Abstract
Oxygen exchange reactions occurring at β-catalytic sites of the FOF1-ATP synthase/F1-ATPase imprint a unique record of molecular events during the catalytic cycle of ATP synthesis/hydrolysis. This work presents a new theory of oxygen exchange and tests it on oxygen exchange data recorded on ATP hydrolysis by mitochondrial F1-ATPase (MF1). The apparent rate constant of oxygen exchange governing the intermediate Pi–HOH exchange accompanying ATP hydrolysis is determined by kinetic analysis over a ~50,000-fold range of substrate ATP concentration (0.1–5000 μM) and a corresponding ~200-fold range of reaction velocity (3.5–650 [moles of Pi/{moles of F1-ATPase}−1 s−1]). Isotopomer distributions of [18O]Pi species containing 0, 1, 2, and 3 labeled oxygen atoms predicted by the theory have been quantified and shown to be in perfect agreement with the experimental distributions over the entire range of medium ATP concentrations without employing adjustable parameters. A novel molecular mechanism of steady-state multisite ATP hydrolysis by the F1-ATPase has been proposed. Our results show that steady-state ATP hydrolysis by F1-ATPase occurs with all three sites occupied by Mg-nucleotide. The various implications arising from models of energy coupling in ATP synthesis/hydrolysis by the ATP synthase/F1-ATPase have been discussed. Current models of ATP hydrolysis by F1-ATPase, including those postulated from single-molecule data, are shown to be effectively bisite models that contradict the data. The trisite catalysis formulated by Nath’s torsional mechanism of energy transduction and ATP synthesis/hydrolysis since its first appearance 25 years ago is shown to be in better accord with the experimental record. The total biochemical information on ATP hydrolysis is integrated into a consistent model by the torsional mechanism of ATP synthesis/hydrolysis and shown to elucidate the elementary chemical and mechanical events within the black box of enzyme catalysis in energy metabolism by F1-ATPase.
Keywords:
rotary ATPases; FOF1-ATP synthase and F1-ATPase; ATP synthesis and hydrolysis; energy metabolism and mitochondria; molecular mechanism, kinetics and conformational change; oxygen exchange and stochastic theory; ligand displacement/substitution and ligand permutation; Boyer’s binding change mechanism of ATP synthesis; Nath’s torsional mechanism of ATP synthesis and two-ion theory of energy coupling; 2-site vs. 3-site models of ATP synthesis/hydrolysis 1. Introduction
Given the fundamental importance of ATP as the cellular fuel in animals, plants, and eubacteria, the working mechanism of ATP synthesis/hydrolysis in energy metabolism by the FOF1-ATP synthase/F1-ATPase has been studied extensively. Structural studies using X-ray crystallography provided the first snapshots of this vital enzyme in animal mitochondria at high resolution and glimpses of its interacting parts [1,2]. The first crystal structure solved contained bound ATP analog AMP–PNP at one of its catalytic sites (designated as βTP), ADP at a second catalytic site (denoted by βDP), while a third site (βE) was empty of bound nucleotide [1] and seemed to be compatible with prevailing models. However, subsequent X-ray structures have shown that the three β-catalytic sites can contain a variety of nucleotide combinations, including one structure that contains the transition state analog, MgADPAlF4−, at two sites with the third site filled with MgADP and sulfate (that mimics the cleaved γ-phosphate) [2]. Recent developments in cryo-EM have been exploited by several groups and have provided more detailed structural information on the mitochondrial enzyme [3,4,5,6,7,8].
Single-molecule imaging of F1-ATPase gave the field spectacular videos of the mechanoenzyme as a rotary device [9,10,11,12,13,14,15]. It was specifically shown that the catalytic dwell in thermophilic F1 (TF1) is ~2 ms in duration, during which ATP hydrolysis occurs with a half-time of ~1 ms and ends with Pi release [11,15]. Further, bovine mitochondrial F1-ATPase (MF1) seemed to differ from bacterial F1 in that it has a Pi release dwell that occurs prior to the catalytic dwell [14].
In addition to the use of the above biophysical techniques, biochemical approaches continued to be employed to probe the rotary ATPases. Prominent among these was the utility of crosslinking tools, which had provided the first evidence for rotation in the ATPases [16,17]. In order to improve our understanding of the energy dynamics of rotary machines, various energy landscape methods were developed and applied to the ATPases [18,19]. At times, single-molecule and energy landscape approaches yielded insights that were in concurrence with each other. For instance, Frasch and coworkers discovered from single-molecule data collected with an unprecedented signal-to-noise ratio at an ultra-rapid frame rate of 200,000 that rotation in F1-ATPase was more complex than previously thought and was accompanied by a series of angular accelerations and decelerations [13] and elastic coupling [10] during the power stroke. Nath reached similar conclusions from his landscape approach that quantified the electrostatic motor torque vs. angle and the angular displacement, angular velocity vs. time relationship over a 120° cycle of the enzyme that incorporated a “wheels-within-wheels” mechanism of twisting [18]. A “wheels-within-wheels” mechanism is now being postulated for the bacterial flagellar motor since it is consistent with recent cryo-EM structures of the “5:2 rotary motors” [20]. However, puzzling dynamical effects were also uncovered by single-molecule recordings. Futai and colleagues found that stochastic fluctuations of catalysis are a general property of the F1-ATPase and concluded that understanding it requires “combining studies of steady-state kinetics with single molecule observation” [12].
Other rotary and linear molecular motor systems were studied both experimentally and computationally [21,22,23,24,25,26,27,28], and molecular dynamics [24,25] and multiscale simulations [26,27,28] were performed. In addition, electrostatic [29,30] and molecular systems biology/engineering [31,32,33] approaches were developed to understand function. Several reviews on the rapid developments in ATP and cell life [33,34,35,36,37,38] and cell death [39,40,41,42,43,44,45,46,47] were written. A crucial central question was the following: How could time-resolved information [48,49,50,51,52] on elementary events occurring in single catalytic sites of enzymes, such as in the β-subunits of the F1-ATPase, be gathered and interpreted? The dynamics of movement of the ε-subunit are incompletely understood and continue to be addressed by structural [8], biochemical [53,54], biophysical [52,55], and systems biology [31] approaches.
An interesting methodology by which time-resolved information on catalytic site events can be collated is by monitoring the oxygen exchange reactions [56,57,58]. As is well known, experimental data on the oxygen exchange reactions in the ATP synthesis mode by the FOF1-ATP synthase are scarce [57]. In particular, the characteristics of the intermediate ATP–HOH exchange, considered as a diagnostic of the oxidative phosphorylation process, were experimentally studied by Nath and coworkers [57]. A stochastic kinetic theory was developed from first principles to support the experiments and to analyze and interpret the experimental data in the synthesis mode. The variables in the concatenated system of differential equations were solved sequentially and shown to lead to a Poisson process as the steady-state solution of the continuous-time Markov chain [57]. However, given that the vast majority of studies have focused on the hydrolysis mode, good data on the exchange reactions occurring during ATP hydrolysis by the F1-ATPase, particularly on the intermediate Pi–HOH exchange (Figure 1), needs to be recorded.
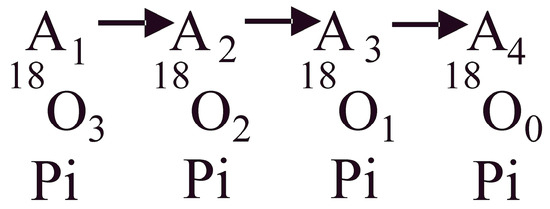
Figure 1.
Schematic of the oxygen exchange reactions within a single β-catalytic site in the F1 portion of the FOF1-ATP synthase/F1-ATPase. The process starts from an initial state of the system with a known distribution of the 18O label in the γ-phosphoryl group of ATP (A0; not shown in the schematic). Following terminal bond cleavage of the bound ATP and release of inorganic phosphate (Pi), the system progresses between enzyme states A1, A2, A3, and A4 containing respectively 3, 2, 1, and 0 atoms of 18O in the released Pi molecule due to intermediate Pi–HOH exchange occurring during ATP hydrolysis, shown by arrows. The 18O3, 18O2, 18O1, and 18O0 Pi distributions are measured, and the estimated distributions obtained by a stochastic kinetic theory are compared with the experimental distributions.
The above works have stimulated the development of at least two different theories or mechanisms of ATP synthesis/hydrolysis. Boyer’s binding change mechanism [58,59,60,61] was primarily arrived at based on the action of uncouplers on the exchange reactions [59]. It postulated reversible catalysis and site-site cooperativity and proposed that external energy input is primarily required for the release of bound ATP from a catalytic site but not for its synthesis [59,60,61]. According to a central tenet of the binding change mechanism, steady-state ATP synthesis/hydrolysis by the enzyme occurs with only two of the three β-catalytic sites filled with bound Mg-nucleotides, i.e., by a bisite mode of catalysis (2-site model of ATP synthesis/hydrolysis) [60,61]. An alternative Nath’s torsional mechanism of energy transduction and ATP synthesis/hydrolysis [18,30,31,32,33,57,62,63,64] considered an irreversible mode of catalysis and asymmetric interactions of the β-catalytic sites with the single copy γ- and ε-subunits of F1 and proposed that every elementary step in ATP synthesis requires energy [65,66,67,68,69,70,71,72,73,74]. According to a central tenet of the torsional mechanism, steady-state physiological ATP synthesis/steady-state Vmax ATP hydrolysis by the FOF1-ATP synthase/F1-ATPase occurs with all three β-catalytic sites filled with bound MgATP or MgADP, i.e., by a trisite mode of catalysis (3-site model of ATP synthesis/hydrolysis) [32,33]. The results presented here are consistent with oxygen exchange occurring at all three catalytic sites during the process of steady-state Vmax hydrolysis by F1-ATPase at high ATP concentrations, as would be predicted by 3-site models of ATP catalysis.
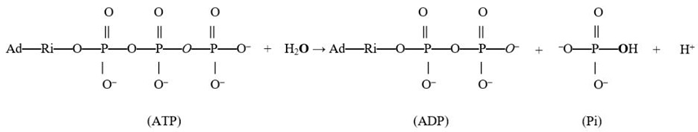
In this work, the intermediate exchange is defined as occurring when a substrate binds to an enzyme catalytic site, undergoes exchanges, and converts to a product, which is then released (from the site into the medium) and contains atoms incorporated by the exchange. For example, if ATP binds, and the released Pi formed upon hydrolysis contains the exchanged oxygen, then it is an intermediate Pi–HOH exchange. The oxygen exchange in such an intermediate exchange may take place before, during, or after the reaction. It should be noted that since oxygen from Pi forms water during ATP synthesis [75], one oxygen atom from medium water gets incorporated into Pi and released during the reverse ATP hydrolysis process (Equation (1)).
The presentation of the material is organized in the following way: The experimental methods employed are described in Section 2. Section 3 formulates a new theory of oxygen exchange for ATP hydrolysis by F1-ATPase. It is divided into two subsections. The first subsection describes how the stochastic kinetic theory of Mehta et al. (2020) (ref. [57]) on oxygen exchange during ATP synthesis by the FOF1-ATP synthase is mathematically adapted to treat the oxygen exchange process occurring during ATP hydrolysis by the F1-ATPase. In particular, kinetic equations for the overall extent of oxygen exchange during ATP hydrolysis by the enzyme are derived in this subsection (Section 3.1). Note that the relevant equations in the two modes, i.e., synthesis and hydrolysis, although possessing a similar form, are not exactly identical to each other [57], Section 3.1. Section 3.2 extends the kinetic theory for the calculation of isotopomer distributions of various [18O] species – containing 0, 1, 2, or 3 labeled oxygens—in the released Pi molecule. The derived expressions show an underlying mathematical similarity to those of stochastic Poisson-type theory. The final equations for predicting the population distributions (Equations (13)–(16) in Section 3.2) are shown to be obtained by direct integration of the kinetic rate equations (Equation (12)).
The results of applying the kinetic theory developed in Section 3 as applied to our oxygen exchange data are shown in Section 4. The apparent rate constant of oxygen exchange during ATP hydrolysis by F1-ATPase is determined over a 50,000-fold concentration range of substrate ATP (Section 4.3). The experimental distributions of [18O]Pi species over the entire ATP concentration range are compared with the theoretical population distributions in Section 4.4 using Equations (13)–(16) of Section 3.2. Comparison of experiment with the predictions of (i) Boyer’s binding change mechanism, which postulates a single site/route of water exchange , and (ii) Nath’s torsional mechanism of energy transduction and ATP synthesis/hydrolysis that invokes multiple sites of oxygen exchange occurring on the MgATP/MgADP.Pi bound at β-subunit interfaces with the α-subunits of F1 , with or , depending on [ATP] concentrations in the hydrolysis mode).
The mechanism of oxygen exchange and mechanistic implications of the results are discussed in Section 5.1, Section 5.2, Section 5.3, Section 5.4 and Section 5.5. A detailed model for multisite Vmax hydrolysis by F1-ATPase that results from these considerations is presented in Section 5.6. Biological implications arising, such as the order of product release steps during ATP hydrolysis and its consonance with structural data (Section 5.7), the identity of site 2 in the high-resolution X-ray structures (Section 5.8), the angle at which the elementary chemical events of binding, hydrolysis, Pi, and ADP release occur during the ATPase catalytic cycle (Section 5.9) are discussed. The consistency of the proposed mechanism with the results of Weber, Senior, and colleagues by direct measurement of catalytic site nucleotide occupancies using optical probes engineered into β-catalytic sites of F1-ATPase and relationship with other models of ATP hydrolysis, including those based on single-molecule observations, is detailed in Section 5.10. The main conclusions arising from our studies of oxygen exchange by F1-ATPase are stated point-wise in Section 6.
2. Materials and Methods
2.1. Materials
MF1-ATPase was isolated using the method of Penefsky [76] and stored as a 50% (NH4)2SO4 suspension at 4 °C, pH 8.0 in a medium containing 125 mM sucrose, 25 mM Tris-HCl (Merck, Darmstadt, Germany), 1 mM EDTA (Sigma-Aldrich, St. Louis, MO, USA), and 2 mM ATP (Merck, Darmstadt, Germany). Then, 0.1 mL aliquots of the suspension were pelleted, and the pellet was washed two times in the above buffer without ATP. The pellet was dissolved at room temperature by adding 0.1 mL of a solution of 50 mM Tris-sulfate, 50 mM K2SO4, and 1 mM EDTA at pH 8.0, followed by centrifugation through a Sephadex G-50 column pre-equilibrated with the buffer solution. Protein concentration was measured by the method of Lowry, and the concentration was converted to dry weight basis as given previously [76]. The specific activity of the ATPase was measured at 105 µmol min−1 mg−1 at pH 7.4, 30 °C. The variation of the ATPase-specific activity over time was measured within 10% of the above value.
2.2. ATP Hydrolysis by F1-ATPase
The ATPase reaction rate was measured as follows. The 3 mL reaction mixture contained ADP and F1-ATPase at the desired concentration, 5 mM magnesium acetate, 0.2 mM potassium phosphoenolpyruvate (Millipore Sigma, Burlington, MA, USA), and 0.05–0.5 mg/mL of pyruvate kinase (Thermo Fisher Scientific, Waltham, MA, USA) in 30 mM Tris-acetate, 30 mM potassium acetate buffer, pH 7.4 at 30 °C. The addition of pyruvate kinase to the reaction mixtures was made before the addition of F1-ATPase, and sufficient time was given for the conversion of almost all the ADP present to be converted to ATP. At various times after the addition of the F1-ATPase, a 0.5 mL sample was withdrawn and added to a cuvette containing 0.27 mL buffer, 0.03 mL of 3 mg mL−1 of NADH (Merck, Darmstadt, Germany), and 0.2 mL of 50 mM sodium EDTA at pH 8.0, which quenched the reaction. A 0.03 mL aliquot containing catalytic amounts of lactate dehydrogenase was added after the cuvette was inserted into the spectrophotometer. The amount of pyruvate formed was determined by the decrease in absorbance at 340 nm owing to NADH oxidation, and the rate of the ATPase reaction was calculated. The quantity of pyruvate produced during the conversion of ADP to ATP before the addition of F1-ATPase was subtracted from the total amount of pyruvate present.
2.3. Oxygen Exchange
Oxygen exchange experiments were performed at pH 7.4 and 30 °C in 30 mM Tris-acetate and 30 mM potassium acetate buffer. The ATPase reactions were started by the addition of varying volumes of F1 and continued for sufficient time to generate 50–100 nmol per ml of sample. The final sample volume measured was 2.5 mL. The F1-ATPase concentration ranged from 70 nM to 10 nM as ATP concentration was increased. The concentration of ATP is the final total concentration of ATP after the reaction is quenched. The quenching was done by vortexing with 2 mL of cold chloroform (Merck, Darmstadt, Germany), and the samples were placed on ice. For each sample, an aliquot of 1 mL was withdrawn for analysis by gas chromatography coupled with mass spectrometry (GC-MS), and the distribution of 18O in the Pi was determined as described below. In another set of experiments, 0.5 mL reaction mixtures containing 5 mM magnesium acetate and varying amounts of [18O]ATP in the Tris-acetate buffer (Merck, Darmstadt, Germany) at 30 °C were made. F1-ATPase of 5–6 mg mL−1 concentration was diluted 10-fold, and 5 µL was added to 50 µL buffer at 30 °C. The reaction was initiated by adding 0.45 mL of reaction mixture to the ATPase. The reaction was quenched by the addition of 0.5 mL of cold chloroform after approximately 100 nmol Pi had been produced.
2.4. Separation of Pi
The Pi formed by the F1-ATPase was isolated by taking aliquots of quenched reaction mixtures adjusted to 1 mL with H2O. Then, 0.5 mL phenol was added and centrifuged to remove the protein with the phenol. The phenol layer was washed with 1 mL of H2O which was added to the original aqueous layer. The phenol present in the combined aqueous layers was extracted into 2 mL of 1:1 isobutenol:benzene by volume. Molybdate was added, the phosphomolybdate extracted with isobutenol:benzene (Sigma-Aldrich, St. Louis, MO, USA), and the Pi was extracted into an alkaline H2O solution. The Pi was separated from the molybdate and converted to H3PO4 using column chromatography techniques [77]. The H3PO4 was lyophilized, converted to triethylphosphate with diazoethane, and analyzed for [18O]Pi species by GC-MS (Agilent Technologies, Santa Clara, CA, USA).
2.5. Determination of 18O in Pi
Lyophilized samples of H3PO4 were derivatized with diazoethane in ethane [77], the excess organic was evaporated, and the resulting triethylphosphate dissolved in dichloromethane (Merck, Darmstadt, Germany) at a concentration of 1 nmol µL−1. Aliquots containing about 1 nmol of sample were analyzed for 18O with a Hewlett-Packard GC-MS system. With the injection port at 250 °C, isothermal elution at 160 °C, and a gas flow rate of 30 mL min−1, the triethylphosphate eluted in around 60 s. The diethylphosphate ion fragment of and its 18O isotopomer cousins characterized by ratios of 157, 159, 161, and 163 were analyzed by selective ion monitoring. These species were designated as 18O0, 18O1, 18O2, as well as 18O3 and 18O4 Pi, respectively.
2.6. Calculations from Oxygen Exchange Measurements
The data were analyzed using the theory developed and customized for ATPase reactions as described in Section 3.
3. Theory
3.1. Kinetic Analysis of the Overall Extent of Oxygen Exchange during ATP Hydrolysis
We now adapt the kinetic theory of Nath and coworkers [57] derived for ATP synthesis by FOF1 to ATP hydrolysis by F1-ATPase. We delineate the concentration of the exchanging species (Pi or the γ-phosphoryl of ATP) at any time as and its initial concentration as . We denote as the fractional extent of 18O incorporated into the Pi released from the catalytic site into the medium after time , and as the fraction of 18O in Pi at time . is, therefore, the time for which oxygen exchange takes place from a single catalytic site during ATP hydrolysis by F1-ATPase, and is the apparent rate constant of the exchange. For 18O exchange from Pi or ATP into water we have
Writing on either side of Equation (2) as , we obtain the first-order ordinary differential equation for the fractional extent as
Separating variables and integrating from time to , we obtain
Upon inserting the initial condition at and determining in Equation (4), we obtain the solution
Following the path of a single 18O atom and making use of Equation (5), we can analyze all the possibilities that may arise for the intermediate Pi–HOH exchange process accompanying ATP hydrolysis. The initial condition for can also be accordingly derived. An interesting case is that of the 18O label initially present on the γ-phosphoryl group of ATP, and the hydrolysis of ATP occurring first on the enzyme catalytic site, followed by the exchange between the 18O oxygens of the bound Pi formed after the hydrolytic cleavage reaction and the 16O of medium HOH (or vice-versa, if initially the 18O label was present in water as H18OH), followed by the release of Pi from the site into the medium. For this case, the initial 18O fractional oxygen of Pi (i.e., the fraction at before the oxygen exchange occurs) has the value . This is because in ATP synthesis, oxygen from Pi forms water, while in the reverse hydrolysis mode, the oxygen atom from 18O-labeled water is incorporated as one of the four oxygen atoms contained in the Pi molecule [75], as shown in Equation (1). Substituting this initial condition in Equation (5) yields the final result
Another interesting case is that of the oxygen exchange between the 18O of the labeled γ-PO3 group of ATP with water (or vice-versa, if H18OH is used) occurring first, followed by the hydrolysis of ATP and thereafter the release of the Pi formed into the medium. In this case, in Equation (5) is zero, which yields an before hydrolysis, of . However, only three-fourths of the exchange takes place by this pathway, with the remaining one-fourths arising from the hydrolytic pathway because one water oxygen atom is incorporated into each Pi by the hydrolysis reaction itself [75], as shown in Equation (1). Hence, for this case, the fractional 18O incorporated in Pi after hydrolysis is given by the weighted sum [(3/4) + (1/4)], which again leads to the same final result when expressed in logarithmic form as given by Equation (6). Hence, Equation (6) is quite general and is valid irrespective of the order of the elementary steps during the oxygen exchange and ATP hydrolysis by F1-ATPase.
3.2. Stochastic Kinetic Theory for Calculation of Isotopomer Distributions of Various [18O] Species in Released Pi during ATP Hydrolysis
The oxygen exchange reactions taking place within a single enzyme catalytic site during ATP hydrolysis are represented by the overall kinetic scheme of Figure 1. A1, A2, A3, and A4 represent the progression of the enzyme system with defined concentrations respectively of [18O3], [18O2], [18O1], and [18O0] species in Pi for a specified initial distribution, A0 of 18O in the γ-PO3 group of substrate ATP, with standing for the percentage of the labels with the 3, 2, 1, and 0 labels in the initial distribution. Let represent states of the enzymatic system containing a certain distribution of the 18O with 3, 2, 1, and 0 labels that progressively change with time due to oxygen exchange.
Following several authors [78,79,80], a stochastic differential equation-based approach was developed for the ATP synthesis/hydrolysis to determine , the probability that the system is in state at time for rate constants that generally depend on . This led to the differential equation for
and for of
Computer simulation of the system with different values of for did not offer any meaningful insights even with the three extra parameters. We then noticed the important point that the differential equations of the general stochastic model (Equations (7) and (8)) reduce to a Poisson process if for all . Incorporating this restriction on our system of exchange reactions leads to a final differential equation model for the Poisson process that also provides novel physical insights. We therefore have
And
The chemical kinetics of the series reaction system for a population of enzyme molecules (Equation (11)) can be directly written as a set of ordinary differential equations using mass action principles (Equation (12)):
Integrating and solving the set of ordinary differential equations (Equation (12)) recursively [57,78,79,80] with the initial conditions (%) for labels with three to zero oxygen atoms in the γ-phosphoryl of ATP leads to the solution
And
The similarity in the form of the above chemical kinetics equations with the mathematical equations of a Poisson-type model of the process is useful because the exchange reactions satisfy the requirement that the average time between oxygen exchange events is set by a particular experimental variable (e.g., substrate ATP concentration) despite the exchange events themselves being stochastic, with the exact timing of the events possessing a random nature. The oxygen exchange events are also independent of each other, and the occurrence of one exchange event does not influence the probability of the occurrence of a second exchange event. Moreover, two or more exchange events do not occur simultaneously at a single catalytic site of the enzyme, and the average rate of oxygen exchange events per unit time in the population of enzyme molecules is constant.
4. Results
This study attempts to analyze and quantify the overall extent of the intermediate Pi–HOH exchange and predict the resulting Pi isotopomer distributions as a function of time during ATP hydrolysis by mitochondrial F1-ATPase.
4.1. Overall Extent of Oxygen Exchange by F1-ATPase as a Function of ATP Concentration
Figure 2 plots the mitochondrial adenosine triphosphatase data of the overall extent of oxygen exchange as a function of ATP concentration. It shows the variation of the 18O/P ratio with changing ATP concentration on a semi-log scale and estimates the average number of 18O water oxygens incorporated into Pi by intermediate Pi–HOH exchange occurring during ATP hydrolysis by the F1-ATPase. At saturating ATP concentrations, 0.01 oxygens are transferred to Pi by exchange, while, on the other hand, at low ATP concentrations, 0.99 fraction of the oxygens in Pi released from the enzyme site into the medium are derived from the 18O in water or the 18O in the γ-phosphoryl group of ATP, depending on where the label is situated.
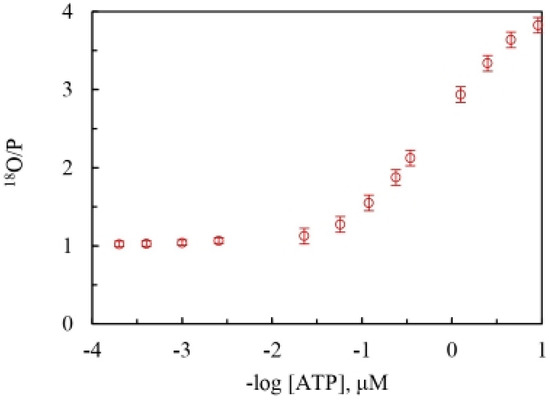
Figure 2.
Experimentally-determined average number of 18O water oxygen atoms present in each molecule of Pi released (18O/P, mean SD, n = 4) during ATP hydrolysis by mitochondrial F1-ATPase (MF1-ATPase) over a ~50,000-fold range of concentrations of medium ATP (~0.1–5000 μM).
4.2. Fractional Extent of Oxygen Exchange as a Function of Time during ATP Hydrolysis by F1-ATPase
The time scale of the oxygen exchange measurements is determined by the reciprocal of the steady-state velocity during ATP hydrolysis that enables the analysis of the modulation of the fractional extent of oxygen exchange, by the ATP concentration, and allows calculation of a time-resolved at each velocity corresponding to a particular medium ATP concentration. This is shown in Figure 3. Equation (6) also reveals that the rate of oxygen exchange, given by the product of the apparent rate constant of exchange and the time of exchange, can be readily determined at each value of .
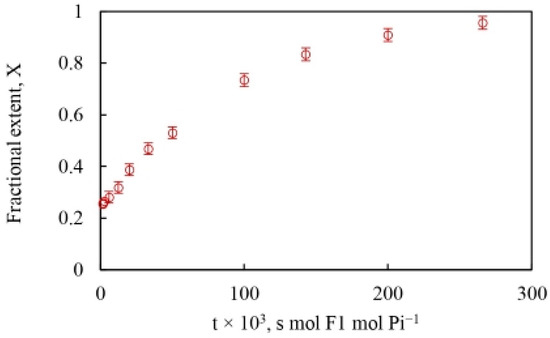
Figure 3.
Fractional extent, X of 18O incorporated into each Pi released from the enzyme catalytic site into the medium during ATP hydrolysis by MF1-ATPase (mean SD, n = 4) based on the stochastic kinetic theory in Section 2. The X-axis plots time, from the reciprocal of the steady state reaction velocity [moles of Pi/{moles of F1-ATPase}−1 s−1].
Figure 3 and Equation (6)—which make no specific assumptions regarding mechanism—show that any difference in the rate of oxygen exchange can either be (i) due to a difference in the value of the apparent rate constant, , of the exchange reaction itself [58,60], or (ii) due to a difference in the lifetime of the exchanging intermediates, i.e., due to the time available for exchange, [32,33,57].
4.3. Evaluation of the Apparent Rate Constant of Oxygen Exchange during ATP Hydrolysis by F1-ATPase
The results plotted in Figure 4 using Figure 3 and Equation (6) yield a close-to-perfect straight line with R2 = 0.999. The data thus reveal a constant slope with a value of the apparent rate constant of oxygen exchange () of 10.5 s−1 over the entire substrate concentration range, i.e., over five decades of ATP concentration during steady-state ATP hydrolysis by mitochondrial F1-ATPase. It ought to be emphasized that, contrary to expectations from a Boyerean model [58], no variation (increase) in the value of the apparent rate constant of oxygen exchange was found as medium ATP concentration was increased from ~0.1 µM–5 mM, i.e., across a 50,000-fold concentration range of [ATP] (Figure 4).
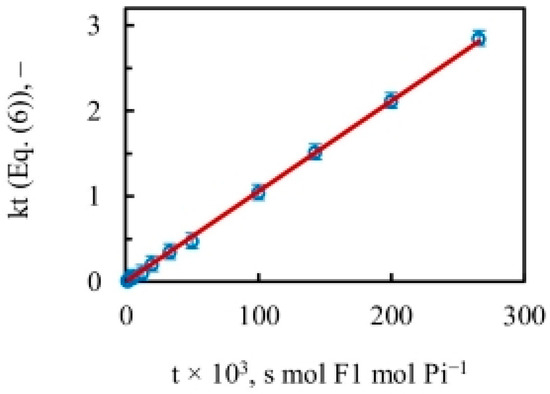
Figure 4.
Amount of intermediate Pi–HOH exchange during ATP hydrolysis by MF1-ATPase (Equation (6)) as a function of time (mean SD, n = 4). The slope of the almost-perfect straight line over five decades of medium ATP concentration () gave a constant value for the apparent rate constant of oxygen exchange () of 10.5 s−1. The X-axis is a measure of time, plotted as the reciprocal of the steady state reaction velocity [moles of Pi/{moles of F1-ATPase}−1 s−1].
4.4. Quantification of the Distributions of [18O]Pi Species at Various ATP Concentrations during ATP Hydrolysis by F1-ATPase
Section 4.1, Section 4.2 and Section 4.3 quantified the average extent of the 18O/P ratios arising from the Pi–HOH exchange during ATP hydrolysis by the F1-ATPase. The experiments also measured the pattern of distribution of the various [18O]Pi species over five decades of ATP concentrations by mass spectrometry (Section 2). In other words, detailed information on the number of 18O atoms (0, 1, 2, 3) transferred to each released Pi molecule from the γ-phosphoryl group of ATP upon intermediate Pi–HOH exchange occurring by ligand permutation about the phosphorus center [81] is available. The experimental and theoretically-predicted distributions of such [18O]Pi species containing 0, 1, 2, or 3 18O atoms as a percentage of the total have been compared using Equations (13)–(16) as a function of medium ATP concentration, given the initial distribution of 18O in the γ–PO3 group of ATP shown in Figure 5.
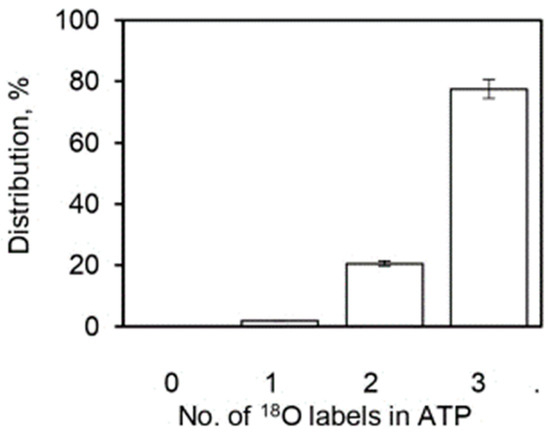
Figure 5.
Initial distribution of the 18O oxygen atoms (%) in the γ-PO3 group of substrate ATP (n = 3). The X-axis represents the number of labeled oxygens (0, 1, 2, 3) in the starting distribution.
The comparison of experimental and theoretical [18O]Pi distributions at each medium ATP concentration or time point generated a vast amount of data, and we sought a cleaner and short-form way to represent the results. A C++ computer program was written that collated data at each temporal point for [18O]P3, [18O]P2, [18O]P1, and [18O]P0, i.e., with [18O3], [18O2], [18O1], and [18O0] species in the released Pi after oxygen exchange by F1-ATPase (Supplementary Materials) for the starting distribution in Figure 5. The code used Equations (13)–(16) of the stochastic kinetic theory to model the predicted distributions at each time point. It should be noted that since the distributions in Equations (13)–(16) are written in terms of , and both and are available from the experimental data (Figure 4), no fitted or adjustable parameters were employed in arriving at the predicted [18O]Pi distributions.
Boyer’s binding change mechanism of ATP synthesis/hydrolysis involves a single site or mode of entry of water oxygen [59,60]. This key assumption of the mechanism has not been subjected to scientific scrutiny previously. Figure 6 illustrates the results of such a test. The results show that an oxygen exchange model with a single site of entry of 18O water and 10.5 s−1 (Figure 4) does not properly predict the observed distributions of the four [18O]Pi species at any substrate ATP concentration (Figure 6). The torsional mechanism of ATP synthesis/hydrolysis, on the other hand, predicts multiple sites/points of entry of water oxygen [32]. This is the number of catalytic sites at which 18O-labeled water can interact with bound ATP or bound ADP.Pi at the same time. A model in which oxygen exchange occurs at the constant value of the apparent rate constant, , of 10.5 s−1 governing the exchange process under the experimental hydrolysis conditions at each ATP concentration at two catalytic sites, essentially doubling the time of exchange, , in Figure 3 and Figure 4—from what is anticipated to occur at a single catalytic site during a given time—is shown in Figure 7.
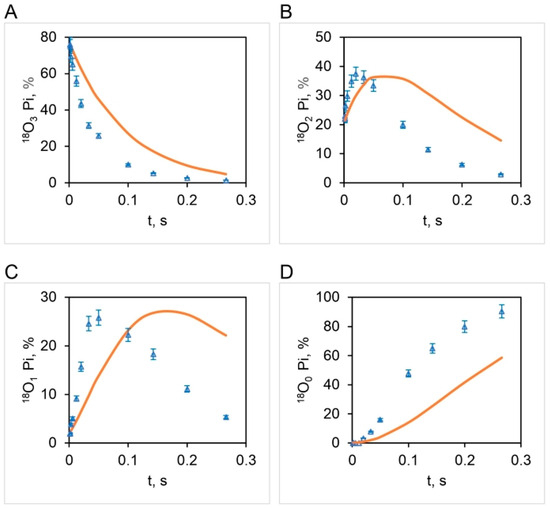
Figure 6.
Experimental (open triangles: mean SD, n = 3) and theoretical distributions (bold curve) of 18O (%) in released Pi as a function of time, , during ATP hydrolysis by mitochondrial F1-ATPase for a single site/point of oxygen exchange/water entry. The starting distribution of 18O in the γ-phosphoryl group of ATP is given in Figure 5. Pi with three labels, 18O3 (A), two labels, 18O2 (B), one label, 18O1 (C), and zero label of 18O in Pi, 18O0 (D).
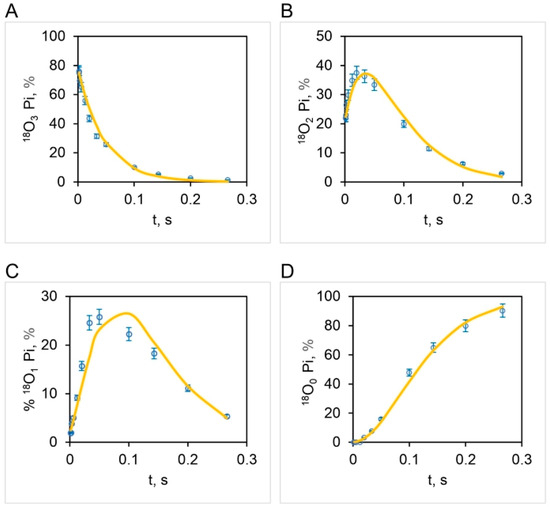
Figure 7.
Comparison of experimental (open circles: mean SD, n = 3) and theoretical isotopomer distributions (bold curve) of 18O (%) in released Pi as a function of time, , during ATP hydrolysis by mitochondrial F1-ATPase. The bold curves simulate oxygen exchange over the entire 50,000-fold range of ATP concentrations (0.1–5000 μM) for two sites/points of oxygen exchange/water entry, given a starting distribution of 18O in the γ-phosphoryl group of ATP of Figure 5. Triple-labeled Pi, 18O3 (A), double-labeled Pi, 18O2 (B), single-labeled Pi, 18O1 (C), and Pi with no label, 18O0 (D).
The correspondence between the observed and theoretical [18O]Pi distributions (based on the stochastic kinetic theory in Section 3) shown in Figure 7 is fairly good but is not perfect. On close inspection, we noticed that the experimental [18O]P3 distribution relaxes at a much faster initial rate than that predicted theoretically (Figure 7A). Furthermore, the data clearly reveal that the [18O]P2 (Figure 7B) and [18O]P1 (Figure 7C) experimental distributions rise much faster at short times (0–33 ms) than that predicted by theory. These trends are very regular, as seen from the experimental data plotted in Figure 7. Hence, although the long-time behavior (33–266 ms) is well predicted, the rate of the short-time decay (for [18O]P3) or rise (for [18O]P2 and [18O]P1) is underestimated by the model. Hence we attempted prediction of the oxygen exchange based on the stochastic kinetic theory developed in Section 3 for three sites of oxygen exchange at short times (0–33 ms) corresponding to high substrate ATP concentrations (4.2–5000 μM) and two sites of oxygen exchange at long times (33–266 ms), i.e., at low substrate ATP concentrations (0.11–4.2 μM), at the experimentally-determined essentially constant value of for oxygen exchange of 10.5 s−1 (Figure 4). The results are plotted in Figure 8 and in further experiments shown in Figure 9 that explore the region of intermediate [ATP] concentration. It should be clearly understood that the measured steady-state hydrolysis activity of F1-ATPase at every ATP concentration (µM to mM) arises entirely from the activity of the (111) species in a trisite mode of catalysis in which rotation occurs with all three catalytic sites occupied by bound Mg-nucleotide (Figure 10 and Figure 11).
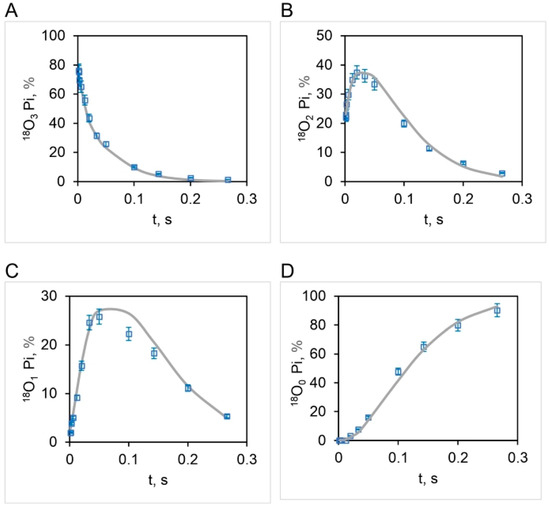
Figure 8.
Comparison of experimental (open squares: mean SD, n = 3) and theoretical isotopomer distributions (bold curve) of 18O (%) in released Pi as a function of time, , over a 50,000-fold range of ATP concentrations (0.11–5000 μM) during ATP hydrolysis by mitochondrial F1-ATPase. The bold curves depict the oxygen exchange based on the stochastic kinetic theory developed in Section 3 for three sites/points of oxygen exchange/water entry at short times (0–33 ms) corresponding to high substrate ATP concentrations (4.2–5000 μM), and two sites/points of exchange/water entry at long times (33–266 ms), i.e. at low substrate ATP concentrations (0.11–4.2 μM) for an initial distribution of 18O in the γ-phosphoryl group of ATP given in Figure 5. Tri-labeled Pi, 18O3 (A), bi-labeled Pi, 18O2 (B), uni-labeled Pi, 18O1 (C), and Pi with no label, 18O0 (D).
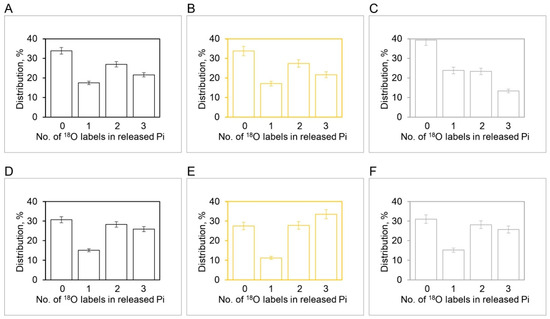
Figure 9.
Experimental and theoretical [18O]Pi distributions (%) during ATP hydrolysis by MF1 at 3 µM ATP (Average 18O/P = 2.10: (A–C)) and 5 µM ATP (Average 18O/P = 1.85: (D–F)) based on experiments in the region of intermediate ATP concentrations (mean SD, n = 6). Experimental distributions at 3 µM ATP (A), theoretical distributions at 3 µM ATP with (B), and theoretical distributions at 3 µM ATP with (C). Experimental distributions at 5 µM ATP (D), theoretical distributions at 5 µM ATP with (E), and theoretical distributions at 5 µM ATP with (F). The X-axis represents the number of 18O labeled oxygens (0, 1, 2, 3) in the released Pi. The initial distribution of the 18O label in the γ-phosphoryl group of ATP in these experiments was measured as follows—3 18O: 57%; 2 18O: 17%; 1 18O: 2%; 0 18O: 24%.
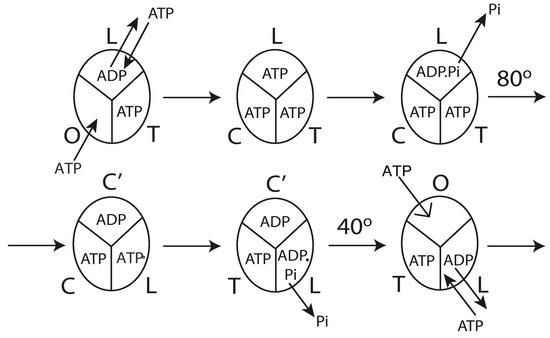
Figure 10.
Model for steady-state multisite hydrolysis of ATP by F1-ATPase from the analysis of experimental oxygen exchange data by the stochastic kinetic theory in Section 3 and mechanistic interpretation based on Nath’s torsional mechanism of ATP synthesis/hydrolysis and the unified theory [18,30,31,32,33,57,62,63,64,81]. The three β-catalytic sites of the F1-ATPase enzyme are depicted; the system is being viewed from the F1-side. T represents the catalytic site of highest affinity for MgATP (site 1); L represents the catalytic site of intermediate affinity (site 2); O represents the site of lowest affinity (site 3). C′ stands for the conformation adopted by a closed catalytic site (which could even be half-closed, C) relative to the open (O) site. The trisite model depicts steady-state Vmax hydrolysis with all three catalytic sites filled by MgATP or MgADP before rotation of the γ-subunit occurs. The open arrow in panel 6 shows a new MgATP molecule binding to site 3 (0°), following which the cycle repeats. See the text in Section 5.6 for details.
The above results clearly show that all the four experimental 18O Pi distributions, i.e., [18O]P3 (Figure 8A), [18O]P2 (Figure 8B), [18O]P1 (Figure 8C), and [18O]P0 (Figure 8D) are perfectly predicted by the stochastic kinetic theory in Section 3 at both short times as well as long times, i.e., throughout the 50,000-fold range of medium ATP concentrations, without employing adjustable parameters. The finding of a constant value of the apparent rate constant of oxygen exchange determined from the measured overall extent of 18O/P (Figure 2 and Figure 4) and of a time of exchange that is twice the value determined from the reciprocal of the steady state hydrolysis rate at each medium ATP concentration at long times, and thrice that value at short times (Figure 8), offers novel mechanistic insights into catalysis by F1-ATPase. These, along with the biological implications arising, are discussed in Section 5. These findings also help in interpreting available high-resolution X-ray structures of F1-ATPase.
5. Discussion
5.1. Limits of Oxygen Exchange during Catalysis by F1-ATPase
An upper limit of the 18O/P ratio of was determined at low ATP concentrations (0.035–0.1 μM), and a lower limit for oxygen exchange based on the 18O/P parameter of was measured at high, saturating medium ATP concentrations (1–5 mM).
5.2. Time-Resolved Analysis of Oxygen Exchange
The principal equations from our theoretical analysis in Section 3, Equations (6) and (13)–(16) enable a time-resolved analysis of oxygen exchange. Since the product of and , i.e., on the right-hand side of Equation (6), is readily evaluated from the 18O/P oxygen exchange data during ATP hydrolysis by F1-ATPase, no fitted parameters are required for the quantification and interpretation of the kinetic information. Equations (13)–(16) implement a Poisson-type model for the distribution of various [18O]Pi isotopomer species during ATP hydrolysis, and since these equations are also in terms of , no fitted parameters are necessary to predict these distributions.
5.3. Mechanistic Implications for Energy Coupling
The stochastic kinetic model of the oxygen exchange process (Equations (6) and (13)–(16)) makes no specific assumptions about the mechanism and is therefore independent of the mechanism. These equations characterize the exchange in terms of the product of the apparent rate constant of exchange, and the lifetime of the exchanging species, . Thus, a change in the 18O of Pi could arise either from an alteration in the rate constant of exchange or from an increase in the time available for exchange. The latter situation would arise due to the differing length of time spent by the Pi/ATP in the β-catalytic site due to a faster/slower turnover of the hydrolytic reaction. For instance, a longer time spent while bound ATP is waiting for rotation of the γ-subunit/conformational change in F1 to take place before activation or before the cleavage chemical step occurs at a catalytic site to form Pi, or a longer time spent while the Pi formed after hydrolysis is waiting for its release from the site etc., will show up as an increased extent of oxygen exchange at a lower ATP concentration.
Different models of energy coupling have been constructed based on their fundamental conception of oxygen exchange, where the parameter chiefly modulates the exchange with changes in medium/substrate concentration. The binding change mechanism of ATP synthesis does not include a timescale for the exchange per se; it focuses on the alteration of the value of the rate constant governing the oxygen exchange, , of nucleotide bound to a catalytic site by the medium substrate concentration. The mechanism proposes long-range transmission of the binding energy of ATP from one β-catalytic site via the ring of β and α subunits to another β-catalytic site (site-site cooperativity) in F1-ATPase and modification in the value of the rate constant of the exchange by such signal and energy transmission [58,60]. In their words, emphasis is placed on “how much rate constants at one catalytic site may be changed by ATP binding at another” [58]. The torsional mechanism of energy transduction and ATP synthesis and the unified theory of ATP synthesis/hydrolysis [31,57] emphasizes the asymmetric interactions of the β-catalytic sites with the single copy γ- and ε-subunits during catalysis [33,62,63,64]. It considers the time available for exchange, , by the bound ATP/Pi at each catalytic site as the appropriate timescale and postulates that this parameter quantitatively determines the extent of the oxygen exchange observed in the experiments when multiplied by an essentially constant apparent rate constant for the exchange process, . According to this theory, changes in the medium substrate concentration alters at each of the multiple sites where the oxygen exchange occurs, which is then responsible for the modulation of the extent and rate of oxygen exchange in accordance with Equations (6) and (13)–(16).
The results shown in Figure 4 and Figure 8 demonstrate that the measured changes in the extent of 18O as a function of medium ATP (from sub-micromolar to millimolar concentration) incorporated into Pi by an intermediate Pi–HOH oxygen exchange accompanying ATP hydrolysis arise entirely from differences in the time available for exchange rather than from any alteration in the value of the rate constant of 18O exchange over the entire 50,000-fold concentration range of substrate.
To sum up, the results prove that consideration of the time available for 18O exchange by bound nucleotide/Pi in a catalytic site at any substrate ATP concentration is sufficient to account quantitatively for the observed extent of oxygen exchange by F1-ATPase, without the need for postulating additional allosteric interactions/site-site cooperativity between catalytic sites. It should be noted that it is imperative fundamentally to account for changes in the timescale arising from substrate concentration changes imposed by the experimentalist. Once this effect is exactly accounted for (Figure 4), there is no leeway for postulating other effects such as site-site cooperativity/allostery.
5.4. Mechanism of Oxygen Exchange
Another fundamental limitation of the model of oxygen exchange proposed by the binding change mechanism was that it involved only a single site or route of entry of water oxygen. Previous research did not report or quantitatively tabulate the absolute rates and rate constants of the exchange of 18O between Pi/ATP and HOH in a model-independent way. Thus, any observed extent of exchange was rationalized by assuming reversible ATP synthesis/hydrolysis at a single catalytic site with an equilibrium constant of , and postulating the number of reversals that must have occurred at the catalytic site to account for the measured extent of exchange, assuming a ¾ probability of loss/gain of the 18O label with each reversal. Thus, at low substrate ATP concentration, more than 400 reversals of the reaction had been postulated to explain the observed extent of oxygen exchange [58]. This view of spontaneous and reversible catalysis has been challenged by several workers [33,34,82]. Moreover, no independent verification of the number of reversals occurring during catalysis has been provided by the proponents of the binding change mechanism. Furthermore, the original postulate of the binding change mechanism of was never supported by the experiment. It was even admitted by the workers that “Measurement of the equilibrium constant for the reversible hydrolysis of bound ATP at high ATP concentrations would be useful but does not appear to be readily accessible experimentally” [58].
A major finding of this work (Section 4.4) shows that there are multiple sites of oxygen exchange during the catalysis of ATP synthesis/hydrolysis, as predicted by the torsional mechanism [32]. We could not match the observed [18O]Pi distributions with a single site/route of entry of 18O water (Figure 6). Hence, our results cannot be explained by the binding change mechanism, and a new mechanism of oxygen exchange is required. We have advanced a mechanism according to which the exchanges do not involve the spontaneous reversal of the formation of ATP from ADP and Pi at a catalytic site but occur instead because the catalytic site of the enzyme lacks absolute spatial selectivity for the oxygen ligands of the phosphorus intermediate that it accepts and binds as substrate [57,81]. Given the above, it is clear that ligands can readily permute at an intermediate state and that the oxygen ligands can interchange their positions about the phosphorus center. We propose here that such ligand permutation is the fundamental cause for the occurrence of the intermediate Pi–HOH (and the intermediate ATP–HOH oxygen exchange during ATP synthesis by the FOF1-ATP synthase [57]), as opposed to multiple spontaneous reversals of ATP synthesis/hydrolysis at a catalytic site [58]. Further, we envisage that such ligand permutation in space about the bound Pi/γ-phosphoryl of ATP occurs under conditions of tight electrostatic interactions with catalytic site groups so that the electrostatic free energy released during the process of ATP hydrolysis can be readily transduced into the torsional energy stored in the γ-subunit of ATP synthase, as per the postulates of the torsional mechanism of energy transduction and ATP synthesis/hydrolysis and the unified theory of ATP synthesis/hydrolysis [30,31,33, 62−64,81]. Mg2+ plays a critical role in this process, as proposed previously [63,64,83,84].
It is clear from the above discussion that the free energy required for the ATP synthesis process is obtained from the torsional energy stored in the γ-subunit, which is ultimately derived from the energy of the anion and proton electrochemical gradients (Δμ̃A− + Δμ̃H+) [85,86,87,88,89]. A unified treatment of coupling [90] and uncoupling [91] during the process of oxidative phosphorylation that employs the same mathematical equations has also been developed.
5.5. Number of Sites of Oxygen Exchange and Their Biological Implications
The results presented in the latter part of Section 4.4 and plotted in Figure 8 pose an interesting mechanistic conundrum. How can we have three catalytic sites participating in oxygen exchange at short times that, however, reduce to two sites mediating intermediate Pi–HOH exchange at long times? In other words, at long times, one catalytic site drops out from the exchange from the maximum value of the number of catalytic sites, mediating oxygen exchange at short times. This fact further imposes severe constraints on possible molecular mechanisms of ATP hydrolysis by F1-ATPase.
Every model of trisite hydrolysis, when looked at in a gross way, postulates for the enzyme as a whole that site 3 (O) changes to site 1 (T), site 1 (T) changes to site 2 (L), and site 2 (L) changes conformation to site 3 (O). Following the conformational change to site 3 (O), the bound nucleotide would be released and, therefore, can no longer participate in the exchange. If two catalytic sites have to contribute equally to exchange at long times by effectively doubling the time of exchange (Figure 8), then these have to be sites 1 and 3 that have ATP bound to them, and they need to continue to exchange oxygen during the O → T and T → L transitions of the respective sites to explain the observed [18O]Pi distributions at long times. If site 2 had contributed at long times, we would obtain a non-integer value for the exchange; moreover, if site 2 contains ADP as expected, then it should not mediate any exchange in the first place. By another element of logic, the site that could contribute to exchange at short times (and subsequently not make a contribution) has to be the site (site 2) that engages in the L → O change.
The above logic, though compelling, still does not solve the conundrum. The L-site initiates the 80° sub-step of γ rotation by activation of the system by ATP binding, or rather, by ADP–ATP exchange in site 2 by ligand displacement as proposed by us [31] (since the site contained bound ADP initially). Now, site 2 contains bound ATP, but if the bound ATP does not exit after the L → O conformational change and continues to exchange oxygen, then always, which is not consistent with the experiment. If the ATP unbinds and exits the site after its L → O transition, and a new ATP does not bind, then is not realized even at short times. If a new molecule of ATP binds in O, then the L-site that had mediated an ADP–ATP exchange by ligand substitution shall only mediate an ATP–HOH exchange but not show up as an intermediate Pi–HOH exchange. Hence, again, we can never get three sites engaged in a Pi–HOH exchange. Again, the conundrum is not solved fully.
Thus, to have three sites mediating an intermediate Pi–HOH exchange at short times, we need to additionally hydrolyze the bound ATP (that had exchanged with bound ADP in L, i.e., site 2 by the phenomenon of ligand displacement) to ADP.Pi, and subsequently, Pi needs to unbind and be released from L. This explains the short-time exchange data (Figure 8). The rate of this activation and ligand displacement phenomenon is itself inversely proportional to medium ATP concentration, as one would expect. Thereafter, L shall only contain bound ADP, which does not mediate oxygen exchange with water, and therefore, the number of catalytic sites mediating exchange, at long times. Subsequently, the bound ADP is released after an L → O transition of the catalytic site, and a new ATP binds to O, which thereafter adopts a closed conformation (relative to O) and participates in oxygen exchange as the site changes to T, and continues to mediate exchange as T changes to L. Following the site’s T → L transition, ATP hydrolyzes to ADP.Pi in L, and after that, Pi continues the exchange process, which ceases only upon the release of Pi from L. This explains both the short and long-time exchange data, predicts the experimental [18O]Pi isotopomer distributions with 3, 2, 1, and 0 18O atoms over the entire range of medium ATP concentration, and resolves the conundrum posed at the beginning of this section.
The above explanation also serves to rationalize our puzzling biochemical observations on MF1 that when normal nonradioactive ATP is pre-loaded in the highest affinity site 1 (T) under sub-stoichiometric ATP:F1 conditions and 5–20 μM radioactive [γ-32P]ATP is used in a cold chase experiment, 92–94% of the promoter [γ-32P]ATP is hydrolyzed in site 2 (L) within 10 s of incubation time, as detected by counting of 32Pi. This also leads to the key insight that ATP hydrolysis and Pi release (and not ATP binding as in previous theories of free energy transduction [59,60]) in site 2 is also required to explain the chase promotion experiments; otherwise, no 32Pi counts should have been registered in the above experiment [92].
Figure 9 shows the results of our experiments on hydrolysis by mitochondrial F1 on the [18O]Pi distributions that further explore the range of intermediate ATP concentrations. The results show that between 5 µM and 3 µM [ATP], the number of catalytic sites mediating oxygen exchange at the same time switches from three (Figure 9D–F) to two (Figure 9A–C), respectively. Alternatively, more precisely, the intermediate Pi–HOH oxygen exchange occurring in site 2 decays to zero and ceases after 33 ms, and hence, that catalytic site drops off from the observed exchange at longer times (33–266 ms). Hence, there is a transition from to between 3 and 5 µM [ATP], and only then can the detailed [18O]Pi distributions shown in Figure 8 and Figure 9 be predicted exactly.
Single-molecule recordings on F1-ATPase do not measure the extensive oxygen exchange that occurs at multiple catalytic sites at the same time before, during, and after ATP cleavage. However, the above results are not in conflict with results from the single-molecule observations. An exchange rate of 10.5 per s (Figure 4) implies that, on average, an exchange occurs once every 100 ms. At the transition concentration between 3 and 5 µM ATP discussed above, the results of Figure 8 and Figure 9 show that the oxygen exchange process in site 2 lasts 33 ms. For this entire duration of 33 ms, the bound MgATP molecules in sites 1 and 3 also participate in the exchange process, and the extent of exchange that occurs is approximately equivalent to that by a single catalytic site involved in the exchange process for 100 ms. Approximately because further exchange occurs at the intermediate stage of the reaction and afterward when MgADP.Pi remains bound, and the Pi is waiting to be unbound and released into the medium. It should be clearly understood that the ATP cleavage reaction takes place at one catalytic site at a time; hydrolysis does not occur simultaneously at two or three sites, although oxygen exchange does. Furthermore, the ATP hydrolysis (bond cleavage) event can occur at a much higher rate, with a of 1 ms during the catalytic dwell, as revealed by molecular dynamics simulations [25] and single-molecule experiments [11,14,93,94,95,96], since much of the oxygen exchange occurs when MgATP is bound at multiple catalytic sites for a characteristic time as a function of medium ATP concentration, and becomes longer as ATP concentration decreases.
5.6. Model for Steady-State Multisite ATP Hydrolysis by F1-ATPase
The detailed molecular mechanism of Vmax ATP hydrolysis by F1-ATPase ([31], pp. 1795–1797 and pp. 1802–1805) can now be refined to make it consistent with the results obtained in Section 4.1, Section 4.2, Section 4.3 and Section 4.4, and the mechanistic implications of biological energy transduction arrived at in Section 5.1, Section 5.2, Section 5.3, Section 5.4 and Section 5.5. Such a model is illustrated in Figure 10.
The molecular mechanism of ATP hydrolysis by F1-ATPase (Figure 10) incorporates a key result arising from this work that it is not sufficient to exchange bound ADP in the catalytic site 2 (L) with medium ATP in order to activate the enzyme and cause an 80° primary rotation of the central γ-subunit (in a clockwise sense when viewed from the F1 side) in F1 (bisite activation) We additionally need to hydrolyze the bound ATP in site 2 (L) (that had exchanged with ADP in the catalytic site) to ADP.Pi and subsequently the Pi need to move away and be released from L, as formulated by a general theory of biological energy transfer [30]. Otherwise, the 18O labels would not have been detected by mass spectrometry in the Pi released into the medium, and we would not be able to explain the distribution of [18O]Pi species (Figure 8). Thus, this L-site is the third catalytic site at which exchange occurs and, therefore, accounts for the observation of at short times. The exchange of bound MgATP for MgADP in site 2 releases an excess binding energy of 9 kJ/mol in the F1-ATPase, i.e., the difference between the binding energy of MgATP in L (36 kJ/mol) and the binding energy of MgADP in L (27 kJ/mol). This 9 kJ/mol energy released weakens to approximately zero the binding of bound Pi formed upon the ATP hydrolysis event in site 2 (i.e., cleavage of the Pβ–O–Pγ terminal phosphoanhydride bond of the ATP originally at a bond distance of 0.3 nm [30]). The effect of ejecting ADP with a certain velocity helps break the 9 kJ/mol γ–βTP interactions, i.e., between γ and site 2. Now, the γ-subunit is free to rotate, and the Pi is free to move away from bound MgADP. As the Pi moves step-wise from 0.3 to 0.4 to 0.6 nm [30], it releases a Coulombic repulsion energy of 9 + 9 = 18 kJ/mol. Another 18 kJ/mol is made available as the Pi is fired out from 0.6 nm to and released into the solution. This 36 kJ/mol electrostatic potential energy rotates the top of the γ-subunit by 80° relative to the stationary β–subunits with an average torque measuring 40 pN-nm generated at the β–γ interface at a radial distance of approximately 1 nm from the central axis of the α3β3 hexamer (Figure 10).
After initiation of the 80° rotation of the top of γ, site 2 (L) changes to a closed conformation C′. Upon the above 80° rotation, the top of γ interacts with the β-catalytic site 1 (T or βDP-like in [31]) and alters its conformation to loose (i.e., site 2, L or βTP in [31]). In other words, the rotation of γtop causes a T L transition of the β-catalytic site, due to which a 9 kJ/mol destabilization (reduction in binding energy of intermediate bound in the site) occurs. Concomitantly, ATP hydrolyzes to ADP.Pi upon the T L transition of the catalytic site. Pi, which is bound to L with 9 kJ/mol binding energy, is now bound in L with ~zero binding energy and is, therefore, free to move away. The 0.3 0.4 0.6 nm movement of Pi away from bound ADP releases 18 kJ/mol, which is transmitted from site 2 to site 3 (O or βE) along the ε-helix and helps break the ε–βE interaction (e.g., between ε–Ser-108 and βE–Glu-381 in the DELSEED loop) [33] along with the 27 kJ/mol binding energy of MgATP in site 3 (O or βE), as already described [31]. The open site O or βE closes (to βC) due to the interactions mentioned above and relieves any steric hindrance that the open site offered to further rotation of the γ-subunit (beyond 80°). The torsional strain in the γ-subunit also helps break the ε–Ser-108–βE and the ε–Met-138–βTP interactions and rotate the ε-subunit (both the N-terminal β-sandwich domain and the C-terminal helical domain) and the bottom of the γ-subunit by 80°—clockwise as seen from the F1 side—and the two-coiled coil α-helices of γ unwind, relieving the torsional strain in the γ-subunit. This is accompanied by the concomitant transition O T of the catalytic site. In the meantime, the release of Pi from the new L-site (i.e., the one that has become site 2 after its T → L transition owing to the 80° rotation of the top of γ already described above) with a velocity upon its movement from 0.6 nm away from ADP to infinity provides the energy for the remaining 40° rotation of the γ- and ε-subunits (Figure 10). This event of Pi release from the new βTP (after the 80° rotation, or at an angular position of 200° after the ATP molecule binds at 0°) into the medium is the one that finally leads to the cessation of the intermediate Pi–HOH exchange. It also accounts for observed at long times arising from oxygen exchange in site C and during C → T, and concomitantly in T and during T → L until Pi is released from L. Upon this 40° rotation, the interaction of ε with C′ changes its conformation to an O-site (open, site 3, the site with the lowest affinity for Mg-nucleotide) from which bound MgADP is released. During steady-state Vmax hydrolysis, the order of conformations that a single catalytic site of F1-ATPase passes through is O T, T L, L C′, and C′ O. Looking at the enzyme as a whole, the order of the conformational changes of the catalytic sites during multi-site hydrolysis by F1-ATPase is L C′, followed by T L, followed by O T, and finally, C′ O, in accordance with our previous predictions, and also shown to be the microscopic reversal of the ATP synthesis cycle ([31], pp. 1801, 1804). The cycle (Figure 10) then repeats.
The catalytic cycle for steady-state Vmax hydrolysis depicted in Figure 10 is in accord with biochemical crosslinking studies [16,17]. These studies inferred from the data that rotation of the γ- and ε-subunits in F1-ATPase is not linked to the unisite hydrolysis of ATP at the highest affinity catalytic site 1 (T) but to ATP binding and/or ATP hydrolysis and product release at the second or third catalytic site on the enzyme (i.e., site 2 or site 3) [16]. The studies also showed that the effect of covalently crosslinking β–Cys-381 to γ–Cys-87, i.e., forming the β–γ crosslink, increased the rate of unisite catalysis to that obtained by the cold chase of ATP of the non-crosslinked enzyme [16]. Since β–γ in the biochemical crosslinking studies corresponds to βTP in the X-ray structure of the enzyme in the Mg-inhibited state [1], we infer that βTP (site 2 or L) is the catalytic site to which ATP binds (and in which it subsequently hydrolyzes) in the native non-crosslinked enzyme. These events are responsible for rotating γ by 80°, changing the conformation of site 1 to site 2, and causing hydrolysis of the bound ATP in the (new) site 2, as shown in Figure 10.
The molecular mechanism shown in Figure 10 also satisfies the fact that Vmax ATP hydrolysis follows trisite catalysis [32,33,34,97], a fact that is considered experimentally proven in our study. This by itself takes it beyond the binding change mechanism, which was necessarily a bisite model [60,61]. However, over the past two decades [29,34] until the present day [7], ATP binding to site 3 (O) has been repeatedly postulated to cause rotation in F1-ATPase. It has been pointed out previously that the O-site (site 3) is open and distorted, and the binding energy of MgATP is only 27 kJ/mol [31,32,97], which is grossly insufficient energetically to change the conformation of the catalytic site from O to closed (C) and also cause a primary rotation of the γ- and ε-subunits by 80° (see p. 1809 of [31]). It ought to also be stressed that the detailed mechanism of steady-state multisite ATP hydrolysis by F1-ATPase presented here (Figure 10) is the microscopic reverse of the molecular mechanism of steady-state ATP synthesis by FOF1-ATP synthase formulated by us in previous publications [31,33,57]. The fundamental constraint of microscopic reversibility has not been shown to be satisfied by other mechanisms. For these compelling reasons, we consider the mechanism shown in Figure 10 to be superior to extant mechanisms in the field.
5.7. Order of Product Release Steps during ATP Hydrolysis and Interpretation in Terms of High-Resolution MF1 X-ray Structures
The order of release of products during ATP hydrolysis by F1-ATPase of Pi followed by ADP in Section 5.6 and Figure 10 is consistent with the finding from the MF1 X-ray structures that ADP is the last of the products of ATP hydrolysis to be released [98] (PDBID: 4ASU). Mechanisms that propose the reverse order, with Pi release following ADP release from a catalytic site, are faced with the difficulty of retaining Pi for an additional 120° after hydrolysis when the catalytic site’s affinity for Pi is already very low. How does the Pi sense the timing of its release? No convincing proposal that overcomes the above difficulties has been put forth. Above all, such a proposal would not be in agreement with the [18O]Pi isotopomer distributions of Figure 8 at long times. Since Pi release necessarily follows ATP bond cleavage in our scheme of coupling (Figure 10), these serious difficulties are avoided by our mechanism (Section 5.6).
Our proposed mechanism and order of product release during ATP hydrolysis is also consistent with our other biochemical findings on mitochondrial F1. For instance, when [γ-32P]ATP is used as a substrate for MF1-ATPase, we found that the ratio of bound 32Pi to that of bound 32Pi and bound [γ-32P]ATP remained constant at 0.333. This distribution of product to substrate was maintained at different incubation times of the reaction mixtures between 1 and 15 min [92]. The same invariant value of the ratio was found for various 1:1 concentrations of MF1 to substrate in the range tested between 0.5 and 1 μM. This observed distribution between the bound product and bound substrate is very difficult to explain using other models.
Finally, the above order of product release is in accord with that found by single-molecule studies on F1-ATPase [11,15] and in other ATP-hydrolyzing systems such as purified myosin II [99] and unconventional myosin V [23] motors, and also with kinetic studies on muscle contraction [100,101,102].
5.8. Identity of Site 2 in the MF1 X-ray Structures
Suppose we were to reverse the identity of sites 1 and 2 in Figure 10 for ATP hydrolysis by F1-ATPase. Let the catalytic site assigned as site 2 or L (βTP) in panel 1 of Figure 10 be identified as site 1, and the site designated as site 1 or T (βDP or βDP-like) in Figure 10 be identified as site 2. Then, in the next conformational change after the 80–90° clockwise rotation of γ, O, i.e., site 3 or βE changes to βDP-like (now assigned as L or site 2), βDP-like changes to βTP (now ascribed to be T or site 1), and βTP changes to βE. That is, in the reversed assignment, O → L, L → T, and T → O. However, this is neither in accordance with our cold chase experiments nor with trisite hydrolysis in which O should change to T, T to L, and L to O. It implies that the reversed identity assumed at the beginning of this paragraph is incorrect. Moreover, the reversed identity of the sites is unable to reproduce the configuration of the enzyme in Figure 3 of the original paper depicting the Leslie–Walker structure in the MgADP-inhibited state [1]. Hence, βTP in the Leslie–Walker structure [1] is site 2, as proposed originally by the proponents [1,103] (PDBID: 1BMF, 1W0J, 1W0K), and also concluded by us from analysis of the architecture of the catalytic sites [32,62,63,64].
Our conclusion that “unisite” hydrolysis occurs in site 2 (βTP) is also in agreement with the finding of an important recent cryo-EM structural study carried out under various reaction conditions [7] (PDBID: 7XKH, 7XKO, 7XKP, 7XKQ, 7XKR).
The important phenomenon of ligand displacement also explains why the βTP site does not contain bound MgADP in the original high-resolution MF1 X-ray structure. Since, according to our mechanism, the ATP (or its analog) displaces bound ADP from site 2 (Section 5.6), βTP should contain bound AMP-PNP, as found [1]. Our mechanism also helps understand why no MF1 structure with ATP or ATP analog bound to the βDP (or βDP-like) subunit has been solved and why most MF1 structures capture an 80–90° rotated state of the enzyme (PDBID: 1BMF, 1H8E) [1,2] (the so-called Mg-inhibited state [1] or the catalytic dwell [11,14,15]) as opposed to a 0° resting state (panel 1 in Figure 10) (the so-called ATP-waiting dwell/state [11,14]). The ADP–ATP exchange and ATP hydrolysis followed by Pi release due to cold chase in site 2 will immediately rotate γ by ~80–90° and transform the βDP (or βDP-like) site to βTP, where the bound ATP gets hydrolyzed, as explained in consummate detail in Section 5.6, which will be captured by the crystal structure.
5.9. Angular Position of ATP Binding, Bond Cleavage, Pi Release, and ADP Release during ATP Hydrolysis by F1-ATPase
Looking at a single catalytic site, MgATP binds to O (site 3 or βE) at 0°, which becomes T (site 1 or βDP-like) [31] after the ε–subunit moves away during the 0 → 120° rotation of γ–ε. The bound MgATP is hydrolyzed at 200° due to its conformation change from βDP-like to βTP (site 2), i.e., owing to a T → L transition of the site. Pi is then released from L at 200°, leading to a 40° rotary sub-step (Figure 10, Section 5.6). The ADP unbinds from L at 240° and is fired out because the ADP is displaced by medium ATP, which now binds in L (ligand substitution). However, the L-site is meant for ADP.Pi, and therefore, the ATP immediately hydrolyzes in site 2 (L), following which Pi is released (“unisite” catalysis in site 2), which gives energy for the 80° rotation sub-step. The L-site now changes to a closed site. The interaction of the C-terminal of the rotated ε–subunit with the closed site induces a conformational change of the catalytic site to its open (O) conformation from which the bound MgADP is unbound and released. A new MgATP binds to O, and the cycle repeats.
To summarize, the elementary chemical processes and the angular position at which they occur during ATP hydrolysis by F1-ATPase that are consistent with our oxygen exchange analysis are as follows: ATP binding—0°, ATP bond cleavage—200°, Pi release—200°, and ADP release—240°.
The above correlation of the timing of elementary chemical processes in the F1-ATPase with rotary angle is in agreement with the latest biochemical study (2023) in which Nishizaka and coworkers generated a hybrid α3β3γ subcomplex in F1 from thermophilic Bacillus PS3 that consisted of one mutant β and two wild type βs [15]. The enzyme carried a β(E190D/F414E/F420E) mutation, which caused extremely slow rates of both ATP cleavage and ATP binding that enabled unequivocal determination of the angular position of the ATP cleavage reaction (200°) after ATP binding at 0° [15]. However, these studies do not explain how and why such a coupling scheme is operative, and of course, they cannot give the detailed molecular mechanism and dynamics of the process as done here.
5.10. Consistency of the Proposed Mechanism with the Results of Catalytic Site Nucleotide Occupancies of Senior and Colleagues Using Tryptophan Fluorescence Quenching [97,104,105,106] and Relationship with Other Models of ATP Hydrolysis [2,7,14,15,31,38,82,93,96,107,108]
Senior, Weber, and colleagues engineered a mutant Escherichia coli F1 in which the tyrosine residue of β-subunits, Tyr-331, was replaced by a tryptophan residue [104]. They showed that the tryptophan fluorescence was quenched proportionally as Mg-nucleotide bound to the β-catalytic sites and that the signal in the βY331W F1 could be reliably used to monitor occupancy of the sites with substrate MgATP. The present author was the first scientist to recognize this experimental breakthrough and laud it in the following words ([33], p. 73), “Recently, in a significant development, Senior and colleagues designed an optical probe by inserting a tryptophan residue to directly monitor, for the first time, the occupancy of the catalytic sites by nucleotides. Their tryptophan fluorescence experiments in the hydrolysis mode established definitively that steady-state Vmax activity is attained by F1-ATPase only when all three catalytic sites are occupied by bound nucleotide, i.e., an enzyme species with all three β subunits occupied is the only catalytically competent species” [97,104,105,106].
The use of a second Trp mutant at β–Phe-148 showed definitively that the fluorescence signals were different with bound MgADP as compared to bound MgATP in catalytic sites. The workers found that, on average, two molecules of ADP and one molecule of ATP were bound to catalytic sites during steady-state hydrolysis by F1-ATPase [105]. Further, the experiments showed that Pi binding to catalytic sites cannot be spontaneous, as in models [60,109] of the binding change mechanism, which must therefore be incorrect (see also pp. 71–75 and Figures 2 and 3 in ref. [33]). Rather, there is a requirement for energy input from the ion gradients to enable Pi binding in the ATP synthesis mode [106] and a fundamental requirement for a mechanism of Pi activation in ATP synthesis [31,57].
Senior and coworkers believed that the exchange results that Boyer obtained [58] were only relevant at the lower ATP concentrations [34]. In their words, “the 18O exchange experiments are probably monitoring a special activity that occurs only at low nucleotide occupancy of catalytic sites, or in the absence of rotation, but is not reflective of physiological steady-state catalysis” [34]. However, this was only a qualitative surmise, and no results were presented in support of their view. The results of our work show that the 18O exchange methodology is indeed capable of providing valuable information on mechanisms over the entire concentration range from low to high [ATP] (Figure 2, Figure 3 and Figure 4 and Figure 8) if a quantitative model of exchange had been developed as done in Section 3 and Reference [57], and the results interpreted correctly using such a stochastic kinetic model. This can be subjected to quantitative testing, as explained below.
In the oxygen exchange experiments presented here over the 50,000-fold range of ATP, we can calculate the extent of nucleotide occupancy at catalytic sites using Senior’s data [97] and determine how that impacts the results and conclusions of this work. Figure 11 shows representative results of this exercise. The open circles show the experimentally determined enzyme activity of MF1 for [ATP] from 0.11 to 5000 μM. The data are normalized by our value for Vmax determined for the mitochondrial ATPase of 640 30 s−1. The calculated curves show the fraction of catalytic sites with trisite filling, i.e., with all three sites occupied by Mg-nucleotide during hydrolysis. These calculations are given for two sets of values of the dissociation constants (Kd) of sites 1, 2, and 3. The bold gray curve uses the Kd values tabulated by Senior and coworkers [97] obtained by their true “equilibrium” (i.e., when Mg-nucleotide binding to catalytic sites has reached a steady state) fluorescence binding technique for sites 1 and 2 of 0.02 and 1.4 µM, respectively—which are for the EF1, not the MF1—and our determined value of Km for MF1 of 99 15 µM, with the very good approximation that Kd3 Km. The bold blue curve shows calculation results with the values of Kd1, Kd2, and Kd3 of 0.018, 1, and 150 µM, respectively, measured by Cross, Nalin, and coworkers on MF1 [110,111]. In general, the finding arises that the binding of MgATP to site 3 is weaker in MF1 compared to that in EF1, as shown by values of the dissociation constant in site 3, Kd3 in the range of 100–150 µM and 23–30 µM for MF1 and EF1 respectively. Figure 11 shows conclusively that a trisite mechanism of ATP hydrolysis is the operative mode of catalysis over five decades of ATP concentration. Models with (110) bisite nucleotide occupancies do not fit the enzymological data of Figure 11 (bold blue circles vs. bold orange curve that uses the experimental Kd values measured for MF1 [110,111]), similar to their inability to predict the oxygen exchange data (for example, compare Figure 6 with Figure 8). The use of all possible bisite enzyme species (i.e., occupancies due to (110) + (101) + (011) species) gave similar results to that modeled by the bold orange curve in Figure 11. Results for the trisite vs. bisite models using experimental data on the E. coli F1-ATPase and Kd values for the three catalytic sites of EF1 measured by Weber and Senior [97] have already been published (see Figure 2 of ref. [32]).
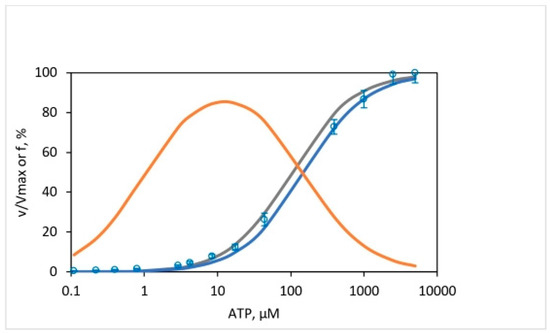
Figure 11.
Fraction, of total ATP hydrolysis activity by F1-ATPase (plotted as %) as a function of medium [ATP] concentration (from 0.11 to 5000 μM) contributed by the activity of (111) enzyme species, i.e., by species with all three catalytic sites occupied by Mg-nucleotide. Open blue circles (○) represent the experimental rates, , normalized by our determined value of Vmax for the mitochondrial ATPase of 640 30 s−1 (n = 3). The gray curve shows the results of calculations using the Kd values of Weber, Senior, and coworkers [97] for sites 1 and 2 determined by their fluorescence binding technique—for the EF1, not the MF1–and our determined value of Km for MF1 of 99 15 µM, along with the good approximation that Kd3 Km. The bold blue curve shows the results of calculations using the experimental values of Kd1, Kd2, and Kd3 determined by Cross, Nalin, and coworkers on the MF1 [110,111]. The bold orange curve shows the results for a bisite model with (110) occupancies of catalytic sites.
The results shown in Figure 11 have major biological implications. First, they show that the results of the 18O exchange are not limited to the low [ATP] range as suggested previously [34] but also apply to the physiologically important limit of mM [ATP] and that a single mechanism operates throughout this range, as also found in a single-molecule study by the Kinosita group [11]. Second, it provides us with another yardstick to discriminate between bisite models, like Boyer’s binding change mechanism, and trisite models, such as Nath’s torsional mechanism of energy transduction and ATP synthesis/hydrolysis, and critically appraise models of ATP hydrolysis by F1-ATPase proposed on the basis of single-molecule experiments.
A large number of technologically sophisticated single-molecule studies on F1 have been carried out by eminent Japanese researchers over the past 25 years [14,15,93,94,95,96], witnessed first-hand by the author during his sabbatical year at Waseda University, Tokyo [92]. Various models of ATP hydrolysis by F1-ATPase have been proposed based on these studies. However, in what must be regarded as a travesty that also points to the inherently elusive nature of the conundrum of mechanochemical coupling, the models are effectively bisite, and none of these models, based on single-molecule data, without exception, incorporates a true trisite mechanism. Therefore, they contradict Senior’s experimental data [97,104,105,106] and our results shown in Figure 8 and Figure 11.
As mentioned above, the models proposed for ATP hydrolysis by F1-ATPase based on single-molecule studies contradict experimental data based on direct, real-time monitoring of catalytic site nucleotide occupancies [97,104,105,106]. In these models, rotation takes place with two sites occupied by Mg-nucleotide; for example, in the model of References [93,95] that postulated for the α3β3γ subcomplex of TF1 from thermophilic Bacillus PS3, both the 80° and the 40° sub-steps of the rotation of the γ-subunit take place in bisite mode. In the model of Reference [96] for the complete human mitochondrial F1-ATPase, all three sub-steps for rotation of γ-ε from 0–65°, 65–90°, and 90–120° postulated to be driven by ATP binding, Pi release, and ATP bond cleavage, respectively [96], occur with two sites containing bound Mg-nucleotide. For bovine MF1, a bisite mode of catalysis is suggested for rotation of γ-ε for the substep from (at least) 10–20° to 80° and for the 80–120° substep [14]. Models that propose concerted ATP binding to a site and ADP release from a different site [14,95,96] as driving rotation cannot be trisite. These models are incorrect, given that the operative mode of catalysis during steady-state ATP hydrolysis by F1-ATPase is trisite. A frequently used definition of ATPase mechanism as trisite, because it alternates between having two and three catalytic sites filled with nucleotide at any time, is inadequate. Based on this imperfect criterion, even the binding change mechanism as modified by Cross is trisite [109]! For a mechanism to be truly trisite, catalysis must occur, and rotation must take place during steady-state Vmax hydrolysis only when all three catalytic sites are occupied by bound Mg-nucleotide [97,104,105,106].
In our view, careful attention needs to be paid to theoretical developments that are based on fundamental physicochemical considerations. For example, it was shown that the electrostatic potential energy obtained upon Pi movement away from bound MgADP and its release from an enzyme catalytic site can be used to weaken a bond, surmount energy barriers in conformational changes, store internal energy in a local domain of the enzyme, or perform useful external mechanical work [30]. Such a fundamental theory can help solve the sturdy problems of molecular mechanisms that have caused major headaches to several researchers in the field. Another approach is to realize that the above anomalies arise primarily from the fact that while single-molecule studies monitor rotation, say of the central γ- or γ-ε subunit on which the optical probe is bound with unprecedented resolution, they do not directly monitor catalytic events in the β-subunits, and therefore they are blind to catalytic site nucleotide occupancies. Hence, a new high-resolution hybrid experimental technique may be required. Most importantly, all the available experimental information needs to be integrated into a single consistent model. This has been the attempt of the torsional mechanism and unified theory.
The converse is also true. Thus, studies that directly monitored nucleotide occupancies of β-subunits proposed models of ATP hydrolysis by F1-ATPase [97,104,105,106] contradict results from decades of single-molecule work [14,15,93,94,95,96]. The former studies correctly inferred that for a trisite mechanism to operate, ATP binding to site 3 with a Kd3 value of 100 µM is too small to provide the required quantum of energy to drive the 80° rotational sub-step. Hence, they proposed that ATP binding to site 3 followed by ATP hydrolysis in site 1 acting in sequence provide energy for the 80° sub-step of γ rotation [34,97]. The problem for these models is that single-molecule experiments on F1 have unequivocally shown that ATP hydrolysis only takes place in site 1 during the catalytic dwell that occurs after the 80° sub-step of γ rotation is complete [14,15,93,94,95,96]. Hence, in ATP hydrolysis, i.e., the bond cleavage step in site 1 cannot be proposed as a driving force for the 80° rotational sub-step in F1-ATPase.
Models based on structural considerations [2] or based on electrostatic or mechanical considerations have also been proposed for both ATP hydrolysis and ATP synthesis [29,31,107,108,112]. A model based on analysis of the fluoroaluminate (ADPAlF4−)2 F1 X-ray structure [2] that also proposed that both binding and hydrolysis steps drive rotation is beset with the same problem discussed above. Moreover, as shown by us previously [31], the model is effectively bisite: it appears trisite only due to the way it is labeled [2]. Models that invoke ATP binding to site 3 as solely or primarily responsible for driving rotation [7,93,94,107,108] are also problematic for the reasons spelled out above. Other models (e.g., see ref. [82]) propose that catalysis (i.e., the hydrolysis event) occurs during the tight to open transition of a catalytic site, which contradicts structural evidence that suggests that opening of the β-subunit occurs after ATP hydrolysis [2]. Several other difficulties with the proposed models of ATP synthesis and hydrolysis have been discussed previously [31,33,34,97].
This is not to say that the experimental data collected by the above single-molecule works [14,15,93,94,95,96] are incorrect. They are correct under the conditions, as are the structural data and the consolidated biochemical and biophysical information in the field. What is required is a molecular mechanism of ATP synthesis/hydrolysis by the ATP synthase/F1-ATPase that is consistent with the entire body of available experimental information.
Attempts to revisit ATP synthesis/hydrolysis catalysis by ATP synthase have been made [30,31,38,57,92,113]. In a recent comprehensive work, Frasch re-analyzed the molecular mechanisms of both ATP synthesis and hydrolysis based on gold nanorod probes and single-molecule microscopy measurements using single-photon counting avalanche photodiodes [38]. Their model of hydrolysis by F1 proposes that Pi release from βDP and ATP binding to βE drives the two phases of the power stroke (Figure 10 in ref. [38]). The model has similar shortcomings as the models discussed above, in that site 3 is open and distorted and cannot support the phase 2 power stroke, as postulated [38]. In fact, all models in which MgADP release precedes Pi release do not satisfy trisite catalysis [14,38,96] because, in these models, rotation does not occur with all three catalytic sites occupied by bound MgATP or bound MgADP.
A true trisite model of ATP hydrolysis that does not have the above shortcomings is one proposed by Nath in Reference [31]. However, even this model is not fully correct if we accept Senior and colleagues’ finding using their optical probes in fluorescence experiments that during steady state Vmax catalysis, the predominant long-lived enzyme species contains two bound MgADP and one bound MgATP [34,105]. Such a state is considered to be one that occurs after an 80–90° rotation from the ATP binding state at 0° [105]. In other words, at high [ATP], product Pi release is rate-limiting. If the model of Reference [31] is revised to include the fact that the ATP molecule in site 2 that binds upon ligand displacement (ADP–ATP exchange in site 2) hydrolyzes and releases Pi from that site [92,113], then the revised model (Figure 10; see panels 4 and 5) is consistent both with the biochemical observations [105] and with the results of the oxygen exchange experiments in this work. A 2023 paper describes in considerable detail the various states/angular positions along the catalytic pathway of F1-ATPase [92] and their correspondence with snapshots represented by relevant high-resolution X-ray and cryo-EM structures according to Nath’s torsional mechanism of energy transduction and ATP synthesis/hydrolysis. The biological significance and implications of the work [92] have been discussed in a Perspective Article by Wray [113].
To sum up, a wealth of novel mechanistic insights, along with a detailed molecular model that also elucidates and explains the how and why in ATP hydrolysis mechanism by F1-ATPase, has been formulated based on stochastic kinetic analysis of the oxygen exchange reactions. Previously, a view had been expressed that the 18O exchange technique can only offer qualitative information on the mechanism [34]. The present work on ATP hydrolysis by F1-ATPase and our previous work on ATP synthesis by the FOF1-ATP synthase [57] has falsified this view. It has shown definitively that given the right probabilistic approach to the analysis of oxygen exchange data (Section 3), unique insights (Section 5.3, Section 5.4, Section 5.5 and Section 5.6) and unprecedented information (Section 5.7, Section 5.8 and Section 5.9) on molecular mechanisms can indeed be obtained. These detailed insights had proved very difficult to obtain previously by other biochemical and biophysical techniques.
6. Conclusions
A quantitative analysis of the intermediate Pi–HOH exchange accompanying ATP hydrolysis by F1-ATPase over a wide substrate ATP concentration range—from sub-micromolar to millimolar—has been carried out. The kinetic data covered the full range of oxygen exchange that can be mediated by the F1-ATPase. Both the overall extent of oxygen exchange as well as the distribution of all [18O]Pi isotopomer species—containing 0, 1, 2, and 3 18O atoms in the Pi released from β-catalytic sites into the medium—were quantified over the 50,000-fold medium ATP range. The analysis of oxygen exchange was performed by application of a stochastic kinetic theory specifically developed for that purpose. The following conclusions were reached in this study based on oxygen exchange methodology:
- A single site of entry of water oxygen—a fundamental limitation of the model of oxygen exchange proposed by Boyer’s binding change mechanism of ATP synthesis/hydrolysis—is inadequate to predict even gross features of the exchange process.
- Multiple sites of water entry, as proposed by Nath’s torsional mechanism of ATP synthesis/hydrolysis since 2003 [32], are essential to explain exchange data.
- A constant value for the apparent rate constant of oxygen exchange () of 10.5 s−1 is found over five decades of ATP concentration during steady-state ATP hydrolysis by mitochondrial F1-ATPase.
- Finding 3 contradicts a fundamental tenet of the binding change mechanism that postulates that medium ATP concentration markedly modifies values of the rate constant(s) governing the oxygen exchange of Pi bound at the catalytic site prior to its release [58,59,60] and neglects consideration of a timescale for the exchange.
- The results are consistent with the torsional mechanism that considers the time available for exchange, , by the bound nucleotide/Pi in the catalytic site as the appropriate timescale and postulates that this parameter quantitatively determined the observed extent of the oxygen exchange when multiplied by an essentially constant apparent rate constant for the exchange process, . According to this theory, changes in the medium substrate concentration alter , which then is responsible for the modulation of the extent and rate of oxygen exchange.
- Ligand permutation is the fundamental cause for the occurrence of the intermediate Pi–HOH exchange (during ATP hydrolysis) (this work) and the intermediate ATP–HOH oxygen exchange (during ATP synthesis) [57], as opposed to multiple spontaneous reversals of ATP synthesis/hydrolysis in a single catalytic site [60].
- The exchanges occur mechanistically because the enzyme catalytic site lacks absolute spatial selectivity for the oxygen ligands of a phosphorus intermediate that it accepts and binds as substrate. Hence, the oxygen ligands readily exchange, i.e., permute their positions about the central phosphorus atom.
- Three catalytic sites contribute to oxygen exchange at short times (high [ATP]) that, however, reduce to two sites mediating intermediate Pi–HOH exchange at long times (low [ATP]), as revealed by the detailed experimental distribution of [18O]Pi isotopomer species found in our work and its stochastic kinetic analysis.
- The finding 8 imposes strong constraints on possible molecular mechanisms of ATP hydrolysis by F1-ATPase.
- None of the existing mechanisms, in their current form, fully explain the experimental observations in 8 and present a solution to the mechanistic conundrum.
- The concept of ligand displacement was proposed in 2008 for bisite activation of the F1-ATPase by ATP binding at a second catalytic site by the torsional mechanism and the unified theory [31], with the additional new element that the enzyme needs to also hydrolyze the bound ATP (that had exchanged with bound ADP in site 2 by substitution chemistry) to ADP.Pi, and subsequently unbinding and releasing the Pi from the site, presently offers the only way to resolve the mechanistic conundrum [92,113].
- βTP in the Leslie–Walker structures [1,2] (PDBID: 1BMF, 1H8E) is definitively identified as site 2 on the basis of our oxygen exchange data.
- Pi release precedes ADP release in the catalytic cycle of steady-state ATP hydrolysis by F1-ATPase.
- The elementary chemical processes and the angular position at which they occur during ATP hydrolysis by the complete F1-ATPase that are consistent with the results of the oxygen exchange experiments in this work are ATP binding—0°, ATP bond cleavage—200°, Pi release—200°, and ADP release [92]—240°.
- F1-ATPase performs its function of steady-state Vmax hydrolysis using a trisite mechanism, i.e., by a mode of operation in which rotation and catalysis occur with 3-site filling of β-catalytic sites by the Mg-nucleotides MgATP or MgADP. This finding contradicts bisite models of steady-state catalysis by ATP synthase/F1-ATPase proposed within Boyer’s binding change mechanism [60] but is fully consistent with Nath’s torsional mechanism of energy transduction and ATP synthesis [31,33] and hydrolysis [31,92].
These results and conclusions need to be taken into account in future work on ATP synthesis/hydrolysis by the FOF1/F1-ATPase [113].
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biom13111596/s1, Computer program for the analysis of kinetic data.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data are contained within the article or the Supplementary Materials.
Acknowledgments
The author thanks R. Balling and A. Zimmer for making all facilities available during the course of this work. He acknowledges the generous financing of his stay as a Visiting Professor at the University of Bonn, Germany, from January to December 2023, during which time this research was completed. Thanks are also due to V. K. Ghosh and I. Schmidt for skillful technical assistance and to S. Gupta for writing a C++ program to implement the results of the stochastic kinetic theory developed in this work.
Conflicts of Interest
The author declares no conflict of interest.
References
- Abrahams, J.P.; Leslie, A.G.W.; Lutter, R.; Walker, J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 1994, 370, 621–628. [Google Scholar] [CrossRef]
- Menz, R.I.; Walker, J.E.; Leslie, A.G.W. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: Implication for the mechanism of rotary catalysis. Cell 2001, 106, 331–341. [Google Scholar] [CrossRef]
- Zhou, A.; Rohou, A.; Schep, D.G.; Bason, J.V.; Walker, J.E.; Rubinstein, J.L. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife 2015, 4, e10180. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.J.; Klusch, N.; Langer, J.; Mills, D.J.; Yildiz, Ö.; Kühlbrandt, W. Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1-Fo coupling. Science 2019, 364, eaaw9128. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhang, L.; Zong, S.; Guo, R.; Liu, T.; Yi, J.; Wang, P.; Zhuo, W.; Yang, M. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science 2019, 364, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Pinke, G.; Zhou, L.; Sazanov, L.A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol. 2020, 27, 1077–1085. [Google Scholar] [CrossRef]
- Nakano, A.; Kishikawa, J.I.; Nakanishi, A.; Mitsuoka, K.; Yokoyama, K. Structural basis of unisite catalysis of bacterial F0F1-ATPase. PNAS Nexus 2022, 1, pgac116. [Google Scholar] [CrossRef]
- Sobti, M.; Zeng, Y.C.; Walshe, J.L.; Brown, S.H.J.; Ishmukhametov, R.; Stewart, A.G. Changes within the central stalk observed of E. coli F1Fo ATP synthase observed after addition of ATP. Commun. Biol. 2023, 6, 26. [Google Scholar] [CrossRef]
- Noji, H.; Yasuda, R.; Yoshida, M.; Kinosita, K. Direct observation of the rotation of F1-ATPase. Nature 1997, 386, 299–302. [Google Scholar] [CrossRef]
- Martin, J.L.; Ishmukhametov, R.; Spetzler, D.; Hornung, T.; Frasch, W.D. Elastic coupling power stroke mechanism of the F1-ATPase molecular motor. Proc. Natl. Acad. Sci. USA 2018, 115, 5750–5755. [Google Scholar] [CrossRef]
- Sakaki, N.; Shimo-Kon, R.; Adachi, K.; Itoh, H.; Furuike, S.; Muneyuki, E.; Yoshida, M.; Kinosita, K. One rotary mechanism for F1-ATPase over ATP concentrations from millimolar down to nanomolar. Biophys. J. 2005, 88, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi-Matsui, M.; Kashiwagi, S.; Hosokawa, H.; Cipriano, D.J.; Dunn, S.D.; Wada, Y.; Futai, M. Stochastic high-speed rotation of Escherichia coli F1 sector: The ε subunit-sensitive rotation. J. Biol. Chem. 2006, 281, 4126–4131. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.L.; Ishmukhametov, R.; Hornung, T.; Ahmad, Z.; Frasch, W.D. Anatomy of F1-ATPase powered rotation. Proc. Natl. Acad. Sci. USA 2014, 111, 3715–3720. [Google Scholar] [CrossRef]
- Kobayashi, R.; Ueno, H.; Li, C.-B.; Noji, H. Rotary catalysis of bovine mitochondrial F1-ATPase studied by single-molecule experiments. Proc. Natl. Acad. Sci. USA 2020, 117, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Hasimoto, Y.; Sugawa, M.; Nishiguchi, Y.; Aeba, F.; Tagawa, A.; Suga, K.; Tanaka, N.; Ueno, H.; Yamashita, H.; Yokota, R.; et al. Direct identification of the rotary angle of ATP cleavage in F1-ATPase from Bacillus PS3. Biophys. J. 2023, 122, 554–564. [Google Scholar] [CrossRef] [PubMed]
- García, J.J.; Capaldi, R.A. Unisite catalysis without rotation of the γ–ε domain in the Escherichia coli F1-ATPase. J. Biol. Chem. 1998, 273, 15940–15945. [Google Scholar] [CrossRef]
- Wilkens, S.; Capaldi, R.A. Solution structure of the ε-subunit of the F1-ATPase from Escherichia coli and the interactions of this subunit with β-subunits in the complex. J. Biol. Chem. 1998, 273, 26645–26651. [Google Scholar] [CrossRef]
- Nath, S. Energy landscapes and dynamics of ion translocation through membrane transporters: A meeting ground for physics, chemistry, and biology. J. Biol. Phys. 2021, 47, 401–433. [Google Scholar] [CrossRef]
- Holmes-Cerfon, M.; Gortler, S.J.; Brenner, M.P. A geometrical approach to computing free-energy landscapes from short-ranged potentials. Proc. Natl. Acad. Sci. USA 2012, 109, E5–E14. [Google Scholar] [CrossRef]
- Rieu, M.; Taylor, N.M.I.; Krutyholowa, R.; Berry, R.M. A new class of biological ion-driven rotary molecular motors with 5:2 symmetry. Front. Microbiol. 2022, 13, 948383. [Google Scholar] [CrossRef]
- Wadhwa, N.; Berg, H.C. Bacterial motility: Machinery and mechanism. Nat. Rev. Microbiol. 2021, 20, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Trybus, K.M. Myosin V from head to tail. Cell. Mol. Life Sci. 2008, 65, 1378–1389. [Google Scholar] [CrossRef]
- Burgess, S.; Walker, M.; Wang, F.; Sellers, J.R.; White, H.D.; Knight, P.J.; Trinick, J. The prepower stroke conformation of myosin V. J. Cell Biol. 2002, 159, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Takada, S. Structural comparison of F1-ATPase: Interplay among enzyme structures, catalysis, and rotations. Structure 2011, 19, 588–598. [Google Scholar] [CrossRef]
- Hayashi, S.; Ueno, H.; Shaikh, A.R.; Umemura, M.; Kamiya, M.; Ito, Y.; Ikeguchi, M.; Komoriya, Y.; Iino, R.; Noji, H. Molecular mechanism of ATP hydrolysis in F1-ATPase revealed by molecular simulations and single-molecule observations. J. Am. Chem. Soc. 2012, 134, 8447–8454. [Google Scholar] [CrossRef]
- Forsyth, A.M.; Wan, J.; Owrutsky, P.D.; Abkarian, M.; Stone, H.A. Multiscale approach to link red blood cell dynamics, shear viscosity, and ATP release. Proc. Natl. Acad. Sci. USA 2011, 108, 10986–10991. [Google Scholar] [CrossRef] [PubMed]
- Singharoy, A.; Chipot, C. Methodology for the simulation of molecular motors at different scales. J. Phys. Chem. B 2017, 121, 3502–3514. [Google Scholar] [CrossRef]
- Jarvis, K.J.; Bell, K.M.; Loya, A.K.; Swank, D.M.; Walcott, S. Force-velocity and tension transient measurements from Drosophila jump muscle reveal the necessity of both weakly-bound cross-bridges and series elasticity in models of muscle contraction. Arch. Biochem. Biophys. 2021, 701, 108809. [Google Scholar] [CrossRef]
- Štrajbl, M.; Shurki, A.; Warshel, A. Converting conformational changes to electrostatic energy in molecular motors: The energetics of ATP synthase. Proc. Natl. Acad. Sci. USA 2013, 100, 14834–14839. [Google Scholar] [CrossRef]
- Nath, S.S.; Nath, S. Energy transfer from adenosine triphosphate: Quantitative analysis and mechanistic insights. J. Phys. Chem. B 2009, 113, 1533–1537. [Google Scholar] [CrossRef]
- Nath, S. The new unified theory of ATP synthesis/hydrolysis and muscle contraction, its manifold fundamental consequences and mechanistic implications and its applications in health and disease. Int. J. Mol. Sci. 2008, 9, 1784–1840. [Google Scholar] [CrossRef] [PubMed]
- Nath, S. Molecular mechanisms of energy transduction in cells: Engineering applications and biological implications. Adv. Biochem. Eng. Biotechnol. 2003, 85, 125–180. [Google Scholar] [PubMed]
- Nath, S. The molecular mechanism of ATP synthesis by F1F0-ATP synthase: A scrutiny of the major possibilities. Adv. Biochem. Eng. Biotechnol. 2002, 74, 65–98. [Google Scholar]
- Senior, A.E.; Nadanaciva, S.; Weber, J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta 2002, 1553, 188–211. [Google Scholar] [CrossRef]
- Xu, L.; Liu, F. The chemo-mechanical coupled model for F1F0-motor. Prog. Biophys. Mol. Biol. 2012, 108, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Nath, S. Analysis of molecular mechanisms of ATP synthesis from the standpoint of the principle of electrical neutrality. Biophys. Chem. 2017, 224, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B. A protet-based, protonic charge transfer model of energy coupling in oxidative and photosynthetic phosphorylation. Adv. Microb. Physiol. 2021, 78, 1–35. [Google Scholar]
- Frasch, W.D.; Bukhari, Z.A.; Yanagisawa, S. F1FO ATP synthase molecular motor mechanisms. Front. Microbiol. 2022, 13, 965620. [Google Scholar] [CrossRef]
- Bruns, K.; Studtrucker, N.; Sharma, A.; Fossen, T.; Mitzner, D.; Eissman, A.; Tessmer, U.; Roder, R.; Henklein, P.; Wray, V.; et al. Structural characterization and oligomerization of PB1–F2, a proapoptotic influenza A virus protein. J. Biol. Chem. 2007, 282, 353–363. [Google Scholar] [CrossRef]
- Hards, K.; McMillan, D.G.G.; Schurig-Briccio, L.A.; Gennis, R.B.; Lill, H.; Bald, D.; Cook, G.M. Ionophoric effects of the antitubercular drug bedaquiline. Proc. Natl. Acad. Sci. USA 2018, 115, 7326–7331. [Google Scholar] [CrossRef]
- Nath, S. Interpretation of the mechanism of action of antituberculosis drug bedaquiline based on a novel two-ion theory of energy coupling in ATP synthesis. Bioeng. Transl. Med. 2019, 4, 164–170. [Google Scholar] [CrossRef]
- Sarathy, J.P.; Grüber, G.; Dick, T. Re-understanding the mechanisms of action of the anti-mycobacterial drug bedaquiline. Antibiotics 2019, 8, 261. [Google Scholar] [CrossRef]
- Niedzwiecka, K.; Baranowska, E.; Panja, C.; Kucharczyk, R. ATP synthase subunit a supports permeability transition in yeast lacking dimerization subunits and modulates yPTP conductance. Cell. Physiol. Biochem. 2020, 54, 211–229. [Google Scholar]
- Nath, S. A novel conceptual model for the dual role of FOF1-ATP synthase in cell life and cell death. Biomol. Concepts 2020, 11, 143–152. [Google Scholar] [CrossRef]
- Bernardi, P.; Carraro, M.; Lippe, G. The mitochondrial permeability transition: Recent progress and open questions. FEBS J. 2022, 289, 7051–7074. [Google Scholar] [CrossRef]
- Nath, S. Supercomplex supercomplexes: Raison d’etre and functional significance of supramolecular organization in oxidative phosphorylation. Biomol. Concepts 2022, 13, 272–288. [Google Scholar] [CrossRef]
- Chu, X.-Y.; Xu, Y.-Y.; Tong, X.-Y.; Wang, G.; Zhang, H.-Y. The legend of ATP: From origin of life to precision medicine. Metabolites 2022, 12, 461. [Google Scholar] [CrossRef] [PubMed]
- Callender, R.; Dyer, R.B. Advances in time-resolved approaches to characterize the dynamical nature of enzymatic catalysis. Chem. Rev. 2006, 106, 3031–3042. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Moerner, W.E. An adaptive anti-Brownian electrokinetic trap with real-time information on single-molecule diffusivity and mobility. ACS Nano 2011, 5, 5792–5799. [Google Scholar] [CrossRef]
- Rob, T.; Wilson, D.J. Time-resolved mass spectrometry for monitoring millisecond time-scale solution-phase processes. Eur. J. Mass. Spectrom. 2012, 18, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.B.; Ziółkowski, M.; Schatz, G.C.; Goodson, T. Two-photon and time-resolved fluorescence spectroscopy as probes for structural determination in amyloid β-peptides and aggregates. J. Phys. Chem. B 2014, 118, 2351–2359. [Google Scholar] [CrossRef]
- Sielaff, H.; Börsch, M. Twisting and subunit rotation in single FOF1-ATP synthase. Philos. Trans. R. Soc. B 2013, 368, 20120024. [Google Scholar] [CrossRef]
- Krah, A.; Vogelaar, T.; De Jong, S.I.; Claridge, J.K.; Bond, P.J.; McMillan, D. ATP binding by an F1Fo ATP synthase ε subunit is pH subunit, suggesting a diversity of ε subunit functional regulation in bacteria. Front. Mol. Biosci. 2023, 10, 1059673. [Google Scholar] [CrossRef]
- Shah, N.B.; Hutcheon, M.L.; Haarer, B.K.; Duncan, T.M. F1-ATPase of Escherichia coli. The ε-inhibited state forms after ATP hydrolysis, is distinct from the ADP-inhibited state, and responds dynamically to catalytic site ligands. J. Biol. Chem. 2013, 288, 9383–9395. [Google Scholar] [CrossRef]
- Zimmermann, B.; Diez, M.; Zarrabi, N.; Gräber, P.; Börsch, M. Movements of the ε-subunit during catalysis and activation in single membrane-bound H+-ATP synthase. EMBO J. 2005, 24, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-L.; Brudwig, G.W. Kinetic modeling of substrate-water exchange in Photosystem II. BBA Adv. 2021, 1, 100014. [Google Scholar] [CrossRef]
- Mehta, R.; Singh, J.; Nath, S. Time-resolved oxygen exchange measurements offer novel mechanistic insights into enzyme-catalyzed ATP synthesis during photophosphorylation. J. Phys. Chem. B. 2020, 124, 5139–5148. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, C.C.; Boyer, P.D. Assessment of the rate of bound substrate interconversion and of ATP acceleration of product release during catalysis by mitochondrial adenosine triphosphatase. J. Biol. Chem. 1984, 259, 5761–5767. [Google Scholar] [CrossRef] [PubMed]
- Boyer, P.D.; Cross, R.L.; Momsen, W. A new concept for energy coupling in oxidative phosphorylation based on a molecular explanation of the oxygen exchange reactions. Proc. Natl. Acad. Sci. USA 1973, 70, 2837–2839. [Google Scholar] [CrossRef]
- Boyer, P.D. The binding change mechanism for ATP synthase—Some probabilities and possibilities. Biochim. Biophys. Acta 1993, 1140, 215–250. [Google Scholar] [CrossRef]
- Boyer, P.D. Catalytic site occupancy during ATP synthase catalysis. FEBS Lett. 2002, 512, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Rohatgi, H.; Saha, A. The torsional mechanism of energy transfer in ATP synthase. Curr. Sci. 1999, 77, 167–169. [Google Scholar]
- Nath, S.; Rohatgi, H.; Saha, A. The catalytic cycle of ATP synthesis by means of a torsional mechanism. Curr. Sci. 2000, 78, 23–27. [Google Scholar]
- Nath, S.; Jain, S. Breakthroughs and views: Kinetic modeling of ATP synthesis by ATP synthase and its mechanistic implications. Biochem. Biophys. Res. Commun. 2000, 272, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Murugavel, R.; Hansen, L.D. ATP synthase and the torsional mechanism: Resolving a 50-year-old mystery. Curr. Sci. 2004, 87, 16–19. [Google Scholar]
- Villadsen, J.; Nielsen, J.; Lidén, G. Bioreaction Engineering Principles, 3rd ed.; Springer: New York, NY, USA, 2011; Chapter 4. [Google Scholar]
- Channakeshava, C. New paradigm for ATP synthesis and consumption. J. Biosci. 2011, 36, 3–4. [Google Scholar] [CrossRef]
- Agarwal, B. A role for anions in ATP synthesis and its molecular mechanistic interpretation. J. Bioenerg. Biomembr. 2011, 43, 299–310. [Google Scholar] [CrossRef]
- Agarwal, B. Revisiting the ‘chemiosmotic theory’: Coupled transport of anion and proton for ATP synthesis. Bioenergetics 2013, 2, e116. [Google Scholar] [CrossRef]
- Rao, N.M. Medical Biochemistry, 2nd ed.; New Age International: New Delhi, India, 2014; pp. 280–281. [Google Scholar]
- Wray, V. Commentary on “Oxidative phosphorylation revisited”. Biotechnol. Bioeng. 2015, 112, 1984–1985. [Google Scholar] [CrossRef]
- Ji, S. The Cell Language Theory: Connecting Mind and Matter; World Scientific: Singapore, 2018; Chapter 3. [Google Scholar]
- Levy, W.B.; Calvert, V.G. Communication consumes 35 times more energy than computation in the human cortex, but both costs are needed to predict synapse number. Proc. Natl. Acad. Sci. USA 2021, 118, e2008173118. [Google Scholar] [CrossRef]
- Juretić, D. Bioenergetics: A Bridge across Life and Universe; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Hill, R.D.; Boyer, P.D. Inorganic orthophosphate activation and adenosine diphosphate as the primary phosphoryl acceptor in oxidative phosphorylation. J. Biol. Chem. 1967, 242, 4320–4323. [Google Scholar] [CrossRef] [PubMed]
- Penefsky, H.S. Preparation of beef heart mitochondrial ATPase. Methods Enzymol. 1979, 55, 304–308. [Google Scholar] [PubMed]
- Hackney, D.D.; Stempel, K.E.; Boyer, P.D. Oxygen–18 probes of enzymic reactions of phosphate compounds. Methods Enzymol. 1980, 64, 60–83. [Google Scholar]
- Feller, W. An Introduction to Probability Theory and its Applications: Volume I, 3rd ed.; John Wiley: New York, NY, USA, 1968; Chapter 17. [Google Scholar]
- Ross, S.M. Introduction to Probability Models, 10th ed.; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Witkov, C.; Zengel, K. Chi-Squared Data Analysis and Model Testing for Beginners; Oxford University Press: Oxford, UK, 2019. [Google Scholar]
- Nath, S. Consolidation of Nath’s torsional mechanism of ATP synthesis and two-ion theory of energy coupling in oxidative phosphorylation and photophosphorylation. Biophys. Chem. 2020, 257, 106279. [Google Scholar] [CrossRef]
- Allison, W.S. F1-ATPase: A molecular motor that hydrolyzes ATP with sequential opening and closing of catalytic sites coupled to rotation of its γ-subunit. Acc. Chem. Res. 1998, 31, 819–826. [Google Scholar] [CrossRef]
- Ko, Y.H.; Hong, S.; Pedersen, P.L. Chemical mechanism of ATP synthase. J. Biol. Chem. 1999, 274, 28853–28856. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Saxena, A.K.; Simcoke, W.N.; Garboczi, D.N.; Pedersen, P.L.; Ko, Y.H. Mitochondrial ATP synthase: Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism. J. Biol. Chem. 2006, 281, 13777–13783. [Google Scholar] [CrossRef]
- Nath, S. Beyond the chemiosmotic theory: Analysis of key fundamental aspects of energy coupling in oxidative phosphorylation in the light of a torsional mechanism of energy transduction and ATP synthesis—Invited review part 1. J. Bioenerg. Biomembr. 2010, 42, 293–300. [Google Scholar] [CrossRef]
- Nath, S. Beyond the chemiosmotic theory: Analysis of key fundamental aspects of energy coupling in oxidative phosphorylation in the light of a torsional mechanism of energy transduction and ATP synthesis—Invited review part 2. J. Bioenerg. Biomembr. 2010, 42, 301–309. [Google Scholar] [CrossRef]
- Nath, S. The torsional mechanism of energy transduction and ATP synthesis as a breakthrough in our understanding of the mechanistic, kinetic and thermodynamic details. Thermochim. Acta 2004, 422, 5–17. [Google Scholar] [CrossRef]
- Nath, S. The thermodynamic efficiency of ATP synthesis in oxidative phosphorylation. Biophys. Chem. 2016, 219, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Nath, S. The need for consistency with physical laws and logic in choosing between competing molecular mechanisms in biological processes: A case study in modeling ATP synthesis. Function 2022, 3, zqac054. [Google Scholar] [CrossRef] [PubMed]
- Nath, S. Molecular mechanistic insights into coupling of ion transport to ATP synthesis. Biophys. Chem. 2018, 241, 20–26. [Google Scholar] [CrossRef]
- Nath, S. Molecular mechanistic insights into uncoupling of ion transport from ATP synthesis. Biophys. Chem. 2018, 242, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Nath, S. Beyond binding change: The molecular mechanism of ATP hydrolysis by F1-ATPase and its biochemical consequences. Front. Chem. 2023, 11, 1058500. [Google Scholar] [CrossRef]
- Yasuda, R.; Noji, H.; Yoshida, M.; Kinosita, K.; Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 2001, 410, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Nishizaka, T.; Oiwa, K.; Noji, H.; Kimura, S.; Muneyuki, E.; Kinosita, K.; Yoshida, M. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 2004, 11, 142–148. [Google Scholar] [CrossRef]
- Adachi, K.; Oiwa, K.; Nishizaka, T.; Furuike, S.; Noji, H.; Ito, H.; Yoshida, M.; Kinosita, K. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 2007, 130, 309–321. [Google Scholar] [CrossRef]
- Suzuki, T.; Tanaka, K.; Wakabayashi, C.; Saita, E.; Yoshida, M. Chemomechanical coupling of human mitochondrial F1-ATPase motor. Nat. Chem. Biol. 2014, 10, 930–936. [Google Scholar] [CrossRef]
- Weber, J.; Senior, A.E. Bi-site catalysis in F1-ATPase: Does it exist? J. Biol. Chem. 2001, 276, 35422–35428. [Google Scholar] [CrossRef]
- Rees, D.M.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc. Natl. Acad. Sci. USA 2012, 109, 11139–11143. [Google Scholar] [CrossRef]
- Rastogi, K.; Puliyakodan, M.S.; Pandey, V.; Nath, S.; Elangovan, R. Maximum limit to the number of myosin II motors participating in processive sliding of actin. Sci. Rep. 2016, 6, 32043. [Google Scholar] [CrossRef]
- Trentham, D.R.; Eccleston, J.F.; Bagshaw, C.R. Kinetic analysis of ATPase mechanisms. Q. Rev. Biophys. 1976, 9, 217–281. [Google Scholar] [CrossRef] [PubMed]
- Goldman, Y.E. Kinetics of the actomyosin ATPase in muscle fibers. Annu. Rev. Physiol. 1987, 49, 637–654. [Google Scholar] [CrossRef]
- Yount, R.G.; Lawson, D.; Rayment, I. Is myosin a “back door” enzyme? Biophys. J. 1995, 68, 44s−49s. [Google Scholar]
- Kagawa, R.; Montgomery, M.G.; Braig, K.; Leslie, A.G.W.; Walker, J.E. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004, 23, 2734–2744. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Wilke-Mounts, S.; Lee, R.S.-F.; Grell, E.; Senior, A.E. Specific placement of tryptophan in the catalytic sites of Escherichia coli F1-ATPase provides a direct probe of nucleotide binding: Maximal ATP hydrolysis occurs with three sites occupied. J. Biol. Chem. 1993, 268, 20126–20133. [Google Scholar] [CrossRef]
- Weber, J.; Bowman, C.; Senior, A.E. Specific tryptophan substitution in catalytic sites of Escherichia coli F1-ATPase allows differentiation between bound substrate ATP and product ADP in steady-state catalysis. J. Biol. Chem. 1996, 271, 18711–18718. [Google Scholar] [CrossRef]
- Löbau, S.; Weber, J.; Senior, A.E. Catalytic site nucleotide binding and hydrolysis in F1Fo-ATP synthase. Biochemistry 1998, 37, 10846–10853. [Google Scholar] [CrossRef]
- Mukherjee, S.; Warshel, A. Electrostatic origin of the mechanochemical rotary mechanism and the catalytic dwell of F1-ATPase. Proc. Natl. Acad. Sci. USA 2011, 108, 20550–20555. [Google Scholar] [CrossRef]
- Oster, G.; Wang, H. Reverse engineering a protein: The mechanochemistry of ATP synthase. Biochim. Biophys. Acta 2000, 1458, 482–510. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.L.; Duncan, T.M. Subunit rotation in F0F1-ATP synthases as a means of coupling proton transport through F0 to the binding changes in F1. J. Bioenerg. Biomembr. 1996, 28, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.L.; Nalin, C.M. Adenine nucleotide binding sites on beef heart F1-ATPase: Evidence for three exchangeable sites that are distinct from three noncatalytic sites. J. Biol. Chem. 1982, 257, 2874–2881. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.L.; Grubmeyer, C.; Penefsky, H.S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase: Rate enhancements resulting from cooperative interactions between multiple catalytic sites. J. Biol. Chem. 1982, 257, 12101–12105. [Google Scholar] [CrossRef]
- Nath, S. Charge transfer across biomembranes: A solution to the conundrum of high desolvation free energy penalty in ion transport. Biophys. Chem. 2021, 275, 106604. [Google Scholar] [CrossRef]
- Wray, V. Elucidating a complex mechanism: Perspective on “Beyond binding change: The molecular mechanism of ATP hydrolysis by F1-ATPase and its biochemical consequences”. Function 2023, 4, zqad051. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).