A Spatial Transcriptome Reveals Changes in Tumor and Tumor Microenvironment in Oral Cancer with Acquired Resistance to Immunotherapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Characteristics
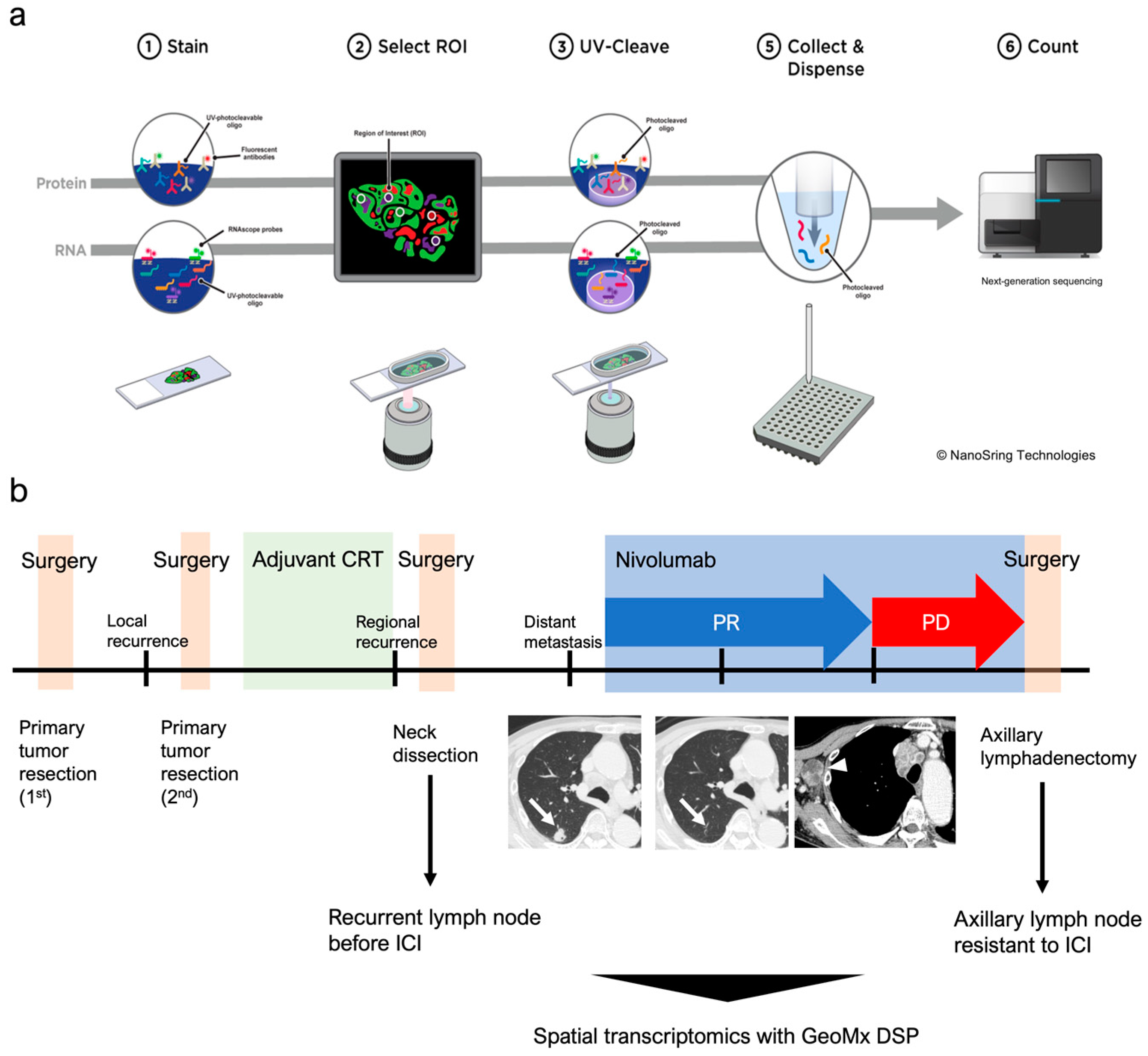
2.2. Tissue Preparation and Digital Spatial Profiling
3. Results
3.1. Fluorescent Staining and Selection of ROIs for Whole Transcriptome
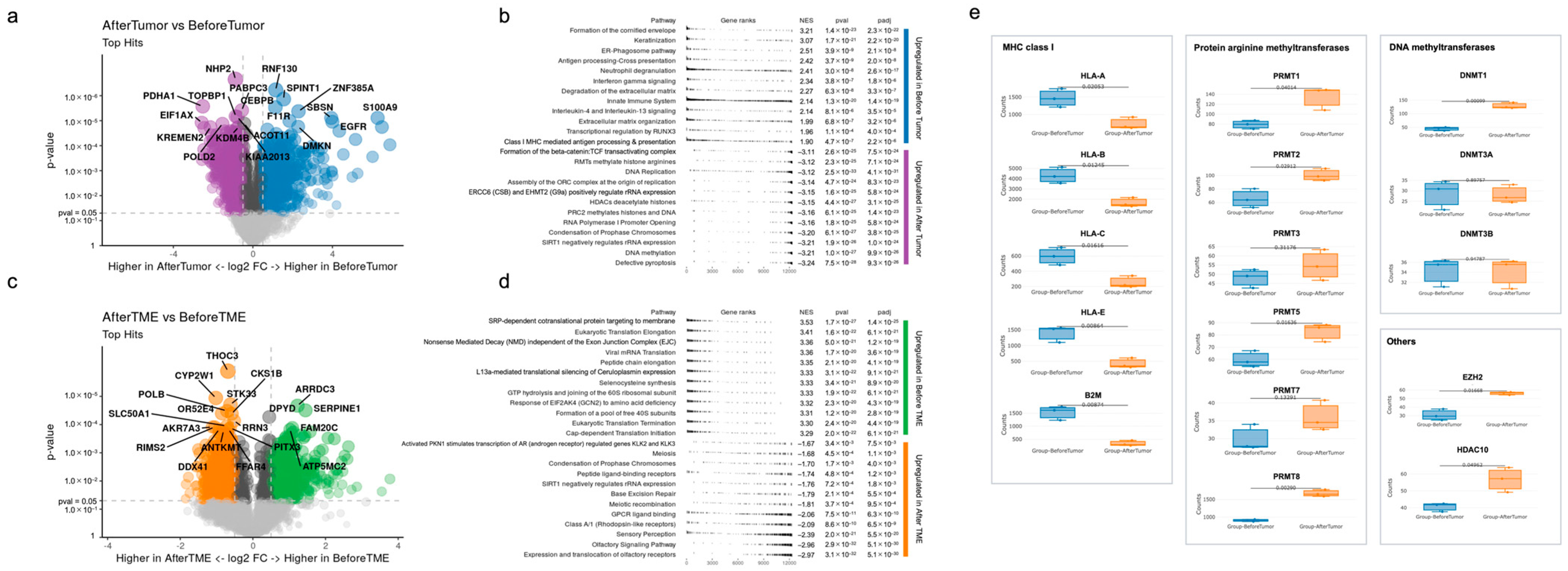
3.2. Differences in Gene Expression before Immunotherapy and after Acquiring Immunotherapy Resistance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chow, L.Q.M. Head and neck cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Burtness, B.; Rischin, D.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Brana, I.; Basté, N.; Neupane, P.; et al. Pembrolizumab alone or with chemotherapy for recurrent/metastatic head and neck squamous cell carcinoma in KEYNOTE-048: Subgroup analysis by programmed death ligand-1 combined positive score. J. Clin. Oncol. 2022, 40, 2321. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.J.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab vs. investigator’s choice in recurrent or metastatic squamous cell carcinoma of the head and neck: 2-year long-term survival update of CheckMate 141 with analyses by tumor PD-L1 expression. Oral Oncol. 2018, 81, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Harrington, K.J.; Burtness, B.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Brana, I.; Basté, N.; Neupane, P.; et al. Pembrolizumab with or without chemotherapy in recurrent or metastatic head and neck squamous cell carcinoma: Updated results of the phase III KEYNOTE-048 Study. J. Clin. Oncol. 2022, 41, 790–802. [Google Scholar] [CrossRef]
- Schoenfeld, A.J.; Hellmann, M.D. Acquired resistance to immune checkpoint inhibitors. Cancer Cell 2020, 37, 443–455. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Monkman, J.; Shah, E.T.; Matigian, N.; Adams, M.N.; O’Byrne, K. Spatial profiling identifies prognostic features of response to adjuvant therapy in triple negative breast cancer (TNBC). Front. Oncol. 2022, 11, 798296. [Google Scholar] [CrossRef]
- Campbell, K.M.; Thaker, M.; Medina, E.; Kalbasi, A.; Singh, A.; Ribas, A.; Nowicki, T.S. Spatial profiling reveals association between WNT pathway activation and T-cell exclusion in acquired resistance of synovial sarcoma to NY-ESO-1 transgenic T-cell therapy. J. Immunother. Cancer 2022, 10, e004190. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Taheri, T.; O’Byrne, K.; Hughes, B.G.M.; Kenny, L.; Punyadeera, C. Highly multiplexed digital spatial profiling of the tumor microenvironment of head and neck squamous cell carcinoma patients. Front. Oncol. 2021, 10, 607349. [Google Scholar] [CrossRef]
- Gavrielatou, N.; Vathiotis, I.; Aung, T.N.; Shafi, S.; Burela, S.; Fernandez, A.I.; Moutafi, M.; Burtness, B.; Economopoulou, P.; Anastasiou, M.; et al. Digital spatial profiling links beta-2-microglobulin expression with immune checkpoint blockade outcomes in head and neck squamous cell carcinoma. Cancer Res. Commun. 2023, 3, 558–563. [Google Scholar] [CrossRef]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Lipson, E.J.; Drake, C.G. Ipilimumab: An anti-CTLA-4 antibody for metastatic melanoma. Clin. Cancer Res. 2011, 17, 6958–6962. [Google Scholar] [CrossRef] [PubMed]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef]
- Gettinger, S.; Choi, J.; Hastings, K.; Truini, A.; Datar, I.; Sowell, R.; Wurtz, A.; Dong, W.; Cai, G.; Melnick, M.A.; et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 2017, 7, 1420–1435. [Google Scholar] [CrossRef]
- Liu, L.; Hu, Q.; Zhang, Y.; Sun, X.; Sun, R.; Ren, Z. Classification molecular subtypes of hepatocellular carcinoma based on PRMT-related genes. Front. Pharmacol. 2023, 14, 1145408. [Google Scholar] [CrossRef] [PubMed]
- Feustel, K.; Falchook, G.S. Protein arginine methyltransferase 5 (PRMT5) inhibitors in oncology clinical trials: A review. J. Immunother. Precis. Oncol. 2022, 5, 58–67. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Feng, Y.; Li, Y.; Tamiya, H.; Tocci, S.; Ronai, Z.E.A. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci. Transl. Med. 2020, 12, eaaz5683. [Google Scholar] [CrossRef]
- Zhou, L.; Mudianto, T.; Ma, X.; Riley, R.; Uppaluri, R. Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents anti–PD-1 resistance in head and neck cancer. Clin. Cancer Res. 2020, 26, 290–300. [Google Scholar] [CrossRef]
- Burkitt, K.; Saloura, V. Epigenetic modifiers as novel therapeutic targets and a systematic review of clinical studies investigating epigenetic inhibitors in head and neck cancer. Cancers 2021, 13, 5241. [Google Scholar] [CrossRef]
- Yoon, S.; Eom, G.H. HDAC and HDAC inhibitor: From cancer to cardiovascular diseases. Chonnam Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.P.; Wu, Q.; Voutsinas, J.; Fromm, J.R.; Jiang, X.; Pillarisetty, V.G.; Lee, S.M.; Santana-Davila, R.; Goulart, B.; Baik, C.S.; et al. A phase II trial of pembrolizumab and vorinostat in recurrent metastatic head and neck squamous cell carcinomas and salivary gland cancer. Clin. Cancer Res. 2020, 26, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Zheng, B.; Wang, J.; Zhao, G.; Yao, Z.; Niu, Z.; He, W. Histone deacetylase 10, a potential epigenetic target for therapy. Biosci. Rep. 2021, 41, BSR20210462. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Liu, M.; Yang, H.; Küçük, C.; You, H. New insights into epigenetic regulation of resistance to PD-1/PD-L1 blockade cancer immunotherapy: Mechanisms and therapeutic opportunities. Exp. Hematol. Oncol. 2022, 11, 101. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Gustafsson, J.R.; Katsioudi, G.; Degn, M.; Ejlerskov, P.; Issazadeh-Navikas, S.; Kornum, B.R. DNMT1 regulates expression of MHC class I in post-mitotic neurons. Mol. Brain 2018, 11, 36. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.L.; Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021, 12, 1786. [Google Scholar] [CrossRef]
- Camuzi, D.; De Amorim, Í.S.S.; Pinto, L.F.R.; Trivilin, L.O.; Mencalha, A.L.; Lima, S.C.S. Regulation is in the air: The relationship between hypoxia and epigenetics in cancer. Cells 2019, 8, 300. [Google Scholar] [CrossRef]
- Vezzani, B.; Carinci, M.; Previati, M.; Giacovazzi, S.; Della Sala, M.; Gafà, R.; Lanza, G.; Wieckowski, M.R.; Pinton, P.; Giorgi, C. Epigenetic regulation: A link between inflammation and carcinogenesis. Cancers 2022, 14, 1221. [Google Scholar] [CrossRef]
- Bourne, C.M.; Mun, S.S.; Dao, T.; Aretz, Z.E.; Molvi, Z.; Gejman, R.S.; Daman, A.; Takata, K.; Steidl, C.; Klatt, M.G.; et al. Unmasking the suppressed immunopeptidome of EZH2-mutated diffuse large B-cell lymphomas through combination drug treatment. Blood Adv. 2022, 6, 4107–4121. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.K.; Kim, S. An insight into GPCR and G-proteins as cancer drivers. Cells 2021, 10, 3288. [Google Scholar] [CrossRef] [PubMed]
- Wu, V.; Yeerna, H.; Nohata, N.; Chiou, J.; Harismendy, O.; Raimondi, F.; Inoue, A.; Russell, R.B.; Tamayo, P.; Gutkind, J.S. Illuminating the Onco-GPCRome: Novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 2019, 294, 11062–11086. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Cho, H.J.; Lee, C.E.; Koo, J.H. Odorant receptors in cancer. BMB Rep. 2022, 55, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Luoma, A.M.; Suo, S.; Wang, Y.; Gunasti, L.; Porter, C.B.M.; Nabilsi, N.; Tadros, J.; Ferretti, A.P.; Liao, S.; Gurer, C.; et al. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 2022, 185, 2918–2935. [Google Scholar] [CrossRef]
- McNamara, K.L.; Caswell-Jin, J.L.; Joshi, R.; Ma, Z.; Kotler, E.; Bean, G.R.; Kriner, M.; Zhou, Z.; Hoang, M.; Beechem, J.; et al. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nat. Cancer 2021, 2, 400–413. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwasa, Y.-i.; Nakajima, T.; Hori, K.; Yokota, Y.; Kitoh, R.; Uehara, T.; Takumi, Y. A Spatial Transcriptome Reveals Changes in Tumor and Tumor Microenvironment in Oral Cancer with Acquired Resistance to Immunotherapy. Biomolecules 2023, 13, 1685. https://doi.org/10.3390/biom13121685
Iwasa Y-i, Nakajima T, Hori K, Yokota Y, Kitoh R, Uehara T, Takumi Y. A Spatial Transcriptome Reveals Changes in Tumor and Tumor Microenvironment in Oral Cancer with Acquired Resistance to Immunotherapy. Biomolecules. 2023; 13(12):1685. https://doi.org/10.3390/biom13121685
Chicago/Turabian StyleIwasa, Yoh-ichiro, Tomoyuki Nakajima, Kentaro Hori, Yoh Yokota, Ryosuke Kitoh, Takeshi Uehara, and Yutaka Takumi. 2023. "A Spatial Transcriptome Reveals Changes in Tumor and Tumor Microenvironment in Oral Cancer with Acquired Resistance to Immunotherapy" Biomolecules 13, no. 12: 1685. https://doi.org/10.3390/biom13121685
APA StyleIwasa, Y.-i., Nakajima, T., Hori, K., Yokota, Y., Kitoh, R., Uehara, T., & Takumi, Y. (2023). A Spatial Transcriptome Reveals Changes in Tumor and Tumor Microenvironment in Oral Cancer with Acquired Resistance to Immunotherapy. Biomolecules, 13(12), 1685. https://doi.org/10.3390/biom13121685






