The Free Energy of Nucleosomal DNA Based on the Landau Model and Topology
Abstract
:1. Introduction
2. Theory
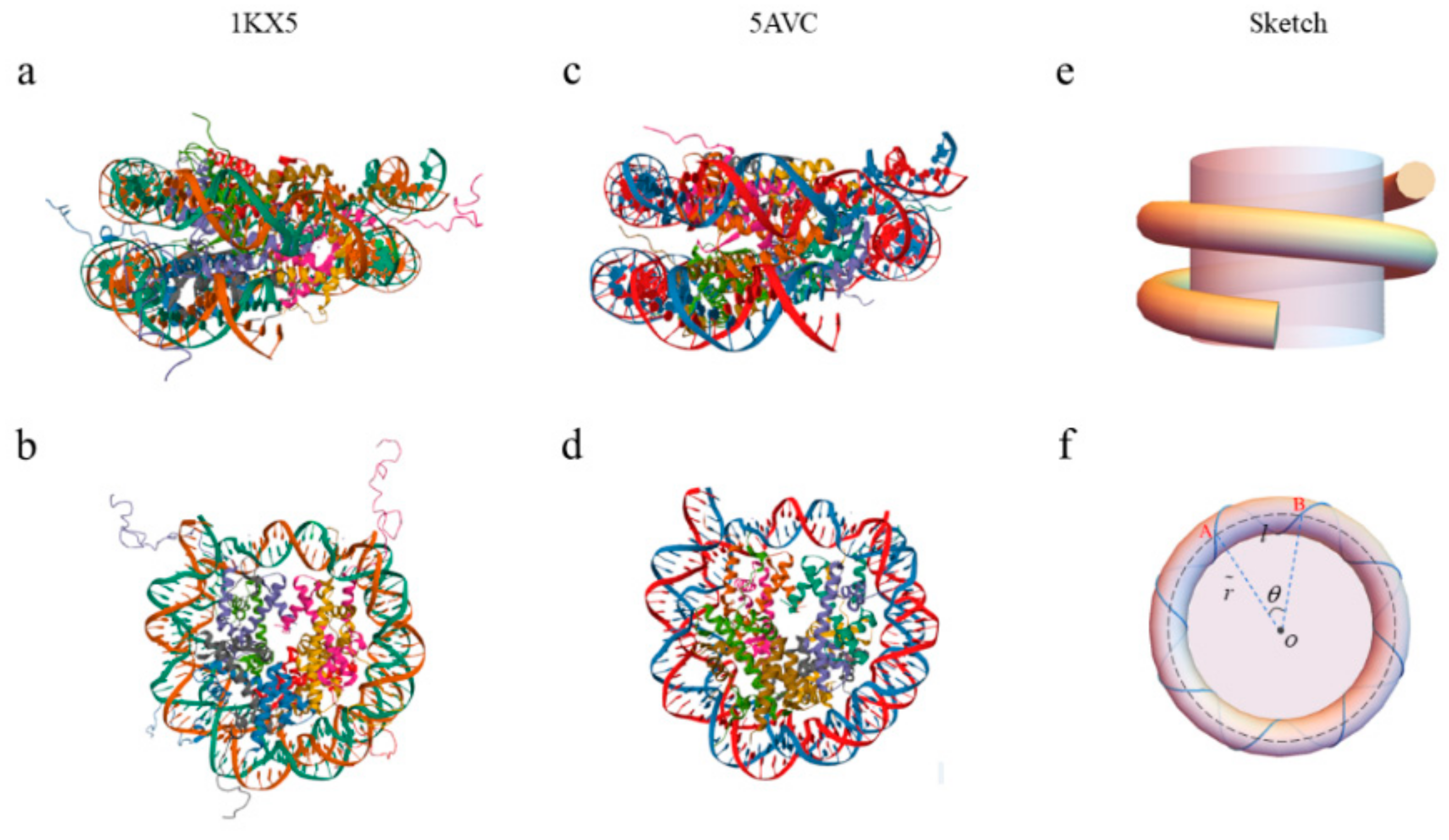
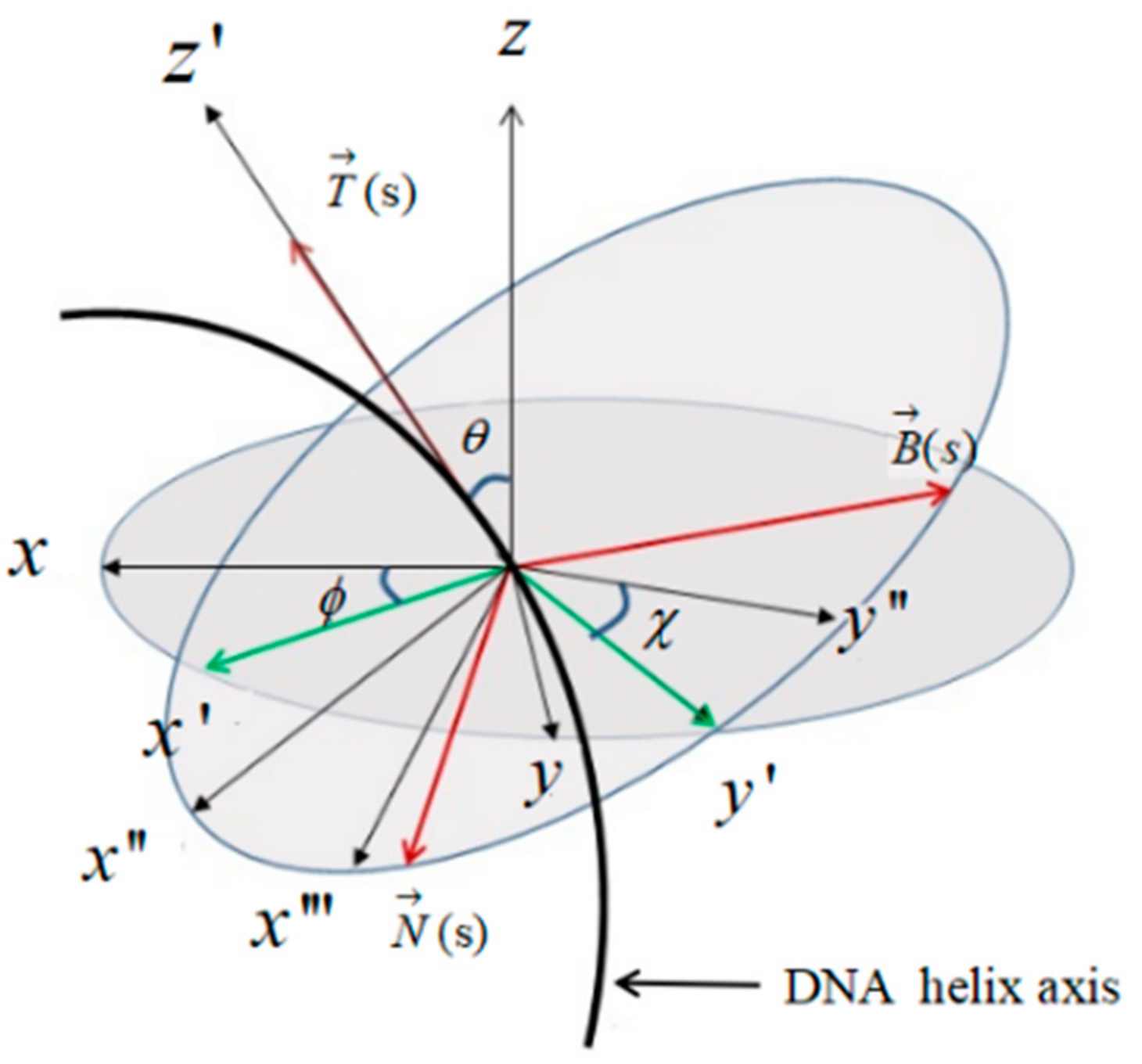
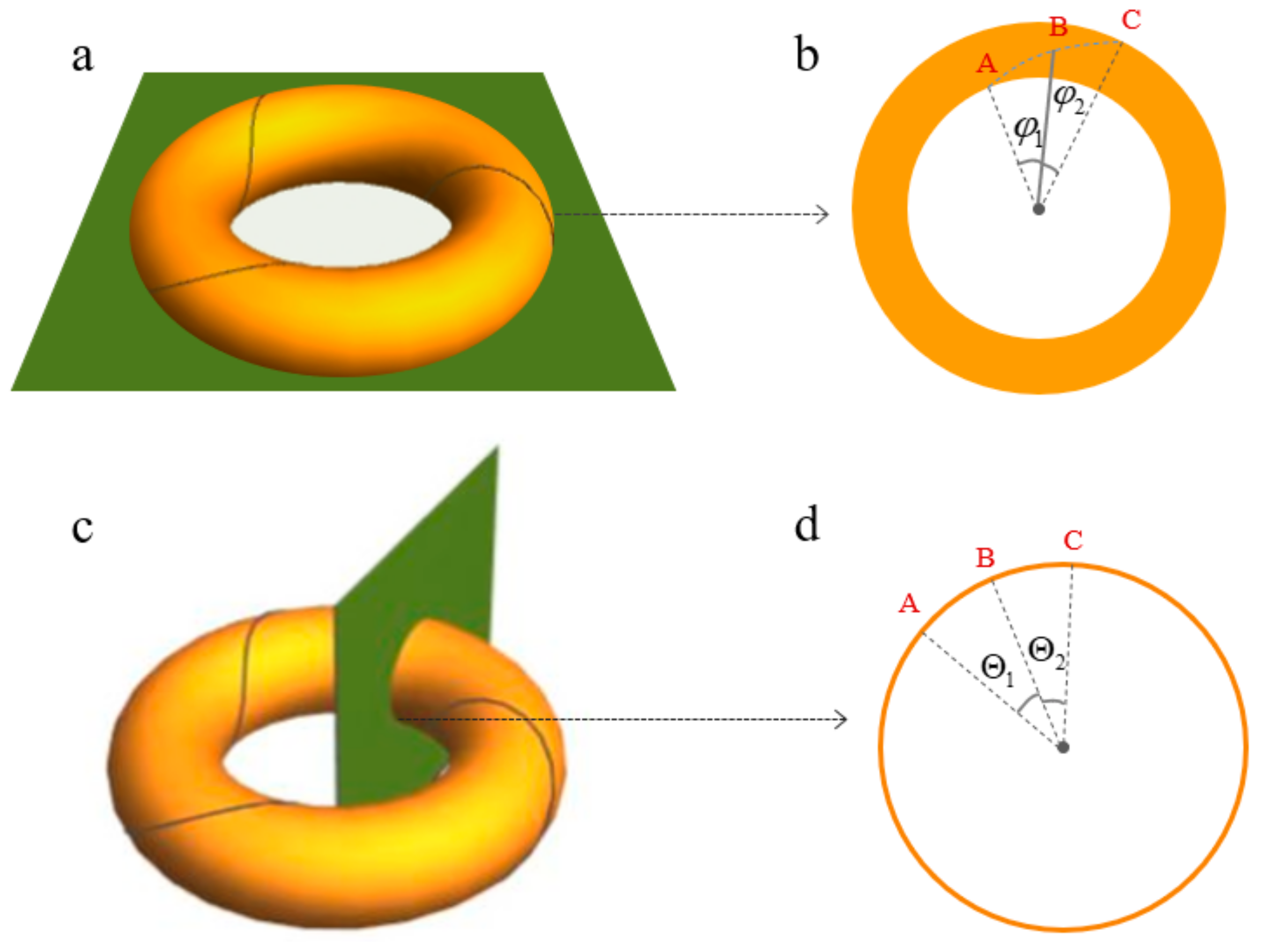
2.1. The Topology and Mechanical Properties of Nucleosomal DNA
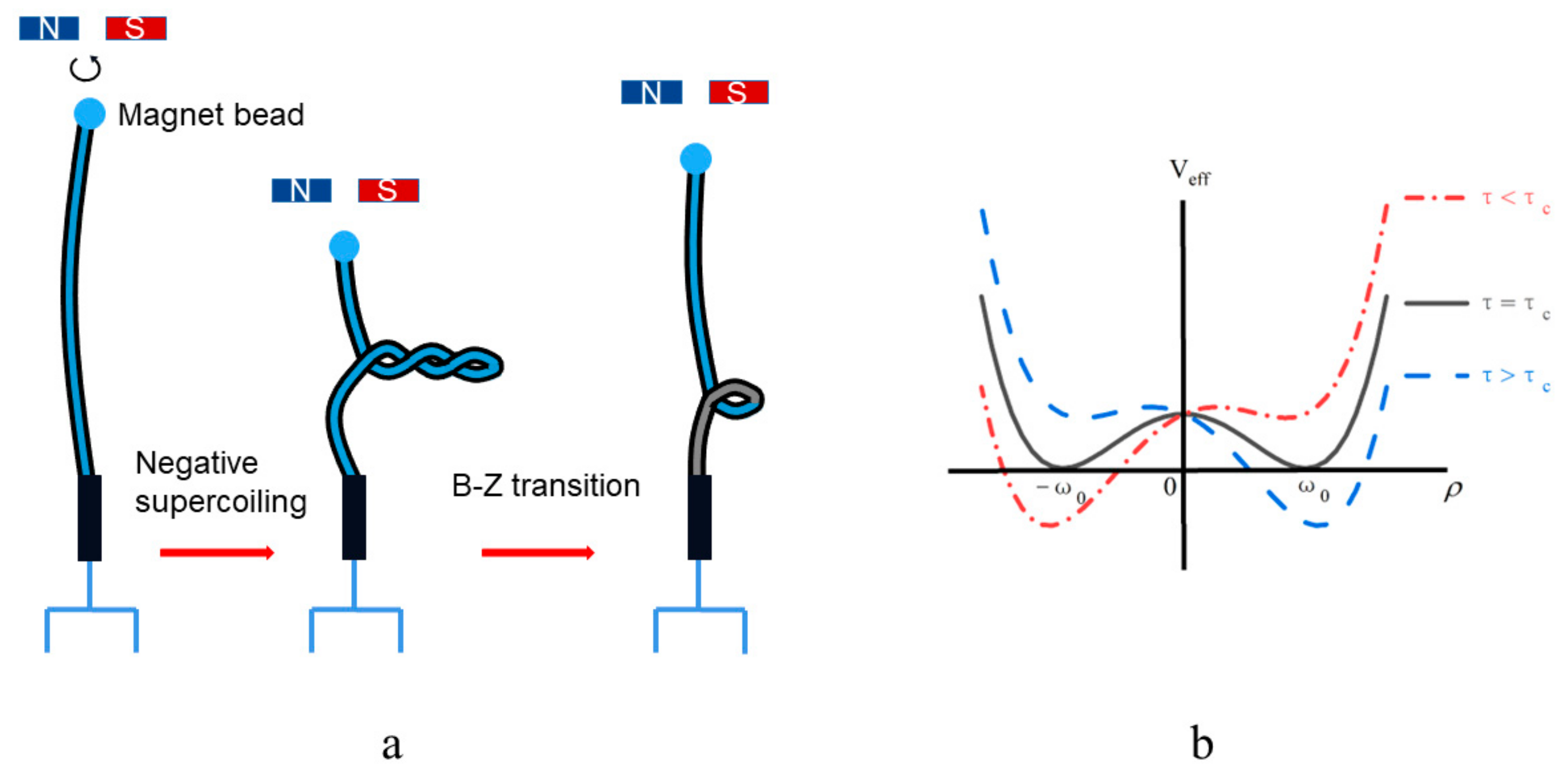
2.2. The DNA Free Energy Relating to Its Topology
3. Results
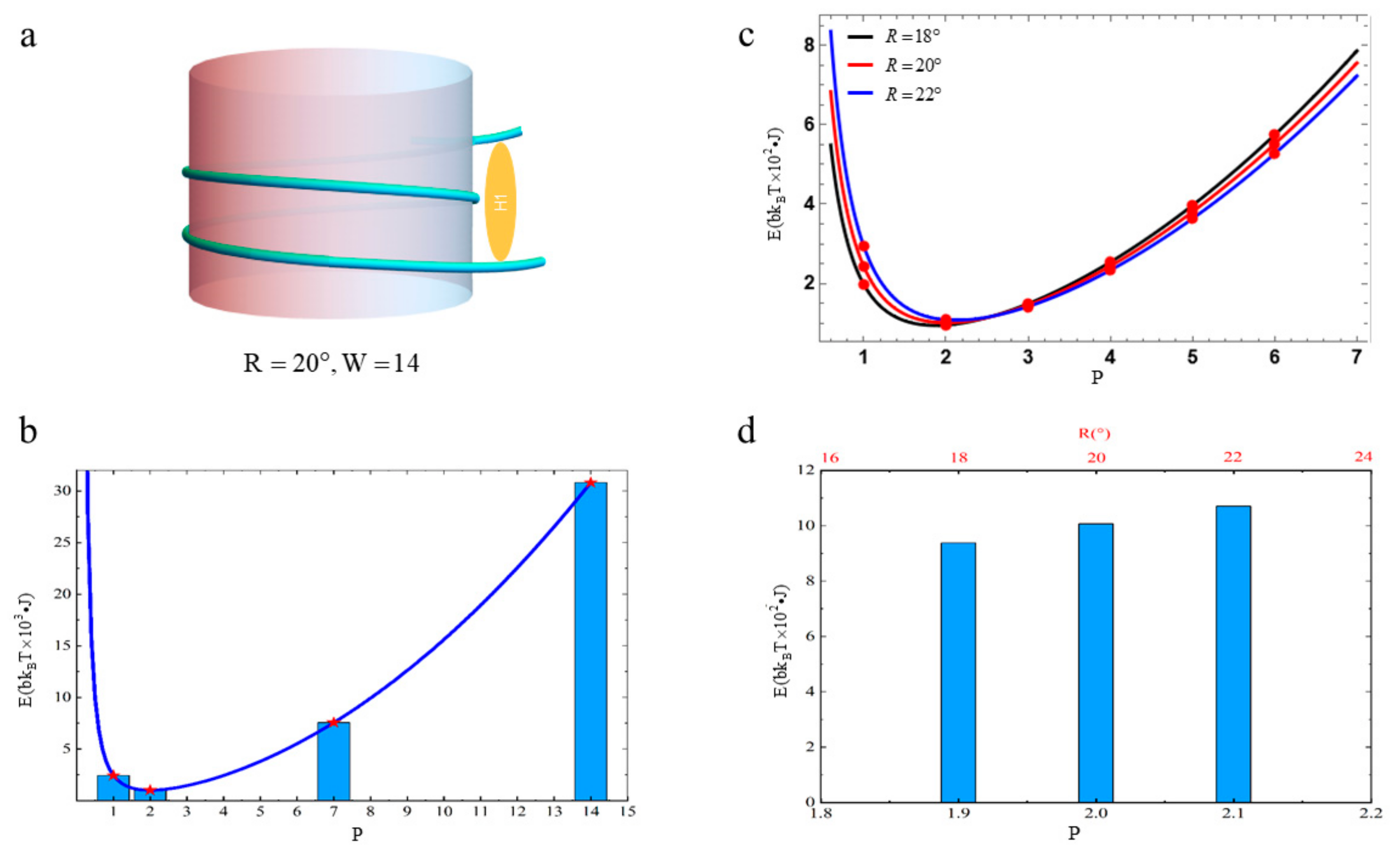
3.1. The Free Energy of Nucleosomal DNA
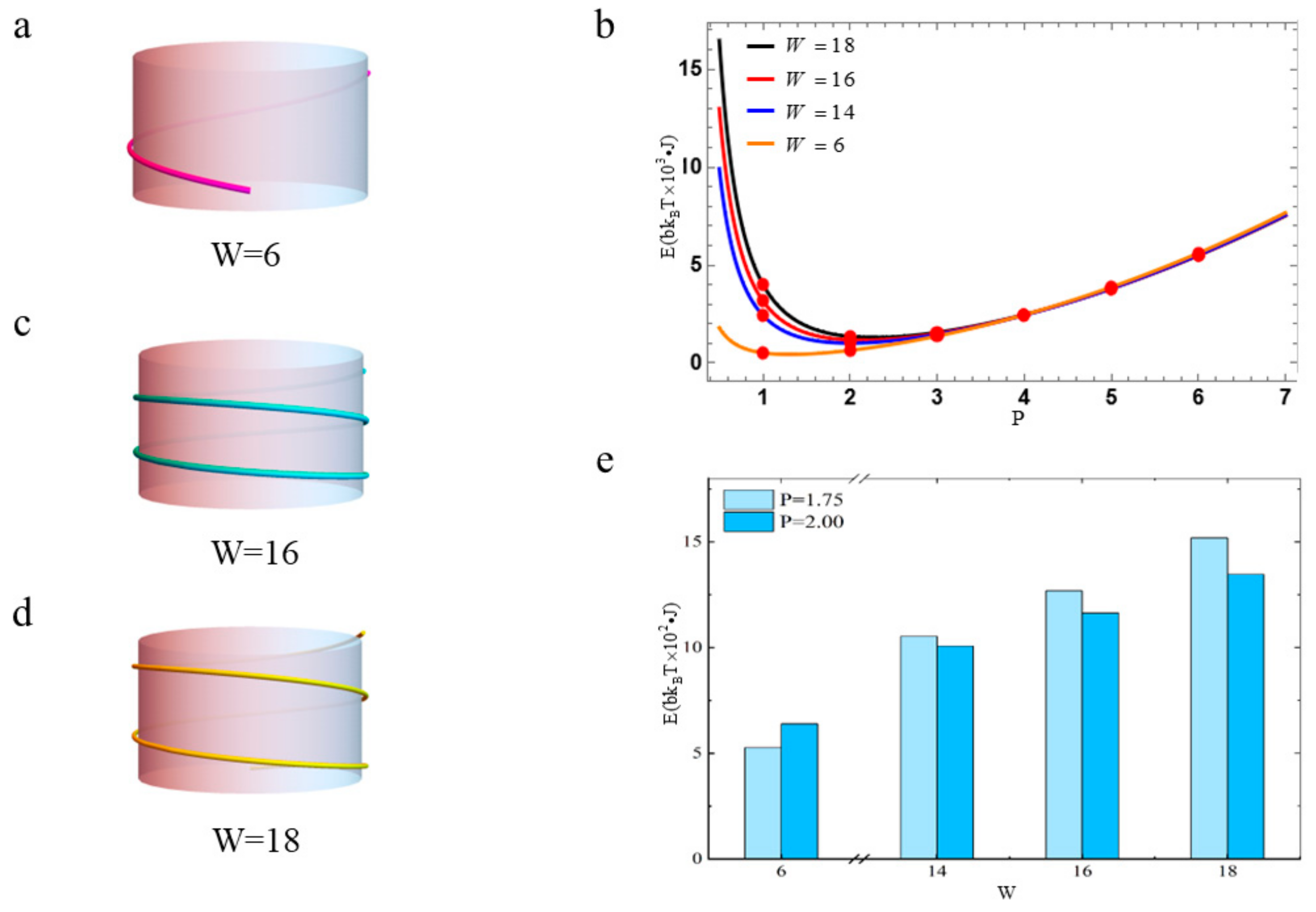
3.2. The Free Energy of Nucleosomal DNA with Different Diameters of Histone Octamers
3.3. The Free Energy of Nucleosomal DNA with Different Linking Numbers
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liverpool, T.B.; Golestanian, R.; Kremer, K. Statistical Mechanics of Double-Stranded Semiflexible Polymers. Phys. Rev. Lett. 1998, 80, 405–408. [Google Scholar] [CrossRef]
- Strick, T.; Allemand, J.-F.; Bensimon, D.; Croquette, V. Behavior of supercoiled DNA. Biophys. J. 1998, 74, 2016–2028. [Google Scholar] [CrossRef]
- Marko, J.F. Course 7 Introduction to single-DNA micromechanics. Les Houches 2005, 82, 211–270. [Google Scholar]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]
- Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; An, P.; et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [PubMed]
- Theologis, A.; Ecker, J.R.; Palm, C.J.; Federspiel, N.A.; Kaul, S.; White, O.; Alonso, J.; Altafi, H.; Araujo, R.; Bowman, C.L.; et al. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 2000, 408, 816–820. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Bednar, J.; Garcia-Saez, I.; Boopathi, R.; Cutter, A.R.; Papai, G.; Reymer, A.; Syed, S.H.; Lone, I.N.; Tonchev, O.; Crucifix, C.; et al. Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Mol. Cell 2017, 66, 384–397.e8. [Google Scholar] [CrossRef]
- McGhee, J.D.; Felsenfeld, G. Nucleosome structure. Annu. Rev. Biochem. 1980, 49, 1115–1156. [Google Scholar] [CrossRef]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Richmond, T.J.; Davey, C.A. The structure of DNA in the nucleosome core. Nature 2003, 423, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Thoma, F.; Koller, T.; Klug, A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979, 83, 403–427. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Gaullier, G.; Luger, K. Nucleosome structure and dynamics are coming of age. Nat. Struct. Mol. Biol. 2019, 26, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Zhao, C.; Ren, J.; Qu, X. Alzheimer’s disease amyloid beta converting left-handed Z-DNA back to right-handed B-form. Chem. Commun. 2010, 46, 7187–7189. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, B.; Barth, R.; Kim, E.; Davidson, I.F.; Bauer, B.; van Laar, T.; Yang, W.; Ryu, J.-K.; van der Torre, J.; Peters, J.-M.; et al. SMC complexes can traverse physical roadblocks bigger than their ring size. Cell Rep. 2022, 41, 111491. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tokuda, J.M.; Topping, T.; Sutton, J.L.; Meisburger, S.P.; Pabit, S.A.; Gloss, L.M.; Pollack, L. Revealing transient structures of nucleosomes as DNA unwinds. Nucleic Acids Res. 2014, 42, 8767–8776. [Google Scholar] [CrossRef]
- Donovan, B.T.; Luo, Y.; Meng, Z.; Poirier, M.G. The nucleosome unwrapping free energy landscape defines distinct regions of transcription factor accessibility and kinetics. Nucleic Acids Res. 2023, 51, 1139–1153. [Google Scholar] [CrossRef]
- Forties, R.A.; North, J.A.; Javaid, S.; Tabbaa, O.P.; Fishel, R.; Poirier, M.G.; Bundschuh, R. A quantitative model of nucleosome dynamics. Nucleic Acids Res. 2011, 39, 8306–8313. [Google Scholar] [CrossRef] [PubMed]
- Todolli, S.; Young, R.T.; Watkins, A.S.; Sha, A.B.; Yager, J.; Olson, W.K. Surprising Twists in Nucleosomal DNA with Implication for Higher-order Folding. J. Mol. Biol. 2021, 433, 167121. [Google Scholar] [CrossRef] [PubMed]
- Cleri, F.; Landuzzi, F.; Blossey, R. Mechanical evolution of DNA double-strand breaks in the nucleosome. PLoS Comput. Biol. 2018, 14, e1006224. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, Y.; Ou-Yang, Z.-C. Elastic theories of single DNA molecules. Physica A 2002, 306, 359–367. [Google Scholar] [CrossRef]
- Morozov, A.V.; Fortney, K.; Gaykalova, D.A.; Studitsky, V.M.; Widom, J.; Siggia, E.D. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009, 37, 4707–4722. [Google Scholar] [CrossRef]
- Shrestha, P.; Yang, D.; Tomov, T.E.; MacDonald, J.I.; Ward, A.; Bergal, H.T.; Krieg, E.; Cabi, S.; Luo, Y.; Nathwani, B.; et al. Single-molecule mechanical fingerprinting with DNA nanoswitch calipers. Nat. Nanotechnol. 2021, 16, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Smith, S.B.; Liphardt, J.; Smith, D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000, 10, 279–285. [Google Scholar] [CrossRef]
- Bell, N.A.W.; Molloy, J.E. Single-molecule force spectroscopy reveals binding and bridging dynamics of PARP1 and PARP2 at DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 2023, 120, e2214209120. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shi, X. Knot soliton in DNA and geometric structure of its free-energy density. J. Biol. Phys. 2018, 44, 81–91. [Google Scholar] [CrossRef]
- Zhang, B.; Zheng, W.; Papoian, G.A.; Wolynes, P.G. Exploring the Free Energy Landscape of Nucleosomes. J. Am. Chem. Soc. 2016, 138, 8126–8133. [Google Scholar] [CrossRef]
- Shi, X. Energy spectrum of the ideal DNA knot on a torus. Eur. Biophys. J. 2023. [Google Scholar] [CrossRef] [PubMed]
- White, J.H. Self-Linking and the Gauss Integral in Higher Dimensions. Am. J. Math. 1969, 91, 693–727. [Google Scholar] [CrossRef]
- Davey, C.A.; Sargent, D.F.; Luger, K.; Maeder, A.W.; Richmond, T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002, 319, 1097–1113. [Google Scholar] [CrossRef]
- Wakamori, M.; Fujii, Y.; Suka, N.; Shirouzu, M.; Sakamoto, K.; Umehara, T.; Yokoyama, S. Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci. Rep. 2015, 5, 17204. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, A. DNA packaging: Nucleosomes and chromatin. Nat. Educ. 2008, 1, 26. [Google Scholar]
- Finch, J.T.; Lutter, L.C.; Rhodes, D.; Brown, R.S.; Rushton, B.; Levitt, M.; Klug, A. Structure of nucleosome core particles of chromatin. Nature 1977, 269, 29–36. [Google Scholar] [CrossRef]
- Daussy, C.; Guinet, M.; Amy-Klein, A.; Djerroud, K.; Hermier, Y.; Briaudeau, S.; Bordé, C.J.; Chardonnet, C. First direct determination of the Boltzmann constant by an optical method. Phys. Rev. Lett. 2007, 98, 250801. [Google Scholar] [CrossRef]
- Mohr, P.J.; Taylor, B.N. CODATA recommended values of the fundamental physical constants: 2002. Rev. Mod. Phys. 2005, 77, 1–107. [Google Scholar] [CrossRef]
- Marko, J.F.; Neukirch, S. Global force-torque phase diagram for the DNA double helix: Structural transitions, triple points, and collapsed plectonemes. Phys. Rev. E 2013, 88, 062722. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Kim, S.H.; Hong, S.C. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc. Natl. Acad. Sci. USA 2010, 107, 4985–4990. [Google Scholar] [CrossRef] [PubMed]
- Okushima, T.; Kuratsuji, H. DNA as a one-dimensional chiral material: Application to the structural transition between B form and Z form. Phys. Rev. E 2011, 84, 021926. [Google Scholar] [CrossRef] [PubMed]
- Schvartzman, J.B.; Hernández, P.; Krimer, D.B.; Dorier, J.; Stasiak, A. Closing the DNA replication cycle: From simple circular molecules to supercoiled and knotted DNA catenanes. Nucleic Acids Res. 2019, 47, 7182–7198. [Google Scholar] [CrossRef]
- Landau, L.D.; Lifshitz, E.M.; Kosevich, A.M.; Pitaevskii, L.P. Theory of Elasticity; Elsevier: Amsterdam, The Netherlands, 1986; Volume 7. [Google Scholar]
- Fuller, F.B. Decomposition of the linking number of a closed ribbon: A problem from molecular biology. Proc. Natl. Acad. Sci. USA 1978, 75, 3557–3561. [Google Scholar] [CrossRef]
- Liverpool, T.B.; Harris, S.A.; Laughton, C.A. Supercoiling and denaturation of DNA loops. Phys. Rev. Lett. 2008, 100, 238103. [Google Scholar] [CrossRef] [PubMed]
- Racko, D.; Benedetti, F.; Dorier, J.; Stasiak, A. Are TADs supercoiled? Nucleic Acids Res. 2019, 47, 521–532. [Google Scholar] [CrossRef]
- Racko, D.; Benedetti, F.; Goundaroulis, D.; Stasiak, A. Chromatin loop extrusion and chromatin unknotting. Polymers 2018, 10, 1126. [Google Scholar] [CrossRef] [PubMed]
- Marko, J.F.; Siggia, E.D. Fluctuations and supercoiling of DNA. Science 1994, 265, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Racko, D.; Benedetti, F.; Dorier, J.; Burnier, Y.; Stasiak, A. Generation of supercoils in nicked and gapped DNA drives DNA unknotting and postreplicative decatenation. Nucleic Acids Res. 2015, 43, 7229–7236. [Google Scholar] [CrossRef] [PubMed]
- Bates, A.D.; Maxwell, A. DNA Topology; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Kobayashi, M.; Nitta, M. Torus knots as Hopfions. Phys. Lett. B 2014, 728, 314–318. [Google Scholar] [CrossRef]
- Winfree, A.T. Persistent tangled vortex rings in generic excitable media. Nature 1994, 371, 233–236. [Google Scholar] [CrossRef]
- Shi, X. The shapes of the knots corresponding to the special Hopfions. Eur. Phys. J. C 2023, 83, 691. [Google Scholar] [CrossRef]
- Hall, M.A.; Shundrovsky, A.; Bai, L.; Fulbright, R.M.; Lis, J.T.; Wang, M.D. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 2009, 16, 124–129. [Google Scholar] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Shi, X. The Free Energy of Nucleosomal DNA Based on the Landau Model and Topology. Biomolecules 2023, 13, 1686. https://doi.org/10.3390/biom13121686
Yang H, Shi X. The Free Energy of Nucleosomal DNA Based on the Landau Model and Topology. Biomolecules. 2023; 13(12):1686. https://doi.org/10.3390/biom13121686
Chicago/Turabian StyleYang, Huimin, and Xuguang Shi. 2023. "The Free Energy of Nucleosomal DNA Based on the Landau Model and Topology" Biomolecules 13, no. 12: 1686. https://doi.org/10.3390/biom13121686
APA StyleYang, H., & Shi, X. (2023). The Free Energy of Nucleosomal DNA Based on the Landau Model and Topology. Biomolecules, 13(12), 1686. https://doi.org/10.3390/biom13121686






