Designing Nanomedicines for Breast Cancer Therapy
Abstract
:1. Introduction
2. Materials and Methods
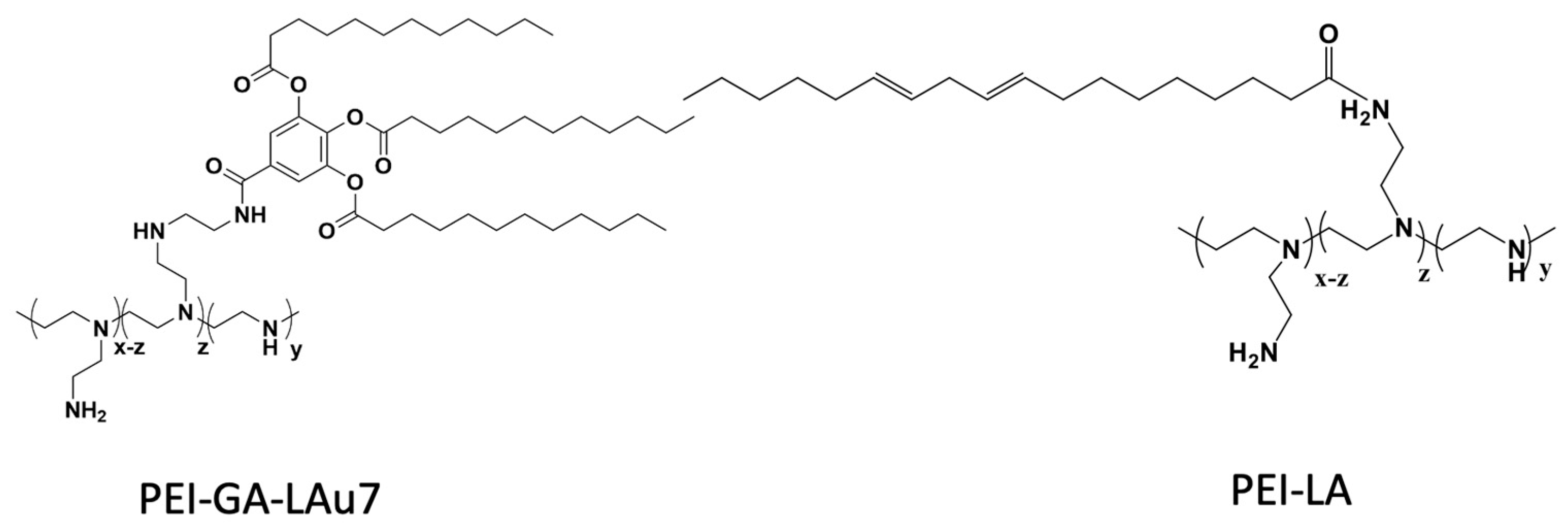
2.1. Materials
2.2. Cell Culture
2.3. siRNA Complex Preparation
2.4. Physiochemical Characterization of siRNA Polyplexes
2.5. GFP Silencing
2.6. Flow Cytometry
2.7. RNA Extraction and Quantitative Reverse Transcription PCR (qRT-PCR)
2.8. MTT Assay
2.9. Statistical Analysis
3. Results
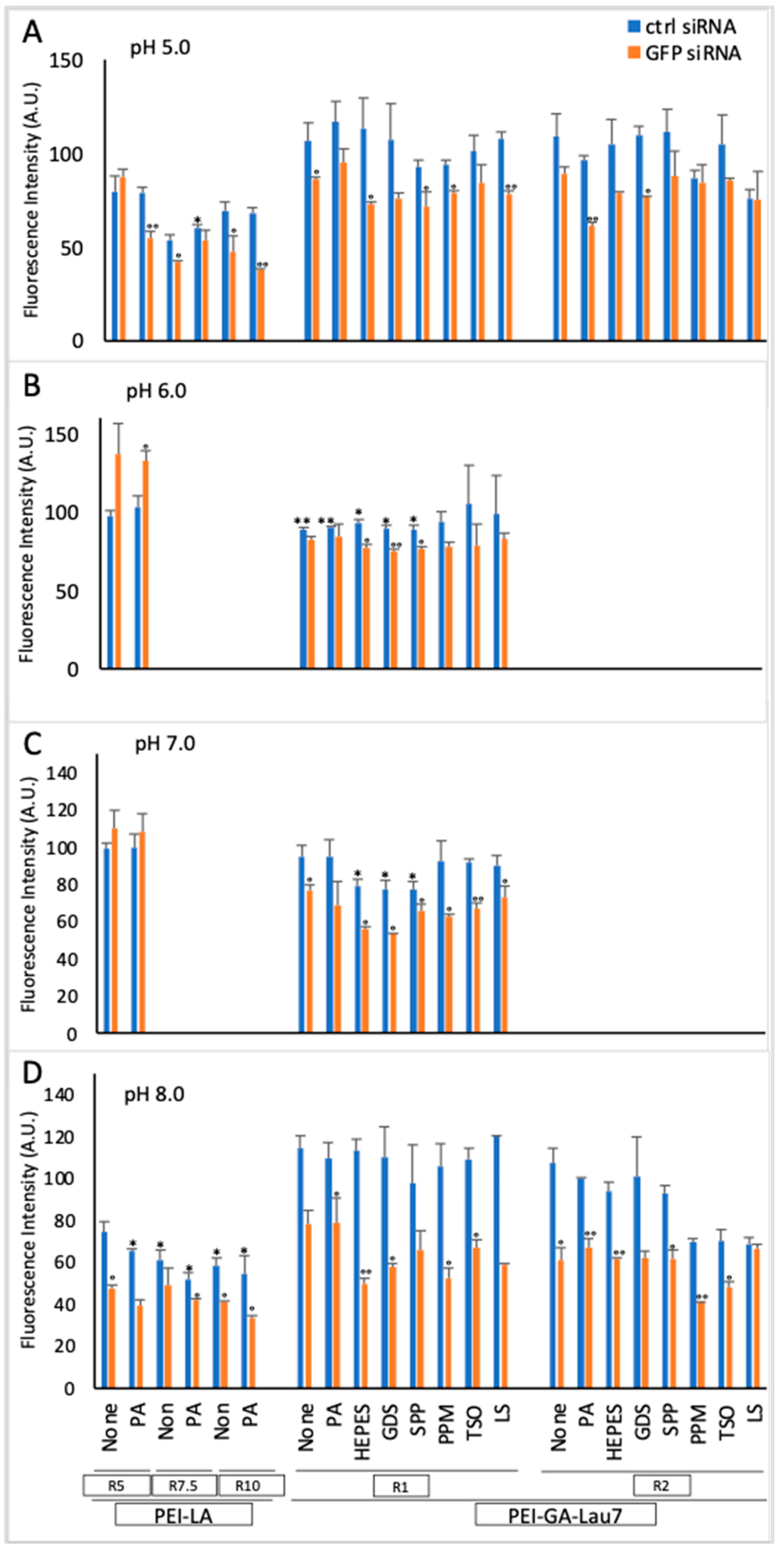
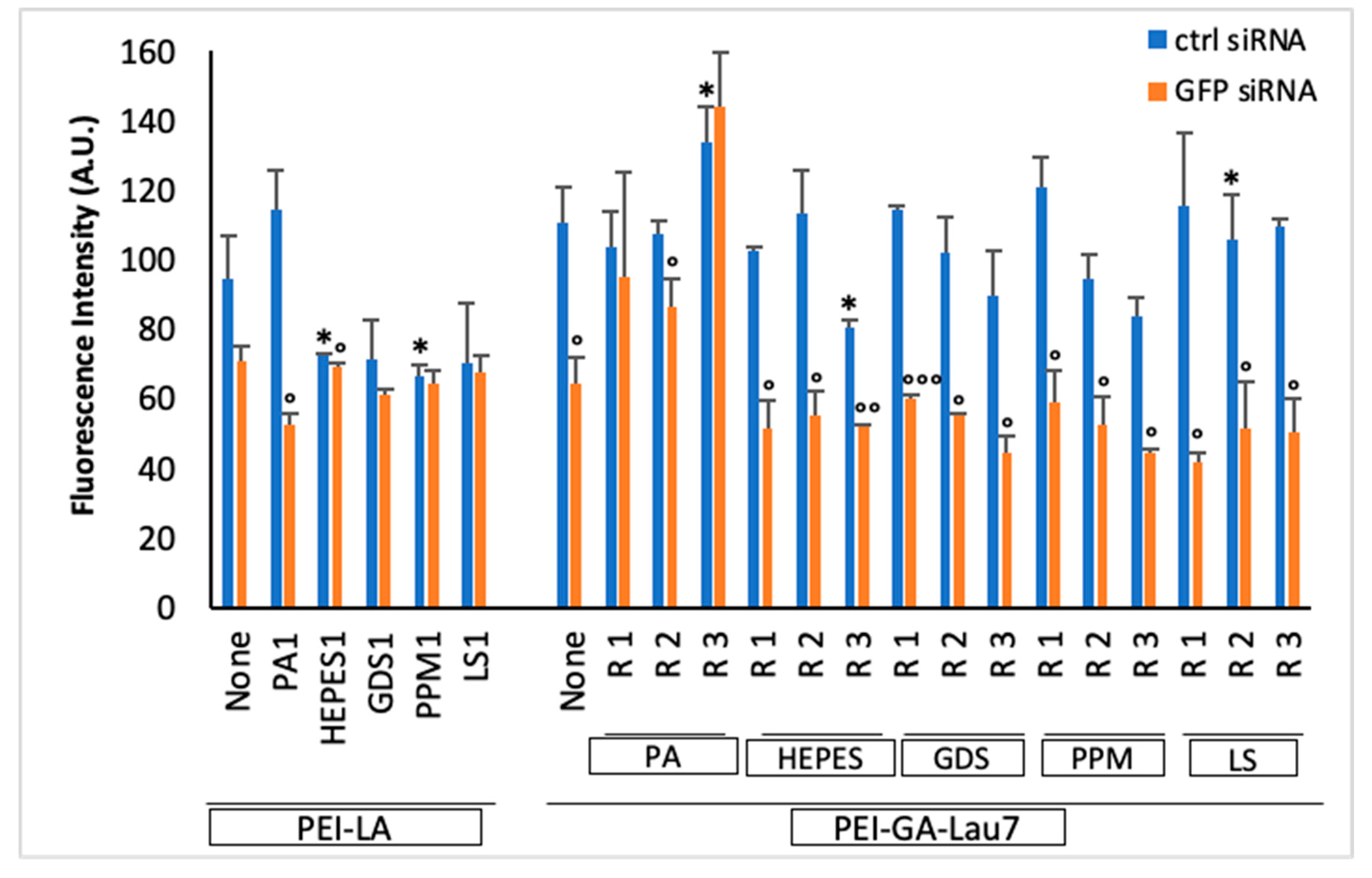
3.1. PEI-GA-Lau7 Can Deliver GFP-siRNA Better Than PEI-LA to MDA-MB-231 Cells
3.2. Ratio 1 of Additive Is Better Than Higher Ratios in MDA-MB-231 GFP+ Cells
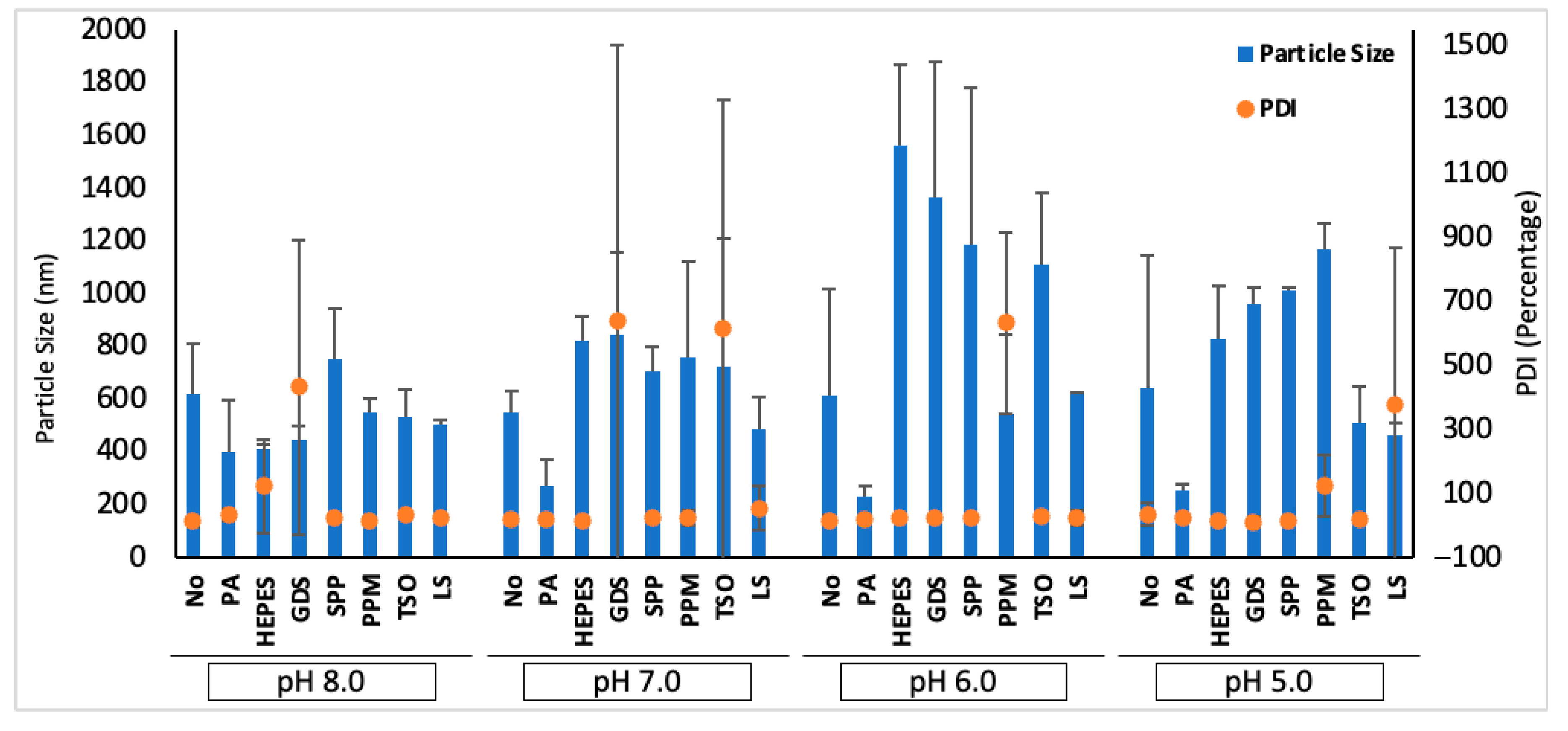
3.3. Physiochemical Characteristics of siRNA Polyplexes
3.4. siRNA-Mediated GFP Silencing Is Persistent for at Least 6 Days in MDA-MB-231 Cells
3.5. Optimized Polyplexes Showed ~95% Cell Uptake in MDA-MB-231 Cells
3.6. Polyplexes Can Silence Endogenous Genes with High Efficiency and Low Cytotoxicity
3.7. Polyplexes Could Effectively Inhibit Cell Growth in MDA-MB-231 Cells
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| siRNA Name | Sense Sequence | Antisense Sequence |
|---|---|---|
| Control | 5′-GCGUAUUAUACGCGA UUAACG-3′ | 5′-CGUUAAUCGCGUAUAAUACGC-3′ |
| GFP | 5′-GAACUUCAGGGUCAGCUUGCCG-3′ | 5′-GCAAGCUGACCCUCUUGUUCAU-3′ |
| FAM-labeled | 5′-/56-FAM/CAGUCGCGUUUGCGACUGGUUTT-3′ | 5′-AACCAGUCGCAAACGCGACUGTT-3′ |
| Survivin | 5′-AGACAGAAUAGAGUGAUAGGAAGCG-3′ | 5′-CGCUUCCUAUCACUCUAUUCUGUCUCC-3′ |
| Primer Name | Forward Sequence | Reverse Sequence |
|---|---|---|
| Survivin | 5′-TGAGAACGAGCCAGACTTGG-3′ | 5′-ATGTTCCTCTATGGGGTCGT-3′ |
| β-Actin | 5′-CCACCCCACTTCTCTCTAAGGA-3′ | 5′-AATTTACACGAAAGCAATGCTATC-3′ |
References
- Cancer. Available online: https://www.who.int/health-topics/cancer (accessed on 11 April 2023).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Nounou, M.I.; ElAmrawy, F.; Ahmed, N.; Abdelraouf, K.; Goda, S.; Syed-Sha-Qhattal, H. Breast Cancer: Conventional Diagnosis and Treatment Modalities and Recent Patents and Technologies. Breast Cancer Basic Clin. Res. 2015, 9 (Suppl. S2), 17–34. [Google Scholar] [CrossRef]
- Dhankhar, R.; Vyas, S.P.; Jain, A.K.; Arora, S.; Rath, G.; Goyal, A.K. Advances in novel drug delivery strategies for breast cancer therapy. Artif. Cells Blood Substit. Biotechnol. 2010, 38, 230–249. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Lim, L.-Y. Paclitaxel-loaded PLGA nanoparticles: Potentiation of anticancer activity by surface conjugation with wheat germ agglutinin. J. Control Release 2005, 108, 244–262. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R. The dawning era of polymer therapeutics. Nat. Rev. Drug Discov. 2003, 2, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M. Cancer nanotechnology: Opportunities and challenges. Nat. Rev. Cancer 2005, 5, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Royal Society. Nanoscience and Nanotechnologies: Opportunities and Uncertainties; Royal Society: London, UK, 2004. [Google Scholar]
- McManus, M.T.; Sharp, P.A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002, 3, 737–747. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67. [Google Scholar] [CrossRef]
- Setten, R.L.; Rossi, J.J.; Han, S. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Sano, M.; Sierant, M.; Miyagishi, M.; Nakanishi, M.; Takagi, Y.; Sutou, S. Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res. 2008, 36, 5812–5821. [Google Scholar] [CrossRef] [PubMed]
- Robbins, M.; Judge, A.; Liang, L.; McClintock, K.; Yaworski, E.; MacLachlan, I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 2007, 15, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Attia, N.; Mashal, M.; Puras, G.; Pedraz, J.L. Mesenchymal Stem Cells as a Gene Delivery Tool: Promise, Problems, and Prospects. Pharmaceutics 2021, 13, 843. [Google Scholar] [CrossRef]
- Sarvari, R.; Nouri, M.; Agbolaghi, S.; Roshangar, L.; Sadrhaghighi, A.; Seifalian, A.M.; Keyhanvar, P. A summary on non-viral systems for gene delivery based on natural and synthetic polymers. Int. J. Polym. Mater. Polym. Biomater. 2022, 71, 246–265. [Google Scholar] [CrossRef]
- Samal, S.K.; Dash, M.; Van Vlierberghe, S.; Kaplan, D.L.; Chiellini, E.; van Blitterswijk, C.; Moroni, L.; Dubruel, P. Cationic polymers and their therapeutic potential. Chem. Soc. Rev. 2012, 41, 7147–7194. [Google Scholar] [CrossRef]
- Aliabadi, H.M.; Landry, B.; Sun, C.; Tang, T.; Uludağ, H. Supramolecular assemblies in functional siRNA delivery: Where do we stand? Biomaterials 2012, 33, 2546–2569. [Google Scholar] [CrossRef]
- Funhoff, A.M.; van Nostrum, C.F.; Koning, G.A.; Schuurmans-Nieuwenbroek, N.M.E.; Crommelin, D.J.A.; Hennink, W.E. Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules 2004, 5, 32–39. [Google Scholar] [CrossRef]
- Aliabadi, H.M.; Landry, B.; Bahadur, R.K.; Neamnark, A.; Suwantong, O.; Uludağ, H. Impact of lipid substitution on assembly and delivery of siRNA by cationic polymers. Macromol. Biosci. 2011, 11, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Kc, R.B.; Kucharski, C.; Uludağ, H. Additive nanocomplexes of cationic lipopolymers for improved non-viral gene delivery to mesenchymal stem cells. J. Mater. Chem. B 2015, 3, 3972–3982. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.P.; Ogundana, O.; Morales, L.C.; Meenakshi Sundaram, D.N.; Kucharski, C.; Kc, R.; Uludağ, H. Transfection Efficacy and Cellular Uptake of Lipid-Modified Polyethyleneimine Derivatives for Anionic Nanoparticles as Gene Delivery Vectors. ACS Appl. Bio Mater. 2023, 6, 1105–1121. [Google Scholar] [CrossRef]
- Akhtar, L.N.; Qin, H.; Muldowney, M.T.; Yanagisawa, L.L.; Kutsch, O.; Clements, J.E.; Benveniste, E.N. Suppressor of cytokine signaling 3 inhibits antiviral IFN-beta signaling to enhance HIV-1 replication in macrophages. J. Immunol. 2010, 185, 2393–2404. [Google Scholar] [CrossRef]
- Young, L.; Sung, J.; Stacey, G.; Masters, J.R. Detection of Mycoplasma in cell cultures. Nat. Protoc. 2010, 5, 929–934. [Google Scholar] [CrossRef]
- Parmar, M.B.; Bahadur K.C., R.; Löbenberg, R.; Uludağ, H. Additive Polyplexes to Undertake siRNA Therapy against CDC20 and Survivin in Breast Cancer Cells. Biomacromolecules 2018, 19, 4193–4206. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.V.; Tan, A.S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): Subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. [Google Scholar] [CrossRef]
- A Dick, T.; Uludağ, H. Mineralized polyplexes for gene delivery: Improvement of transfection efficiency as a consequence of calcium incubation and not mineralization. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 129, 112419. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Chen, X.; Zhang, S. Long non-coding RNAs: From disease code to drug role. Acta Pharm. Sin. B 2021, 11, 340–354. [Google Scholar] [CrossRef]
- Fang, L.; Zhao, Z.; Wang, J.; Xiao, P.; Sun, X.; Ding, Y.; Zhang, P.; Wang, D.; Li, Y. Light-controllable charge-reversal nanoparticles with polyinosinic-polycytidylic acid for enhancing immunotherapy of triple negative breast cancer. Acta Pharm. Sin. B 2022, 12, 353–363. [Google Scholar] [CrossRef]
- Yuen, J.G.; Fesler, A.; Hwang, G.-R.; Chen, L.-B.; Ju, J. Development of 5-FU-modified tumor suppressor microRNAs as a platform for novel microRNA-based cancer therapeutics. Mol. Ther. J. Am. Soc. Gene Ther. 2022, 30, 3450–3461. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Hart, S.L.; Du, Z. Assembly strategy of liposome and polymer systems for siRNA delivery. Int. J. Pharm. 2021, 592, 120033. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Leroueil, P.R.; Janus, E.K.; Peters, J.L.; Kober, M.-M.; Islam, M.T.; Orr, B.G.; Baker, J.R.; Banaszak Holl, M.M. Interaction of polycationic polymers with supported lipid bilayers and cells: Nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem. 2006, 17, 728–734. [Google Scholar] [CrossRef]
- Lungwitz, U.; Breunig, M.; Blunk, T.; Göpferich, A. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm. 2005, 60, 247–266. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.-K.; Suh, D.; Kim, J.M.; Choi, H.-G.; Shin, K.; Ko, J.J. Polyethylenimine-mediated cellular uptake, nucleus trafficking and expression of cytokine plasmid DNA. Gene Ther. 2002, 9, 1627–1632. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.; Paik, H.; Hwang, B.K. Ionization of poly (ethylenimine) and poly (allylamine) at various pH′ s. Bioorganic Chem. 1994, 22, 318–327. [Google Scholar] [CrossRef]
- Montazeri Aliabadi, H.; Landry, B.; Mahdipoor, P.; Uludağ, H. Induction of apoptosis by survivin silencing through siRNA delivery in a human breast cancer cell line. Mol. Pharm. 2011, 8, 1821–1830. [Google Scholar] [CrossRef]
- Santadkha, T.; Skolpap, W.; KC, R.; Ansari, A.; Kucharski, C.; Atz Dick, T.; Uludağ, H. Improved delivery of Mcl-1 and survivin siRNA combination in breast cancer cells with additive siRNA complexes. Investig. New Drugs 2022, 40, 962–976. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, M.; Liu, Y.; Li, C.; Zhang, Q.; Oupicky, D.; Sun, M. Reversible Covalent Cross-Linked Polycations with Enhanced Stability and ATP-Responsive Behavior for Improved siRNA Delivery. Biomacromolecules 2018, 19, 3776–3787. [Google Scholar] [CrossRef]
- Li, H.; Liu, L.; Chang, H.; Zou, Z.; Xing, D. Downregulation of MCL-1 and upregulation of PUMA using mTOR inhibitors enhance antitumor efficacy of BH3 mimetics in triple-negative breast cancer. Cell Death Dis. 2018, 9, 137. [Google Scholar] [CrossRef]
- Liao, Z.-X.; Ho, Y.-C.; Chen, H.-L.; Peng, S.-F.; Hsiao, C.-W.; Sung, H.-W. Enhancement of efficiencies of the cellular uptake and gene silencing of chitosan/siRNA complexes via the inclusion of a negatively charged poly(γ-glutamic acid). Biomaterials 2010, 31, 8780–8788. [Google Scholar] [CrossRef]
- Jin, Y.; Adams, F.; Möller, J.; Isert, L.; Zimmermann, C.M.; Keul, D.; Merkel, O.M. Synthesis and Application of Low Molecular Weight PEI-Based Copolymers for siRNA Delivery with Smart Polymer Blends. Macromol. Biosci. 2023, 23, e2200409. [Google Scholar] [CrossRef]
- Lou, B.; Beztsinna, N.; Mountrichas, G.; van den Dikkenberg, J.B.; Pispas, S.; Hennink, W.E. Small nanosized poly(vinyl benzyl trimethylammonium chloride) based polyplexes for siRNA delivery. Int. J. Pharm. 2017, 525, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Morris, V.B.; Labhasetwar, V. Effectiveness of Small Interfering RNA Delivery via Arginine-Rich Polyethylenimine-Based Polyplex in Metastatic and Doxorubicin-Resistant Breast Cancer Cells. J. Pharmacol. Exp. Ther. 2019, 370, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Lazebnik, M.; Keswani, R.K.; Pack, D.W. Endocytic Transport of Polyplex and Lipoplex siRNA Vectors in HeLa Cells. Pharm. Res. 2016, 33, 2999–3011. [Google Scholar] [CrossRef] [PubMed]
- Hausig-Punke, F.; Dekevic, G.; Sobotta, F.H.; Solomun, J.I.; Richter, F.; Salzig, D.; Traeger, A.; Brendel, J.C. Efficient Transfection via an Unexpected Mechanism by Near Neutral Polypiperazines with Tailored Response to Endosomal pH. Macromol. Biosci. 2023, 23, e2200517. [Google Scholar] [CrossRef]
- Du, J.; Tang, L.; Yuan, Y.; Wang, J. Phosphoester modified poly (ethylenimine) as efficient and low cytotoxic genevectors. Sci. China Chem. 2011, 54, 351–358. [Google Scholar] [CrossRef]
- Kurosaki, T.; Kitahara, T.; Kawakami, S.; Nishida, K.; Nakamura, J.; Teshima, M.; Nakagawa, H.; Kodama, Y.; To, H.; Sasaki, H. The development of a gene vector electrostatically assembled with a polysaccharide capsule. Biomaterials 2009, 30, 4427–4434. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.; Li, Y.; Ahlemeyer, B.; Krieglstein, J.; Kissel, T. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials 2003, 24, 1121–1131. [Google Scholar] [CrossRef]
- Romdhane, A.; Aurousseau, M.; Guillet, A.; Mauret, E. Effect of pH and Ionic Strength on the Electrical Charge and Particle Size Distribution of Starch Nanocrystal Suspensions. Starch 2015, 67, 319–327. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/star.201400181?casa_token=HIC18tgJxLcAAAAA:iNnEOUHnI8MizTQ9pWum6njGJtcAQ8WLjwf18qSBFZPxwLDNnjG7excElfy2D5d6DrYC0eXIf4WDT0Vc (accessed on 11 June 2023). [CrossRef]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Reed, J.C. Dysregulation of apoptosis in cancer. J. Clin. Oncol. 1999, 17, 2941–2953. [Google Scholar] [CrossRef]
- Zaffaroni, N.; Daidone, M.G. Survivin expression and resistance to anticancer treatments: Perspectives for new therapeutic interventions. Drug Resist. Updates 2002, 5, 65–72. [Google Scholar] [CrossRef]
- Yamamoto, H.; Ngan, C.Y.; Monden, M. Cancer cells survive with survivin. Cancer Sci. 2008, 99, 1709–1714. [Google Scholar] [CrossRef] [PubMed]
- Sah, N.K.; Khan, Z.; Khan, G.J.; Bisen, P.S. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006, 244, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997, 3, 917–921. [Google Scholar] [CrossRef]
- Salvesen, G.S.; Duckett, C.S. IAP proteins: Blocking the road to death’s door. Nat. Rev. Mol. Cell Biol. 2002, 3, 401–410. [Google Scholar] [CrossRef]
- Deveraux, Q.L.; Reed, J.C. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999, 13, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Altieri, D.C. The molecular basis and potential role of survivin in cancer diagnosis and therapy. Trends Mol. Med. 2001, 7, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Giodini, A.; Kallio, M.J.; Wall, N.R.; Gorbsky, G.J.; Tognin, S.; Marchisio, P.C.; Symons, M.; Altieri, D.C. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 2002, 62, 2462–2467. [Google Scholar]
- Jha, K.; Shukla, M.; Pandey, M. Survivin expression and targeting in breast cancer. Surg. Oncol. 2012, 21, 125–131. [Google Scholar] [CrossRef]
- Plescia, J.; Salz, W.; Xia, F.; Pennati, M.; Zaffaroni, N.; Daidone, M.G.; Meli, M.; Dohi, T.; Fortugno, P.; Nefedova, Y.; et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell 2005, 7, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, T.; Kita, A.; Yamanaka, K.; Mori, M.; Amino, N.; Takeuchi, M.; Tominaga, F.; Hatakeyama, S.; Kinoyama, I.; Matsuhisa, A.; et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007, 67, 8014–8021. [Google Scholar] [CrossRef]
- Chang, C.-C.; Heller, J.D.; Kuo, J.; Huang, R.C.C. Tetra-O-methyl nordihydroguaiaretic acid induces growth arrest and cellular apoptosis by inhibiting Cdc2 and survivin expression. Proc. Natl. Acad. Sci. USA 2004, 101, 13239–13244. [Google Scholar] [CrossRef]
- Smolewski, P. Terameprocol, a novel site-specific transcription inhibitor with anticancer activity. IDrugs Investig. Drugs J. 2008, 11, 204–214. [Google Scholar]
- Ai, Z.; Yin, L.; Zhou, X.; Zhu, Y.; Zhu, D.; Yu, Y.; Feng, Y. Inhibition of survivin reduces cell proliferation and induces apoptosis in human endometrial cancer. Cancer 2006, 107, 746–756. [Google Scholar] [CrossRef]
- Lu, Y.-H.; Luo, X.-G.; Tao, X. Survivin gene RNA interference induces apoptosis in human HL60 leukemia cell lines. Cancer Biother. Radiopharm. 2007, 22, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Nakao, K.; Hamasaki, K.; Ichikawa, T.; Arima, K.; Eguchi, K.; Ishii, N. Survivin downregulation by siRNA sensitizes human hepatoma cells to TRAIL-induced apoptosis. Oncol. Rep. 2006, 16, 389–392. [Google Scholar] [CrossRef]
- Paduano, F.; Villa, R.; Pennati, M.; Folini, M.; Binda, M.; Daidone, M.G.; Zaffaroni, N. Silencing of survivin gene by small interfering RNAs produces supra-additive growth suppression in combination with 17-allylamino-17-demethoxygeldanamycin in human prostate cancer cells. Mol. Cancer Ther. 2006, 5, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Semizarov, D.; Frost, L.; Sarthy, A.; Kroeger, P.; Halbert, D.N.; Fesik, S.W. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. USA 2003, 100, 6347–6352. [Google Scholar] [CrossRef] [PubMed]
- Persengiev, S.P.; Zhu, X.; Green, M.R. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA 2004, 10, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Thapa, B.; Plianwong, S.; Remant Bahadur, K.; Rutherford, B.; Uludağ, H. Small hydrophobe substitution on polyethylenimine for plasmid DNA delivery: Optimal substitution is critical for effective delivery. Acta Biomater. 2016, 33, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.; Ke, W.; Dirisala, A.; Toh, K.; Tanaka, M.; Li, J. Stealth and pseudo-stealth nanocarriers. Adv. Drug Deliv. Rev. 2023, 198, 114895. [Google Scholar] [CrossRef] [PubMed]
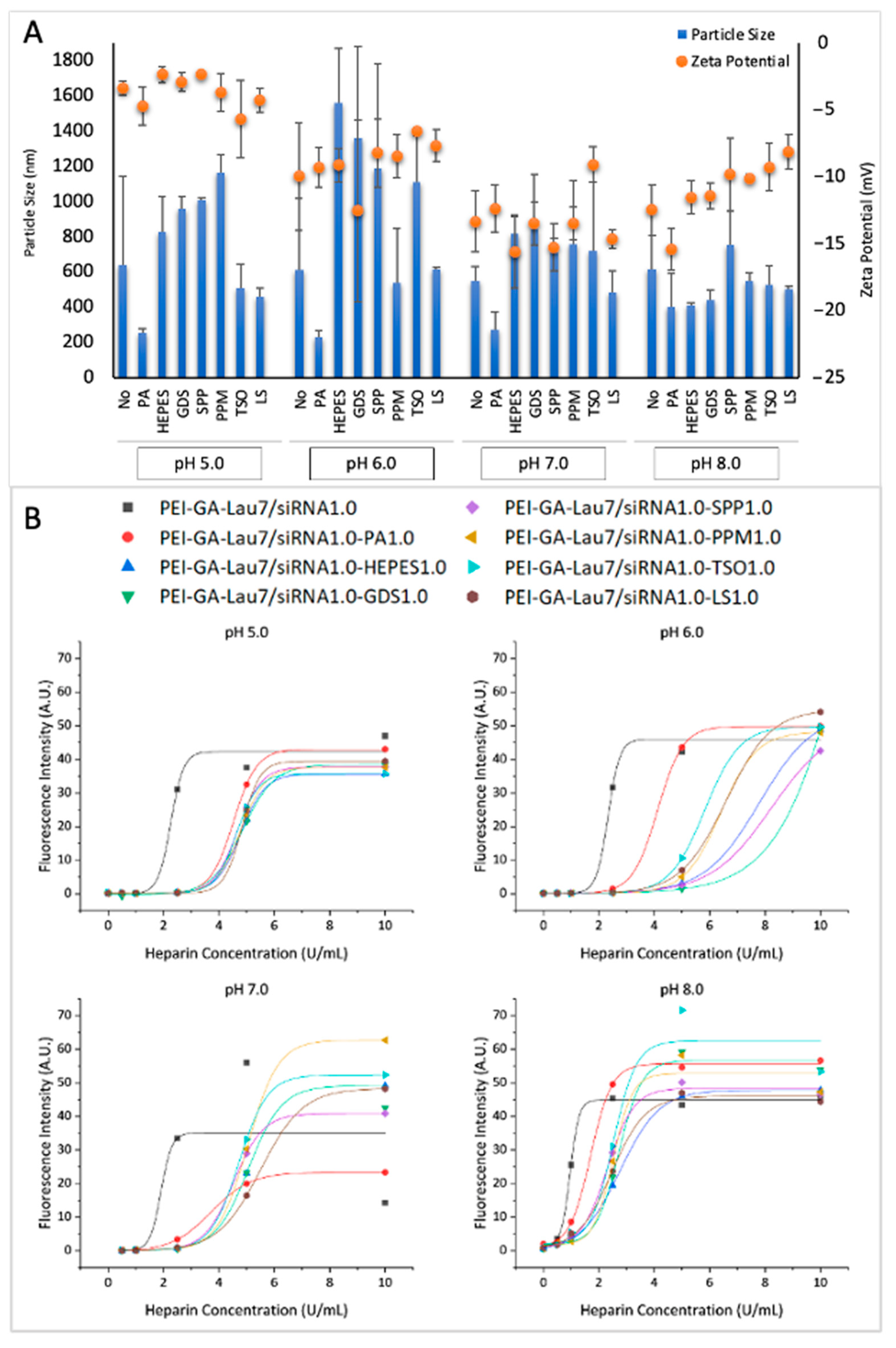

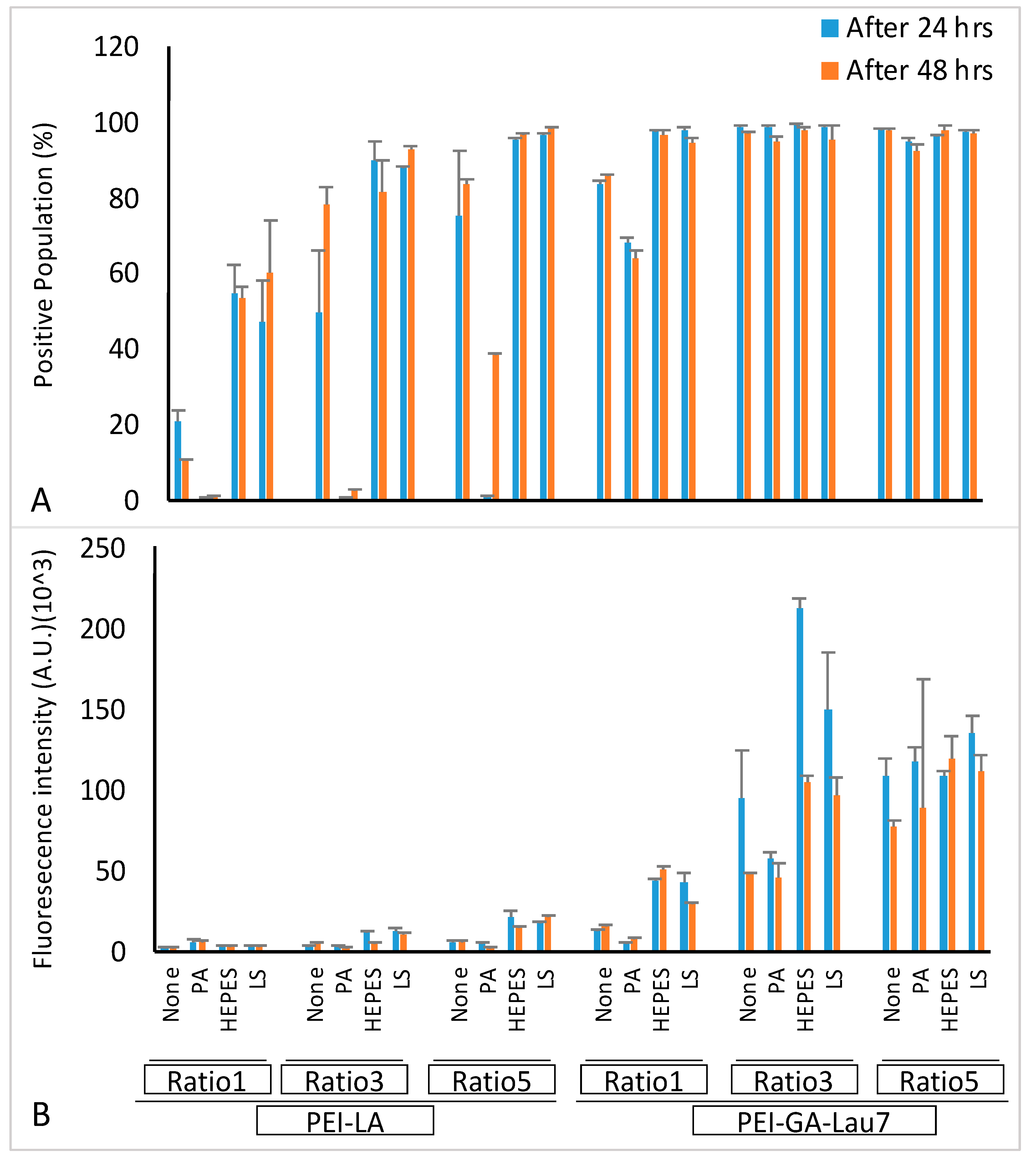
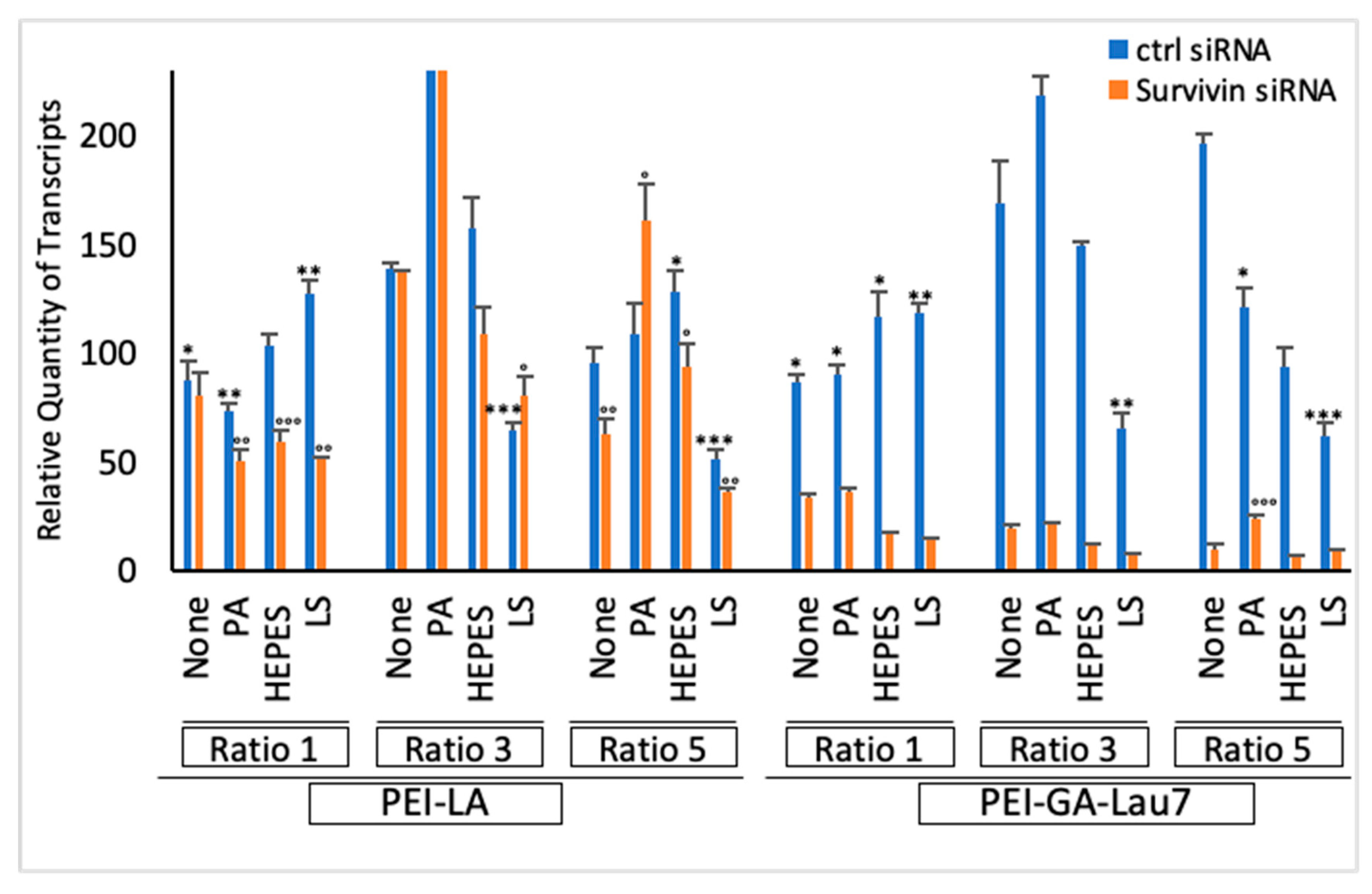
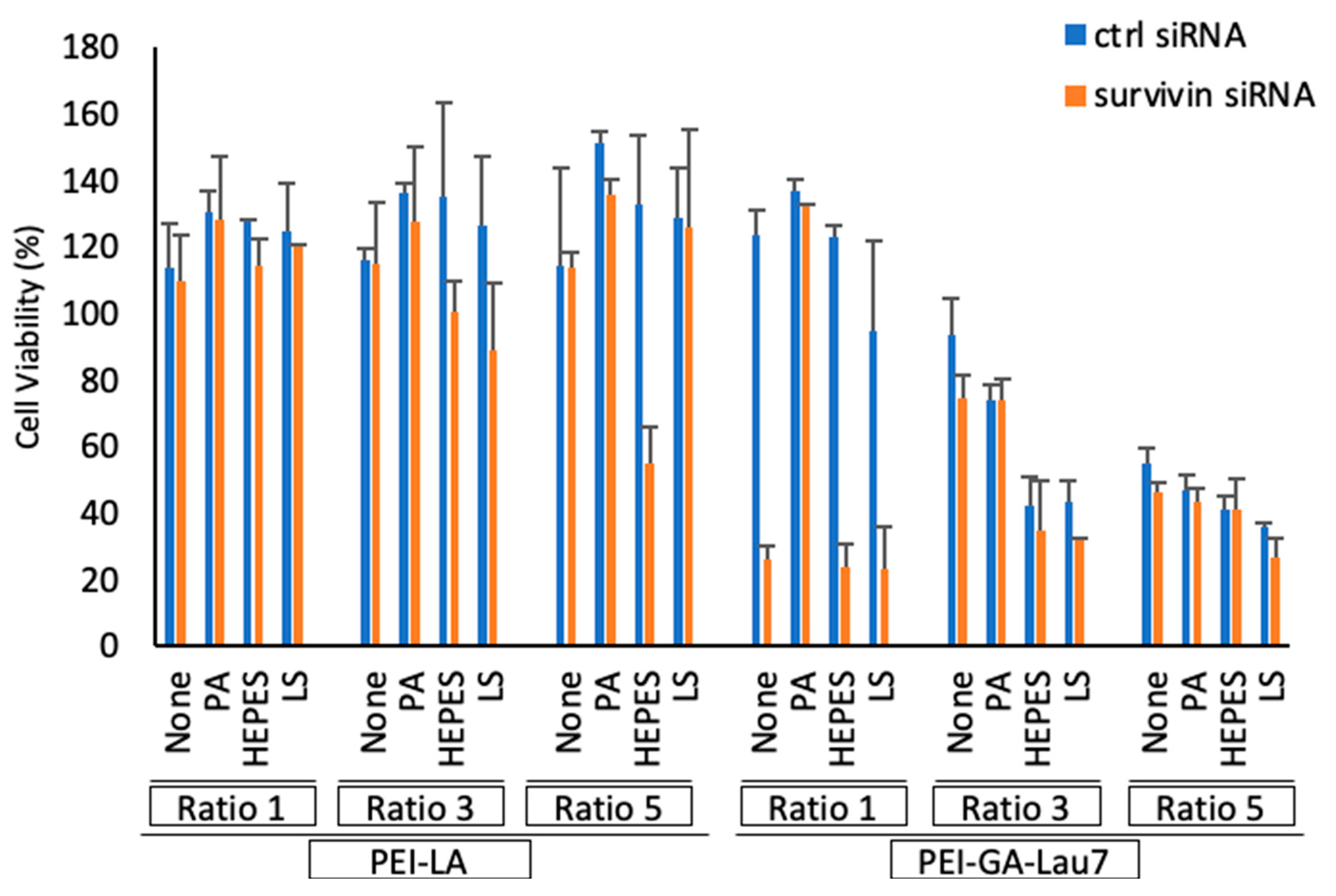
| Carrier | Ratio (w/w) | Media (uL) | siRNA (uL) | Additive (uL) | Transfection Reagent (uL) | Total Volume (uL) |
|---|---|---|---|---|---|---|
| NT | 0 | 200 | 0 | 0 | 0 | 200 |
| Buffer + siRNA | 0 | 196 | 4 | 0 | 0 | 200 |
| PEI-LA/SiRNA5.0 | 5 | 193.2 | 4 | 0 | 2.8 | 200 |
| PEI-LA/SiRNA5.0-PA1.0 | 5 | 186.4 | 4 | 4 | 5.6 | 200 |
| PEI-LA/SiRNA7.5 | 7.5 | 191.8 | 4 | 0 | 4.2 | 200 |
| PEI-LA/SiRNA7.5-PA1.0 | 7.5 | 183.6 | 4 | 4 | 8.4 | 200 |
| PEI-LA/SiRNA10.0 | 10 | 190.4 | 4 | 0 | 5.6 | 200 |
| PEI-LA/SiRNA10.0-PA1.0 | 10 | 180.8 | 4 | 4 | 11.2 | 200 |
| PEI-GA-Lau7/SiRNA1.0 | 1 | 194.88 | 4 | 0 | 1.12 | 200 |
| PEI-GA-Lau7/SiRNA1.0-PA1.0 | 1 | 189.76 | 4 | 4 | 2.24 | 200 |
| PEI-GA-Lau7/SiRNA1.0-HEPES1.0 | 1 | 189.76 | 4 | 4 | 2.24 | 200 |
| PEI-GA-Lau7/SiRNA1.0-GDS1.0 | 1 | 189.76 | 4 | 4 | 2.24 | 200 |
| PEI-GA-Lau7/SiRNA1.0-SPP1.0 | 1 | 189.76 | 4 | 4 | 2.24 | 200 |
| PEI-GA-Lau7/SiRNA1.0-PPM1.0 | 1 | 189.76 | 4 | 4 | 2.24 | 200 |
| PEI-GA-Lau7/SiRNA1.0-TSO1.0 | 1 | 189.76 | 4 | 4 | 2.24 | 200 |
| PEI-GA-Lau7/SiRNA1.0-LS1.0 | 1 | 189.76 | 4 | 4 | 2.24 | 200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbasi Dezfouli, S.; Rajendran, A.P.; Claerhout, J.; Uludag, H. Designing Nanomedicines for Breast Cancer Therapy. Biomolecules 2023, 13, 1559. https://doi.org/10.3390/biom13101559
Abbasi Dezfouli S, Rajendran AP, Claerhout J, Uludag H. Designing Nanomedicines for Breast Cancer Therapy. Biomolecules. 2023; 13(10):1559. https://doi.org/10.3390/biom13101559
Chicago/Turabian StyleAbbasi Dezfouli, Saba, Amarnath P. Rajendran, Jillian Claerhout, and Hasan Uludag. 2023. "Designing Nanomedicines for Breast Cancer Therapy" Biomolecules 13, no. 10: 1559. https://doi.org/10.3390/biom13101559
APA StyleAbbasi Dezfouli, S., Rajendran, A. P., Claerhout, J., & Uludag, H. (2023). Designing Nanomedicines for Breast Cancer Therapy. Biomolecules, 13(10), 1559. https://doi.org/10.3390/biom13101559






