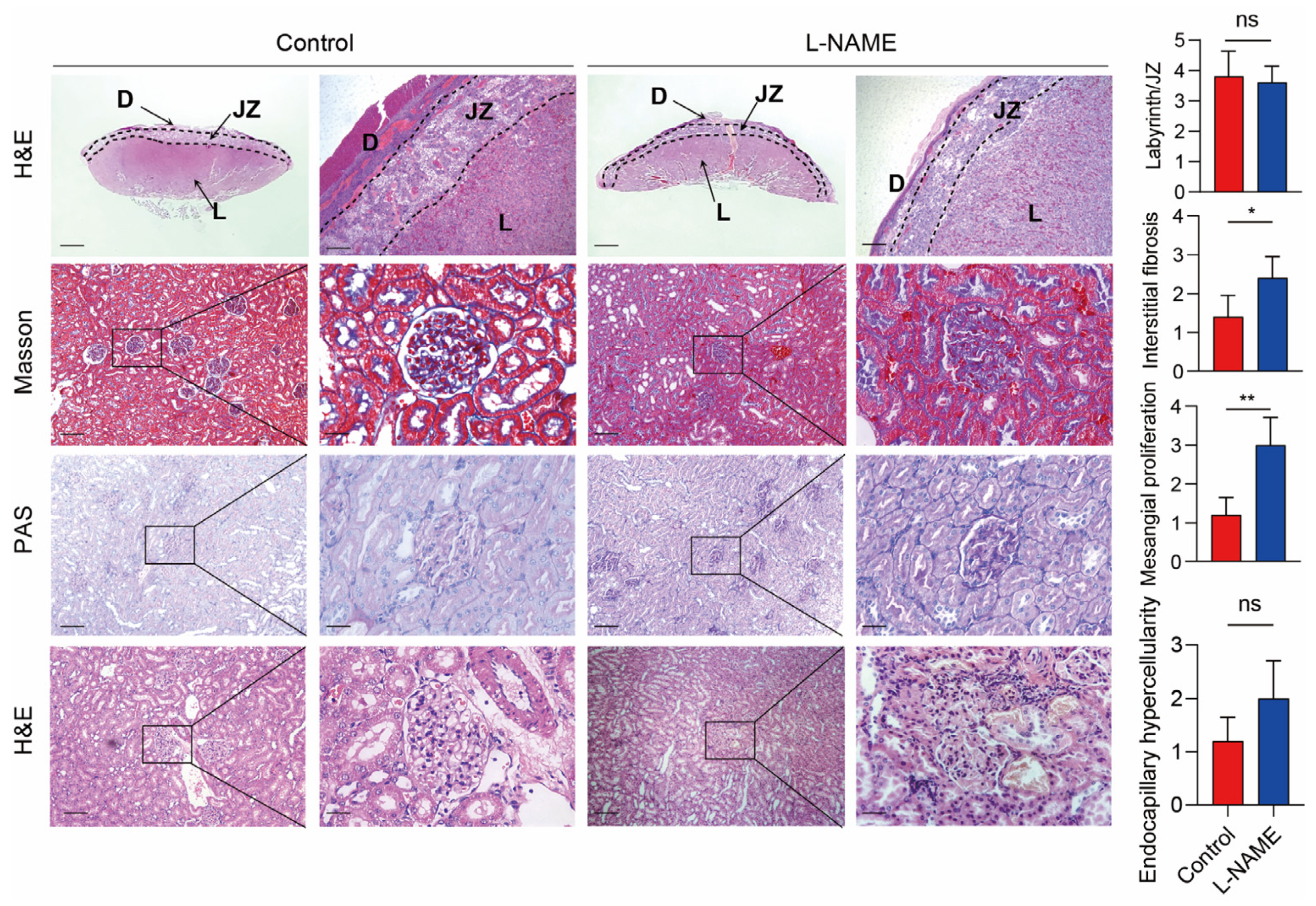
3.1. Serum SERPINA5 Increased in the PE Model Induced by L-NAME
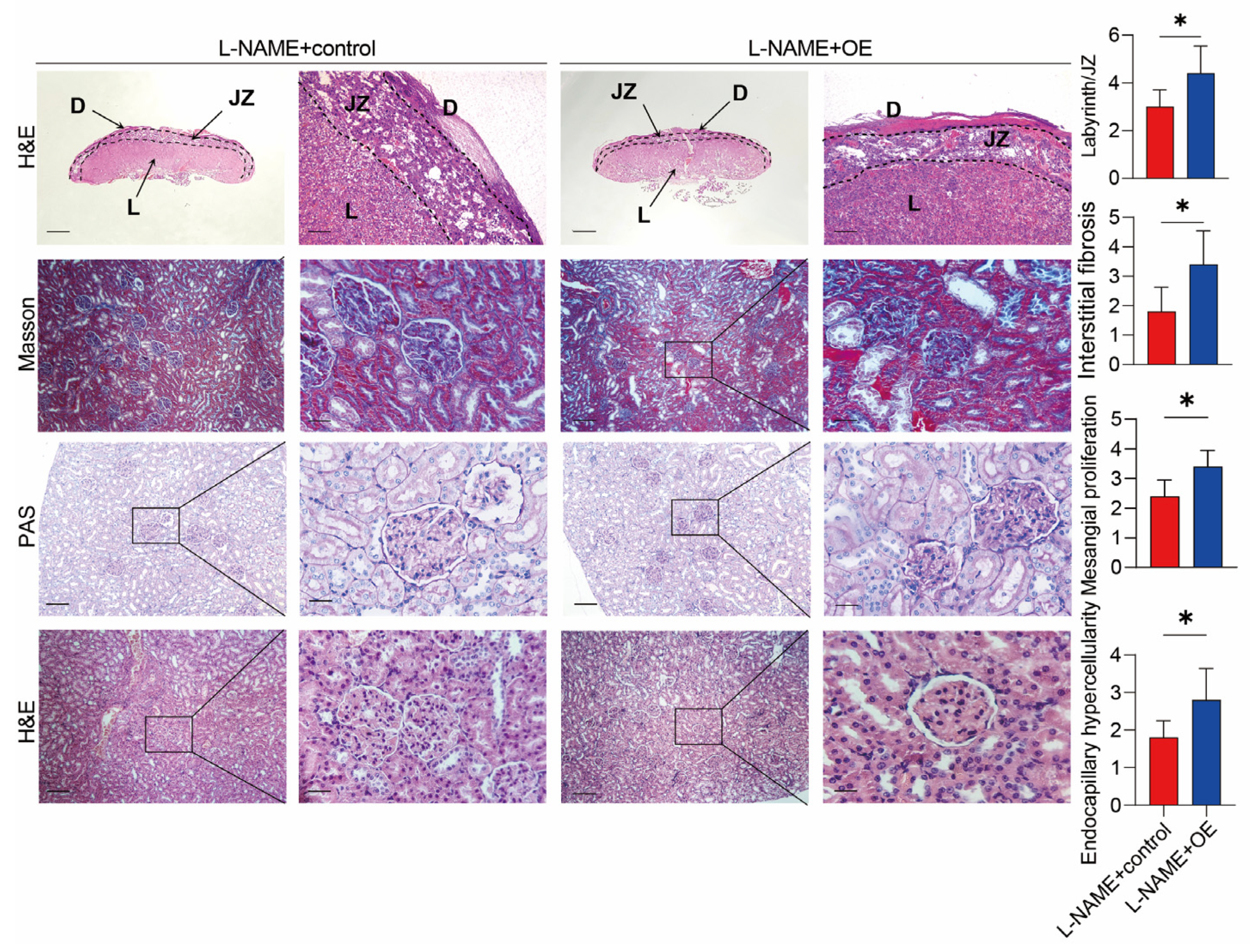
3.2. SERPINA5 Overexpression Aggravates L-NAME-Induced PE-like Conditions
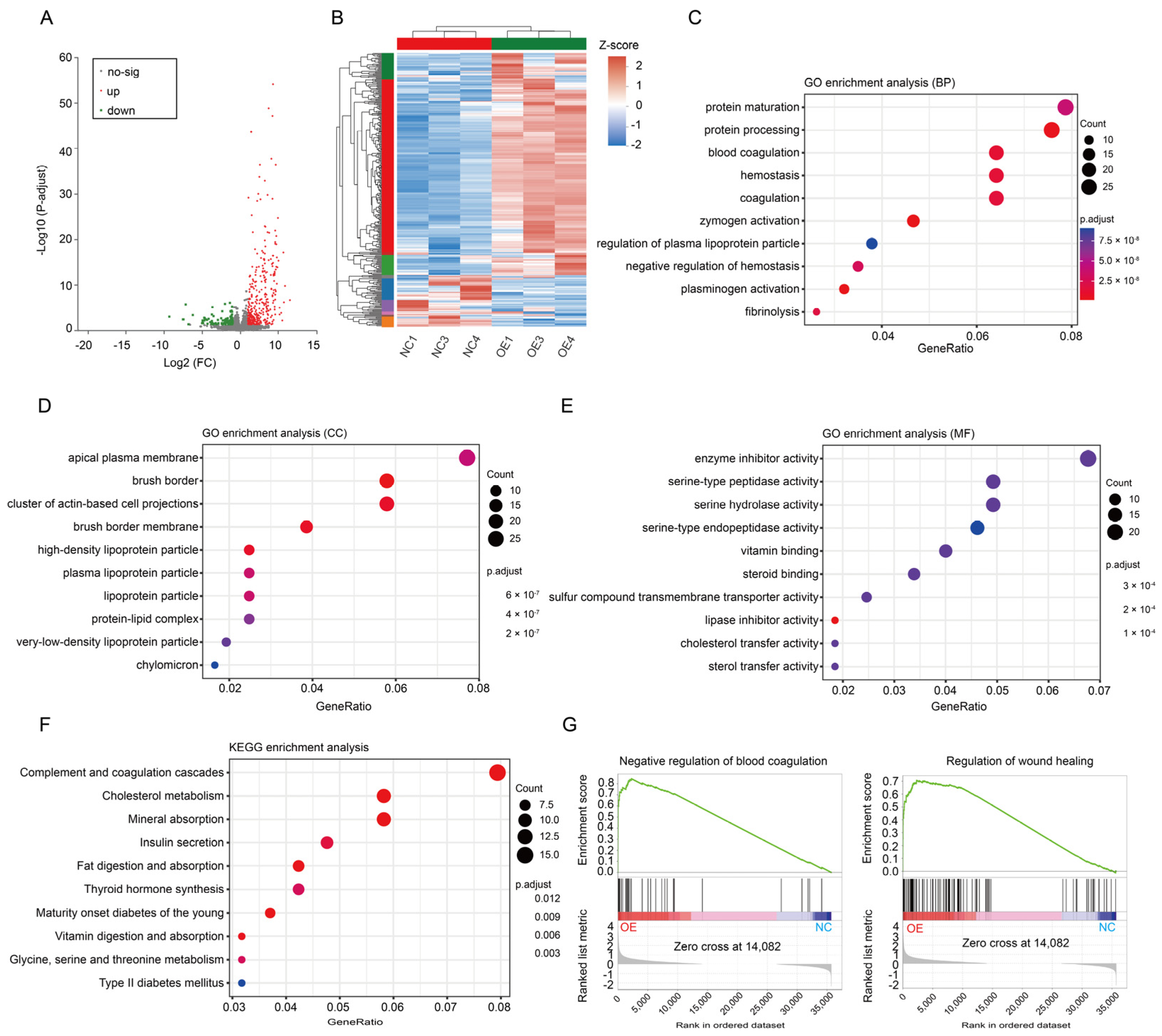
3.3. Changes in the Coagulation Pathway Mainly Led to PE in SERPINA5-Overexpressing Rats
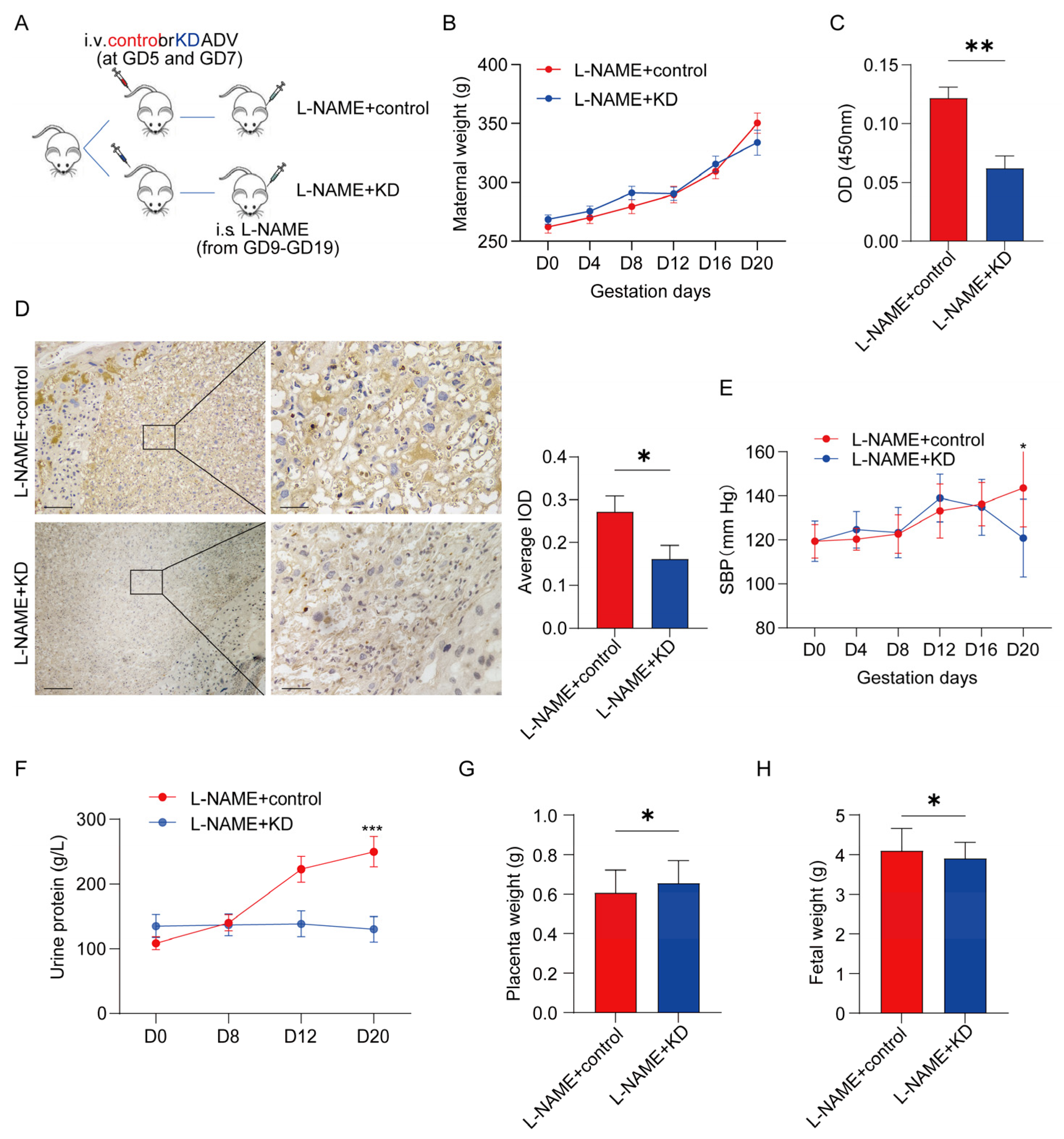
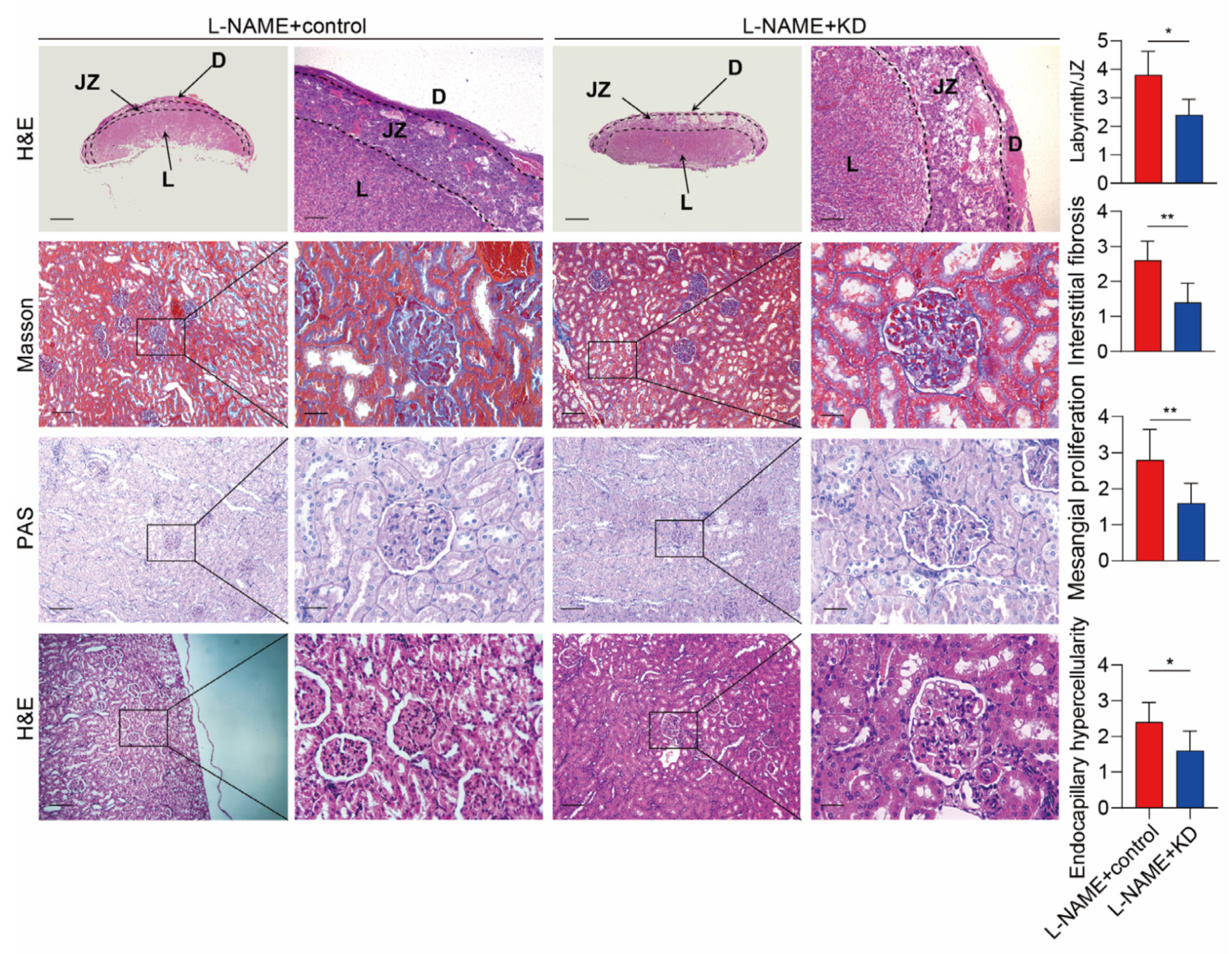
3.4. Inhibiting SERPINA5 Expression Alleviates PE Features
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ACOG Practice Bulletin No. 202: Gestational Hypertension and Pre-eclampsia. Obstet. Gynecol. 2019, 133, 1. [CrossRef]
- Burton, G.J.; Redman, C.W.; Roberts, J.M.; Moffett, A. Pre-eclampsia: Pathophysiology and clinical implications. BMJ 2019, 366, l2381. [Google Scholar] [CrossRef]
- Mol, B.W.J.; Roberts, C.T.; Thangaratinam, S.; Magee, L.A.; de Groot, C.J.M.; Hofmeyr, G.J. Pre-eclampsia. Lancet Lond. Engl. 2016, 387, 999–1011. [Google Scholar] [CrossRef]
- Markovitz, A.R.; Stuart, J.J.; Horn, J.; Williams, P.L.; Rimm, E.B.; Missmer, S.A.; Tanz, L.J.; Haug, E.B.; Fraser, A.; Timpka, S.; et al. Does pregnancy complication history improve cardiovascular disease risk prediction? Findings from the HUNT study in Norway. Eur. Heart J. 2019, 40, 1113–1120. [Google Scholar] [CrossRef]
- Ghulmiyyah, L.; Sibai, B. Maternal mortality from pre-eclampsia/eclampsia. Semin. Perinatol. 2012, 36, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Grandi, S.M.; Filion, K.B.; Yoon, S.; Ayele, H.T.; Doyle, C.M.; Hutcheon, J.A.; Smith, G.N.; Gore, G.C.; Ray, J.G.; Nerenberg, K.; et al. Cardiovascular Disease-Related Morbidity and Mortality in Women With a History of Pregnancy Complications. Circulation 2019, 139, 1069–1079. [Google Scholar] [CrossRef]
- Henderson, J.T.; Thompson, J.H.; Burda, B.U.; Cantor, A. Pre-eclampsia Screening: Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2017, 317, 1668–1683. [Google Scholar] [CrossRef]
- Wójtowicz, A.; Zembala-Szczerba, M.; Babczyk, D.; Kołodziejczyk-Pietruszka, M.; Lewaczyńska, O.; Huras, H. Early- and Late-Onset Preeclampsia: A Comprehensive Cohort Study of Laboratory and Clinical Findings according to the New ISHHP Criteria. Int. J. Hypertens. 2019, 2019, 4108271. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Gupta, A. Hypertensive Disorders of Pregnancy. Cardiol. Clin. 2019, 37, 345–354. [Google Scholar] [CrossRef]
- Zeng, S.; Han, M.; Jiang, M.; Liu, F.; Hu, Y.; Long, Y.; Zhu, C.; Zeng, F.; Gan, Q.; Ye, W.; et al. Serum complement proteomics reveal biomarkers for hypertension disorder of pregnancy and the potential role of Clusterin. Reprod. Biol. Endocrinol. RBE 2021, 19, 56. [Google Scholar] [CrossRef]
- Silva, J.F.; Serakides, R. Intrauterine trophoblast migration: A comparative view of humans and rodents. Cell Adhes. Migr. 2016, 10, 88–110. [Google Scholar] [CrossRef] [PubMed]
- Staff, A.C. The two-stage placental model of pre-eclampsia: An update. J. Reprod. Immunol. 2019, 134–135, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.M.; Hubel, C.A. The two stage model of pre-eclampsia: Variations on the theme. Placenta 2009, 30, S32–S37. [Google Scholar] [CrossRef]
- Moufarrej, M.N.; Vorperian, S.K.; Wong, R.J.; Campos, A.A.; Quaintance, C.C.; Sit, R.V.; Tan, M.; Detweiler, A.M.; Mekonen, H.; Neff, N.F.; et al. Early prediction of pre-eclampsia in pregnancy with cell-free RNA. Nature 2022, 602, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Zhao, L.; Shi, J.; Yang, H. Plasma SerpinA5 in conjunction with uterine artery pulsatility index and clinical risk factor for the early prediction of pre-eclampsia. PLoS ONE 2021, 16, e0258541. [Google Scholar] [CrossRef]
- Long, Y.; Zeng, S.; Gao, F.; Liu, F.; Zhang, Y.; Zhou, C.; Zhu, C.; Zhao, X.; Han, M.; Gan, Q.; et al. SERPINA5 may promote the development of pre-eclampsia by disruption of the uPA/uPAR pathway. Transl. Res. J. Lab. Clin. Med. 2023, 251, 14–26. [Google Scholar] [CrossRef]
- Yallampalli, C.; Garfield, R.E. Inhibition of nitric oxide synthesis in rats during pregnancy produces signs similar to those of pre-eclampsia. Am. J. Obstet. Gynecol. 1993, 169, 1316–1320. [Google Scholar] [CrossRef]
- Kopincová, J.; Púzserová, A.; Bernátová, I. L-NAME in the cardiovascular system-nitric oxide synthase activator? Pharmacol. Rep. PR 2012, 64, 511–520. [Google Scholar] [CrossRef]
- Ramesar, S.V.; Mackraj, I.; Gathiram, P.; Moodley, J. Sildenafil citrate decreases sFlt-1 and sEng in pregnant l-NAME treated Sprague–Dawley rats. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011, 157, 136–140. [Google Scholar] [CrossRef]
- Bakrania, B.A.; George, E.M.; Granger, J.P. Animal models of pre-eclampsia: Investigating pathophysiology and therapeutic targets. Am. J. Obstet. Gynecol. 2022, 226, S973–S987. [Google Scholar] [CrossRef]
- Benova, M.; Herichova, I.; Stebelova, K.; Paulis, L.; Krajcirovicova, K.; Simko, F.; Zeman, M. Effect of L-NAME-induced hypertension on melatonin receptors and melatonin levels in the pineal gland and the peripheral organs of rats. Hypertens. Res. Off. J. Jpn Soc. Hypertens. 2009, 32, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, N.; Wang, B.; Niu, X.; Cai, W.; Li, Y.; Li, Y.; Chen, S. Effect and mechanism of prophylactic use of tadalafil during pregnancy on l-NAME-induced preeclampsia-like rats. Placenta 2020, 99, 35–44. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, H.; Zhang, Y.; Shi, J.; Chen, R.; Xiao, X. CircSFXN1 regulates the behaviour of trophoblasts and likely mediates pre-eclampsia. Placenta 2020, 101, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. IMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Zeng, S.; Pan, Y.; Liu, F.; Yin, J.; Jiang, M.; Long, Y.; Zhao, X.; Lash, G.E.; Yang, H. Role of clusterin in the regulation of trophoblast development and pre-eclampsia. Biochem. Biophys. Res. Commun. 2021, 583, 128–134. [Google Scholar] [CrossRef]
- Xia, Y.; Herlitz, L.C.; Gindea, S.; Wen, J.; Pawar, R.D.; Misharin, A.; Perlman, H.; Wu, L.; Wu, P.; Michaelson, J.S.; et al. Deficiency of fibroblast growth factor-inducible 14 (Fn14) preserves the filtration barrier and ameliorates lupus nephritis. J. Am. Soc. Nephrol. 2015, 26, 1053–1070. [Google Scholar] [CrossRef]
- Chelbi, S.T.; Mondon, F.; Jammes, H.; Buffat, C.; Mignot, T.-M.; Tost, J.; Busato, F.; Gut, I.; Rebourcet, R.; Laissue, P.; et al. Expressional and epigenetic alterations of placental serine protease inhibitors: SERPINA3 is a potential marker of preeclampsia. Hypertension 2007, 49, 76–83. [Google Scholar] [CrossRef]
- Yang, H.; Geiger, M. Cell penetrating SERPINA5 (ProteinC inhibitor, PCI): More questions than answers. Semin. Cell Dev. Biol. 2017, 62, 187–193. [Google Scholar] [CrossRef]
- Wahlmüller, F.C.; Yang, H.; Furtmüller, M.; Geiger, M. Regulation of the Extracellular SERPINA5 (Protein C Inhibitor) Penetration Through Cellular Membranes. Adv. Exp. Med. Biol. 2017, 966, 93–101. [Google Scholar] [CrossRef]
- Leavey, K.; Benton, S.J.; Grynspan, D.; Kingdom, J.C.; Bainbridge, S.A.; Cox, B.J. Unsupervised Placental Gene Expression Profiling Identifies Clinically Relevant Subclasses of Human Preeclampsia. Hypertension 2016, 68, 137–147. [Google Scholar] [CrossRef]
- De Rijk, E.P.C.T.; van Esch, E.; Flik, G. Pregnancy dating in the rat: Placental morphology and maternal blood parameters. Toxicol. Pathol. 2002, 30, 271–282. [Google Scholar] [CrossRef]
- Mao, J.; Jain, A.; Denslow, N.D.; Nouri, M.-Z.; Chen, S.; Wang, T.; Zhu, N.; Koh, J.; Sarma, S.J.; Sumner, B.W.; et al. Bisphenol A and bisphenol S disruptions of the mouse placenta and potential effects on the placenta-brain axis. Proc. Natl. Acad. Sci. USA 2020, 117, 4642–4652. [Google Scholar] [CrossRef]
- Gillis, E.E.; Williams, J.M.; Garrett, M.R.; Mooney, J.N.; Sasser, J.M. The Dahl salt-sensitive rat is a spontaneous model of superimposed pre-eclampsia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 309, R62–R70. [Google Scholar] [CrossRef]
- Maynard, S.E.; Min, J.-Y.; Merchan, J.; Lim, K.-H.; Li, J.; Mondal, S.; Libermann, T.A.; Morgan, J.P.; Sellke, F.W.; Stillman, I.E.; et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in pre-eclampsia. J. Clin. Investig. 2003, 111, 649–658. [Google Scholar] [CrossRef]
- Song, Z.; Ma, N.; Hayashi, T.; Gabazza, E.C.; Sugimura, Y.; Suzuki, K. Intracellular localization of protein C inhibitor (PCI) and urinary plasminogen activator in renal tubular epithelial cells from humans and human PCI gene transgenic mice. Histochem. Cell Biol. 2007, 128, 293–300. [Google Scholar] [CrossRef]
- Wakita, T.; Hayashi, T.; Yuasa, H.; Nishioka, J.; Kawamura, J.; Suzuki, K. Molecular cloning, tissue distribution and androgen regulation of rat protein C inhibitor. FEBS Lett. 1998, 429, 263–268. [Google Scholar] [CrossRef]
- Han, C.; Huang, P.; Lyu, M.; Dong, J. Oxidative Stress and Preeclampsia-Associated Prothrombotic State. Antioxidants 2020, 9, 1139. [Google Scholar] [CrossRef]
- Gris, J.-C.; Bouvier, S.; Cochery-Nouvellon, É.; Mercier, É.; Mousty, È.; Pérez-Martin, A. The role of haemostasis in placenta-mediated complications. Thromb. Res. 2019, 181 (Suppl. S1), S10–S14. [Google Scholar] [CrossRef] [PubMed]
- Phipps, E.A.; Thadhani, R.; Benzing, T.; Karumanchi, S.A. Pre-eclampsia: Pathogenesis, novel diagnostics and therapies. Nat. Rev. Nephrol. 2019, 15, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Cerdeira, A.S.; Wenger, J.; Salahuddin, S.; Lim, K.-H.; Ralston, S.J.; Thadhani, R.I.; Karumanchi, S.A. Plasma Concentrations of Soluble Endoglin versus Standard Evaluation in Patients with Suspected Pre-eclampsia. PLoS ONE 2012, 7, e48259. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hao, H.; Leeper, N.J.; Zhu, L. Recent Highlights of ATVB Thrombotic regulation from the endothelial cell perspectives. Arter. Thromb. Vasc. Biol. 2018, 38, e90–e95. [Google Scholar] [CrossRef] [PubMed]
- Bijsmans, I.T.G.W.; Smits, K.M.; de Graeff, P.; Wisman, G.B.A.; van der Zee, A.G.J.; Slangen, B.F.; de Bruïne, A.P.; van Engeland, M.; Sieben, N.L.; Van de Vijver, K.K. Loss of SerpinA5 protein expression is associated with advanced-stage serous ovarian tumors. Mod. Pathol. 2011, 24, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Asanuma, K.; Yoshikawa, T.; Hayashi, T.; Akita, N.; Nakagawa, N.; Hamada, Y.; Nishioka, J.; Kamada, H.; Gabazza, E.C.; Ido, M.; et al. Protein C inhibitor inhibits breast cancer cell growth, metastasis and angiogenesis independently of its protease inhibitory activity. Int. J. Cancer 2007, 121, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K. The multi-functional serpin, protein C inhibitor: Beyond thrombosis and hemostasis. J. Thromb. Haemost. JTH 2008, 6, 2017–2026. [Google Scholar] [CrossRef]
- Akita, N.; Ma, N.; Okamoto, T.; Asanuma, K.; Yoshida, K.; Nishioka, J.; Shimaoka, M.; Suzuki, K.; Hayashi, T. Host protein C inhibitor inhibits tumor growth, but promotes tumor metastasis, which is closely correlated with hypercoagulability. Thromb. Res. 2015, 135, 1203–1208. [Google Scholar] [CrossRef]
- Meijers, J.C.M.; Herwald, H. Protein C inhibitor. Semin. Thromb. Hemost. 2011, 37, 349–354. [Google Scholar] [CrossRef]
- Duley, L.; Henderson-Smart, D.J.; Meher, S.; King, J.F. Antiplatelet agents for preventing pre-eclampsia and its complications. Cochrane Database Syst. Rev. 2007, 2, CD004659. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, S.; Liu, Z.; Yin, J.; Li, S.; Jiang, M.; Yang, H.; Long, Y. Improvement in Clinical Features of L-NAME-Induced Preeclampsia-like Rats through Reduced SERPINA5 Expression. Biomolecules 2023, 13, 1792. https://doi.org/10.3390/biom13121792
Zeng S, Liu Z, Yin J, Li S, Jiang M, Yang H, Long Y. Improvement in Clinical Features of L-NAME-Induced Preeclampsia-like Rats through Reduced SERPINA5 Expression. Biomolecules. 2023; 13(12):1792. https://doi.org/10.3390/biom13121792
Chicago/Turabian StyleZeng, Shanshui, Zimeng Liu, Jiaye Yin, Shu Li, Min Jiang, Hongling Yang, and Yan Long. 2023. "Improvement in Clinical Features of L-NAME-Induced Preeclampsia-like Rats through Reduced SERPINA5 Expression" Biomolecules 13, no. 12: 1792. https://doi.org/10.3390/biom13121792
APA StyleZeng, S., Liu, Z., Yin, J., Li, S., Jiang, M., Yang, H., & Long, Y. (2023). Improvement in Clinical Features of L-NAME-Induced Preeclampsia-like Rats through Reduced SERPINA5 Expression. Biomolecules, 13(12), 1792. https://doi.org/10.3390/biom13121792





