Association of Cytokine Gene Polymorphisms and Their Impact on Active and Latent Tuberculosis in Brazil’s Amazon Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection and Storage
2.3. DNA Extraction
2.4. Genotyping
2.5. Plasma Cytokine Measurement
2.6. Tuberculin Skin Test (TST)
2.7. Statistical Analysis
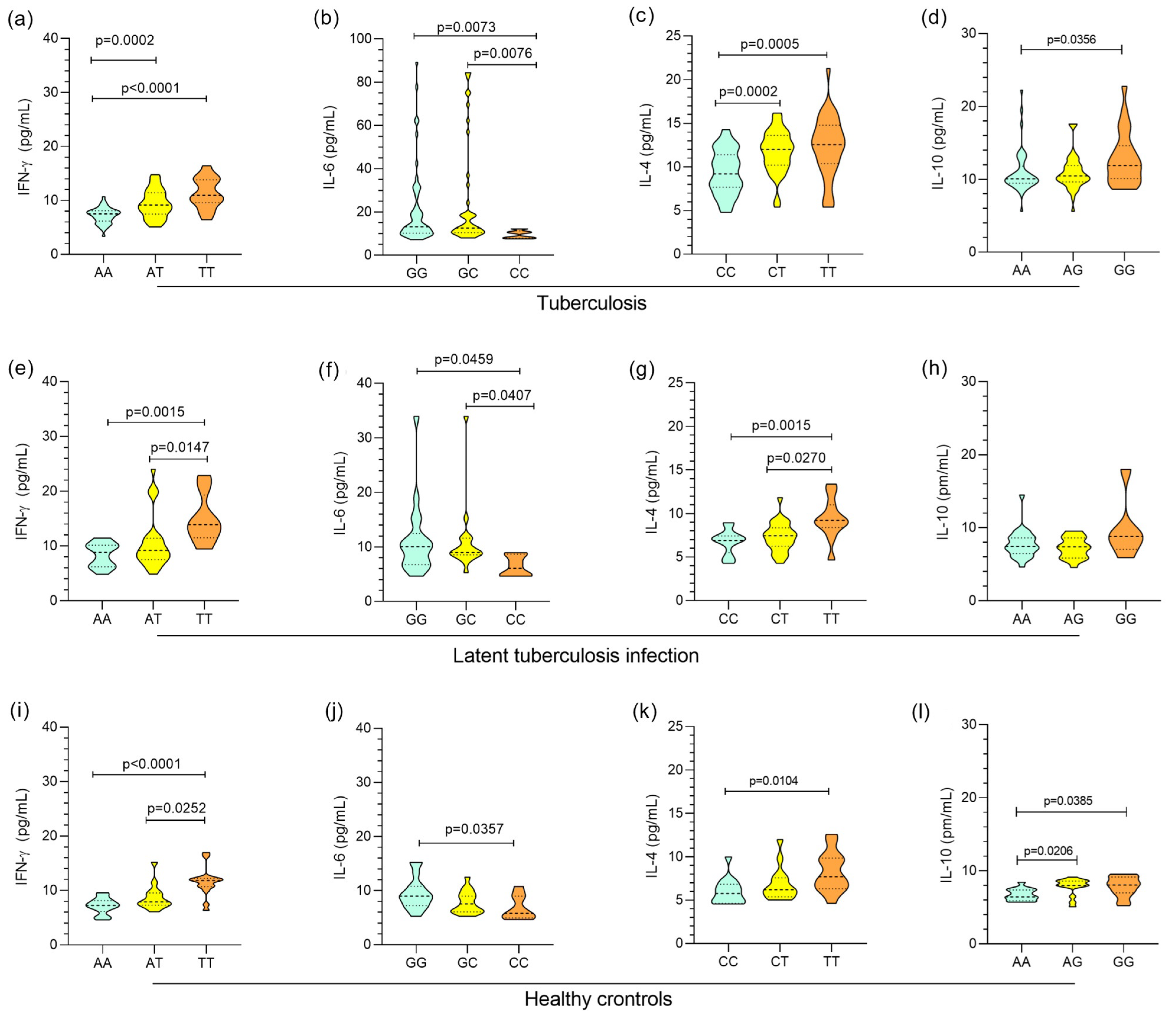
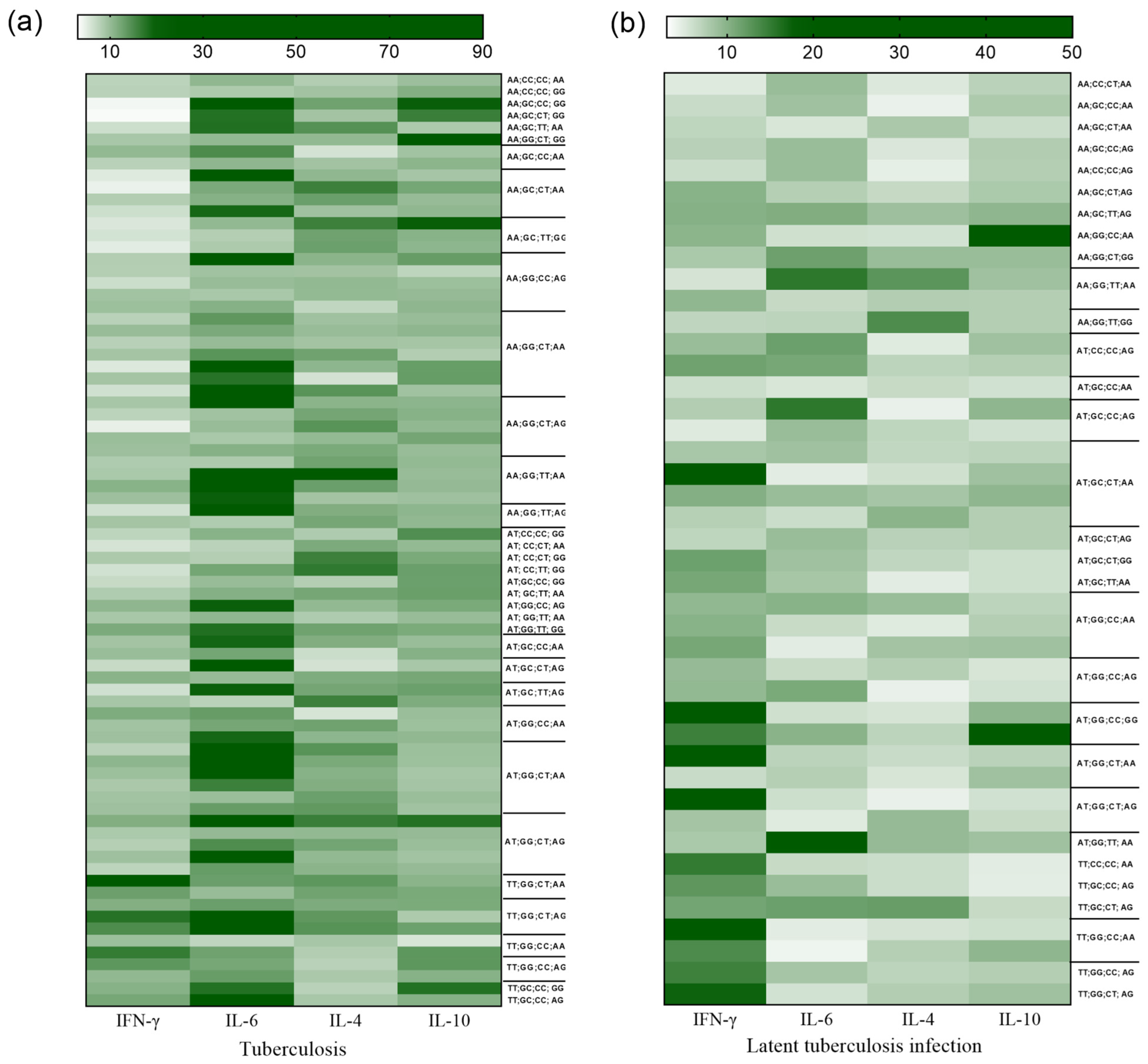
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, A.; Mathiasen, V.D.; Schön, T.; Wejse, C. The global prevalence of latent tuberculosis: A systematic review and meta-analysis. Eur. Respir. J. 2019, 54, 1900655. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Global Tuberculosis Report 2022. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022 (accessed on 2 March 2023).
- Schorey, J.S.; Schlesinger, L.S. Innate Immune Responses to Tuberculosis. Microbiol. Spectr. 2016, 4, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.E., 3rd; Boshoff, H.I.; Dartois, V.; Dick, T.; Ehrt, S.; Flynn, J.; Schnappinger, D.; Wilkinson, R.J.; Young, D. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 2009, 7, 845–855. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Latent Tuberculosis Infection: Updated and Consolidated Guidelines for Programmatic Management 2022. Available online: https://www.who.int/tb/publications/2018/latent-tuberculosis-infection/en/ (accessed on 21 December 2022).
- Getahun, H.; Matteelli, A.; Chaisson, R.E.; Raviglione, M. Latent Mycobacterium tuberculosis infection. N. Engl. J. Med. 2015, 372, 2127–2135. [Google Scholar] [CrossRef] [PubMed]
- Shea, K.M.; Kammerer, J.S.; Winston, C.A.; Navin, T.R.; Horsburgh, C.R., Jr. Estimated rate of reactivation of latent tuberculosis infection in the United States, overall and by population subgroup. Am. J. Epidemiol. 2014, 179, 216–225. [Google Scholar] [CrossRef]
- Flynn, J.L. Immunology of tuberculosis and implications in vaccine development. Tuberculosis. 2004, 84, 93–101. [Google Scholar] [CrossRef]
- Walzl, G.; Ronacher, K.; Hanekom, W.; Scriba, T.J.; Zumla, A. Immunological biomarkers of tuberculosis. Nat. Rev. Immunol. 2011, 11, 343–354. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, Q.; Shi, J. Immune checkpoint modulating T cells and NK cells response to Mycobacterium tuberculosis infection. Microbiol. Res. 2023, 273, 127393. [Google Scholar] [CrossRef]
- Amelio, P.; Portevin, D.K.; Reither, F.; Mhimbira, M.; Mpina, A.; Tumbo, B.; Nickel, H.; Marti, S.; Knopp, S.; Ding, A.; et al. Babu. Mixed Th1 and Th2 Mycobacterium tuberculosis-specific CD4 T cell responses in patients with active pulmonary tubercu-losis from Tanzania. PLoS Negl. Trop. Dis. 2017, 11, e0005817. [Google Scholar] [CrossRef]
- Alvarez, A.H. Revisiting tuberculosis screening: An insight to complementary diagnosis and prospective molecular approaches for the recognition of the dormant TB infection in human and cattle hosts. Microbiol. Res. 2021, 252, 126853. [Google Scholar] [CrossRef]
- Pravica, V.; Perrey, C.; Stevens, A.; Lee, J.H.; Hutchinson, I.V. A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: Absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum. Immunol. 2000, 61, 863–866. [Google Scholar] [CrossRef] [PubMed]
- Ankley, L.; Thomas, S.; Olive, A.J. Fighting persistence: How chronic infections with Mycobacterium tuberculosis evade T cell-mediated clearance and new strategies to defeat them. Infect. Immun. 2020, 88, e00916-19. [Google Scholar] [CrossRef] [PubMed]
- Boni, F.G.; Hamdi, I.; Koundi, L.M.; Shrestha, K.; Xie, J. Cytokine storm in tuberculosis and IL-6 involvement. Infect. Genet. Evol. 2022, 97, 105166. [Google Scholar] [CrossRef]
- Mao, X.; Ke, Z.; Liu, S.; Tang, B.; Wang, J.; Huang, H.; Chen, S. IL-1β+3953C/T, -511T/C and IL-6 -174C/G polymorphisms in association with tuberculosis susceptibility: A meta-analysis. Gene 2015, 573, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Pooran, A.; Davids, M.; Nel, A.; Shoko, A.; Blackburn, J.; Dheda, K. IL-4 subverts mycobacterial containment in Mycobacterium tuberculosis-infected human macrophages. Eur. Respir. J. 2019, 54, 1802242. [Google Scholar] [CrossRef]
- Moreira-Teixeira, L.; Redford, P.S.; Stavropoulos, E.; Ghilardi, N.; Maynard, C.L.; Weaver, C.T.; Freitas do Rosário, A.P.; Wu, X.; Langhorne, J.; O’Garra, A. T Cell-Derived IL-10 Impairs Host Resistance to Mycobacterium tuberculosis Infection. J. Immunol. 2017, 199, 613–623. [Google Scholar] [CrossRef]
- Singh, A.K.; Yadav, A.B.; Garg, R.; Misra, A. Single nucleotide polymorphic macrophage cytokine regulation by Mycobacterium tuberculosis and drug treatment. Pharmacogenomics 2014, 15, 497–508. [Google Scholar] [CrossRef]
- He, S.; Yang, S.; Zhao, Q.; Wang, L.; Liu, H.; Sheng, Y.; Yuan, D.; Jin, T. Association of IL4, IL6, and IL10 polymorphisms with pulmonary tuberculosis in a Tibetan Chinese population. Oncotarget 2018, 9, 16418–16426. [Google Scholar] [CrossRef][Green Version]
- Hoffmann, S.C.; Stanley, E.M.; Cox, E.D.; DiMercurio, B.S.; Koziol, D.E.; Harlan, D.M.; Kirk, A.D.; Blair, P.J. Ethnicity greatly influences cytokine gene polymorphism distribution. Am. J. Transplant. 2002, 2, 560–567. [Google Scholar] [CrossRef]
- van Tong, H.; Velavan, T.P.; Thye, T.; Meyer, C.G. Human genetic factors in tuberculosis: An update. Trop. Med. Int. Health 2017, 22, 1063–1071. [Google Scholar] [CrossRef]
- Tso, H.W.; Ip, W.K.; Chong, W.P.; Tam, C.M.; Chiang, A.K.; Lau, Y.L. Association of interferon gamma and interleukin 10 genes with tuberculosis in Hong Kong Chinese. Genes. Immun. 2005, 6, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Ministério Da Saúde (Brasil). Boletim Epidemiológico Tuberculose 2020. Available online: http://www.aids.gov.br/pt-br/pub/2020/boletim-epidemiologico-de-turbeculose-2020 (accessed on 15 July 2023).
- Domingo-Gonzalez, R.; Prince, O.; Cooper, A.; Khader, S.A. Cytokines and Chemokines in Mycobacterium tuberculosis Infection. Microbiol. Spectr. 2016, 4, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.V. The immunogenetics of human infectious diseases. Annu. Rev. Immunol. 1998, 16, 593–617. [Google Scholar] [CrossRef]
- Rossouw, M.; Nel, H.J.; Cooke, G.S.; van Helden, P.D.; Hoal, E.G. Association between tuberculosis and a polymorphic NFkappaB binding site in the interferon gamma gene. Lancet 2003, 361, 1871–1872. [Google Scholar] [CrossRef] [PubMed]
- Vallinoto, A.C.; Graça, E.S.; Araújo, M.S.; Azevedo, V.N.; Cayres-Vallinoto, I.; Machado, L.F.; Ishak, M.O.; Ishak, R. IFNG +874T/A polymorphism and cytokine plasma levels are associated with susceptibility to Mycobacterium tuberculosis infection and clinical manifestation of tuberculosis. Hum. Immunol. 2010, 71, 692–696. [Google Scholar] [CrossRef]
- Wei, Z.; Wenhao, S.; Yuanyuan, M.; Yang, L.; Daming, Z.; Jiangchun, X.; Jijun, J. A single nucleotide polymorphism in the interferon-γ gene (IFNG +874 T/A) is associated with susceptibility to tuberculosis. Oncotarget 2017, 8, 50415–50429. [Google Scholar] [CrossRef][Green Version]
- Ministério Da Saúde (Brasil). Manual de Recomendações para o Controle da Tuberculose no Brasil. 2nd ed. Brasília, DF: Ministério da Saúde 2019. Available online: http://bvsms.saude.gov.br/bvs/publicacoes/manual_recomendacoes_controle_tuberculose_brasil_2_ed.pdf (accessed on 15 September 2023).
- Ministério Da Saúde (Brasil). Relatório de Recomendação. Teste de Liberação Interferon-Gama (Interferon Gamma Release Assay—IGRA) Para Detecção da Tuberculose em Pacientes Imunocomprometidos 2020. Available online: https://www.gov.br/conitec/pt-br/midias/consultas/relatorios/2020/20201113_relatorio_de_recomendacao_573_igra.pdf (accessed on 9 October 2023).
- Flynn, J.L.; Chan, J.; Triebold, K.J.; Dalton, D.K.; Stewart, T.A.; Bloom, B.R. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 1993, 178, 2249–2254. [Google Scholar] [CrossRef] [PubMed]
- Schnettger, L.; Rodgers, A.; Repnik, U.; Lai, R.P.; Pei, G.; Verdoes, M.; Wilkinson, R.J.; Young, D.B.; Gutierrez, M.G. A Rab20-Dependent Membrane Trafficking Pathway Controls M. tuberculosis Replication by Regulating Phagosome Spaciousness and Integrity. Cell Host Microbe 2017, 21, 619–628.e5. [Google Scholar] [CrossRef]
- Souza, I.d.P.; Amoras, E.d.S.G.; Sousa, F.D.M.d.; Sousa, P.V.N.R.d.; Lima, S.S.; Cayres-Vallinoto, I.M.V.; Ishak, R.; Vallinoto, A.C.R.; Queiroz, M.A.F. Plasma Levels of sFas-sFasL and FASL Gene Expression Are Associated with Tuberculosis. Biomolecules 2023, 13, 80. [Google Scholar] [CrossRef]
- Queiroz, M.A.F.; Lima, S.S.; Amoras, E.D.S.G.; Sousa, F.D.M.D.; Souza, I.D.P.; Nunes, J.A.L.; Brasil-Costa, I.; Cayres-Vallinoto, I.M.V.; Ishak, R.; Vallinoto, A.C.R. Epidemiological and Cytokine Profile of Patients with Pulmonary and Extrapulmonary Tuberculosis in a Population of the Brazilian Amazon. Microorganisms 2022, 10, 2075. [Google Scholar] [CrossRef]
- Wang, H.; Pang, C.; Zeng, N.; Wan, C.; Shen, Y.; Wen, F. Association between the IL-6 gene polymorphism and tuberculosis risk: A meta-analysis. Infect. Drug Resist. 2017, 10, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Milano, M.; Moraes, M.O.; Rodenbusch, R.; Carvalho, C.X.; Delcroix, M.; Mousquer, G.; Laux da Costa, L.; Unis, G.; Dalla Costa, E.R.; Rossetti, M.L. Single Nucleotide Polymorphisms in IL17A and IL6 Are Associated with Decreased Risk for Pulmonary Tuberculosis in Southern Brazilian Population. PLoS ONE 2016, 11, e0147814. [Google Scholar] [CrossRef] [PubMed]
- Joshi, L.; Chelluri, L.K.; Valluri, V.; Gaddam, S. Association of TNF-α, IL-10 and IL-6 promoter polymorphisms in pulmonary tuberculosis patients and their household contacts of younger age group. Comp. Immunol. Microbiol. Infect. Dis. 2018, 56, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Beltrán, S.; Carreto-Binaghi, L.E.; Carranza, C.; Torres, M.; Gonzalez, Y.; Muñoz-Torrico, M.; Juárez, E. Oxidative Stress and Inflammatory Mediators in Exhaled Breath Condensate of Patients with Pulmonary Tuberculosis. A Pilot Study with a Biomarker Perspective. Antioxidants 2021, 10, 1572. [Google Scholar] [CrossRef]
- Tang, S.; Cui, H.; Yao, L.; Hao, X.; Shen, Y.; Fan, L.; Sun, H.; Zhang, Z.; Huang, J.A. Increased Cytokines Response in Patients with Tuberculosis Complicated with Chronic Obstructive Pulmonary Disease. PLoS ONE 2013, 8, e62385. [Google Scholar] [CrossRef]
- Liu, Q.; Li, W.; Chen, Y. Association of IL-4 rs2243250 polymorphism with susceptibility to tuberculosis: A meta-analysis involving 6794 subjects. Microb. Pathog. 2021, 158, 104959. [Google Scholar] [CrossRef]
- Romero-Adrian, T.B.; Leal-Montiel, J.; Fernández, G.; Valecillo, A. Role of cytokines and other factors involved in the Mycobacterium tuberculosis infection. World J. Immunol. 2015, 5, 16–50. [Google Scholar] [CrossRef]
- Liang, B.; Guo, Y.; Li, Y.; Kong, H. Association between IL-10 gene polymorphisms and susceptibility of tuberculosis: Evidence based on a meta-analysis. PLoS ONE 2014, 9, e88448. [Google Scholar] [CrossRef]
- Redford, P.S.; Murray, P.J.; O’Garra, A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 2011, 4, 261–270. [Google Scholar] [CrossRef]
- O’Leary, S.; O’Sullivan, M.P.; Keane, J. IL-10 blocks phagosome maturation in mycobacterium tuberculosis-infected human macrophages. Am. J. Respir. Cell Mol. Biol. 2011, 45, 172–180. [Google Scholar] [CrossRef]
- Liu, Q.; Li, W.; Li, D.; Feng, Y.; Tao, C. The association of interleukin-10-1082, -819, -592 polymorphisms and tuberculosis risk. Saudi Med. J. 2015, 36, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wu, L.; Li, D.; Zhao, Q.; Jiang, W.; Xu, B. Association between cytokine gene polymorphisms and tuberculosis in a Chinese population in Shanghai: A case-control study. BMC Immunol. 2015, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Grigsby, S.J.; Philips, J.A. Immune evasion and provocation by Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2022, 20, 750–766. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, M.A.F.; Azevedo, V.N.; Amoras, E.D.S.G.; Moura, T.C.F.; Guimarães Ishak, M.O.; Ishak, R.; Vallinoto, A.C.R.; Martins Feitosa, R.N. IFNG +874A/T Polymorphism among Asymptomatic HTLV-1-Infected Individuals Is Potentially Related to a Worse Prognosis. Front. Microbiol. 2018, 9, 795. [Google Scholar] [CrossRef] [PubMed]
- Nagabhushanam, V.; Solache, A.; Ting, L.M.; Escaron, C.J.; Zhang, J.Y.; Ernst, J.D. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-γ. J. Immunol. 2003, 171, 4750–4757. [Google Scholar] [CrossRef]
- Martinez, A.N.; Mehra, S.; Kaushal, D. Role of interleukin 6 in innate immunity to Mycobacterium tuberculosis infection. J. Infect. Dis. 2013, 207, 1253–1261. [Google Scholar] [CrossRef]
- Rao, M.; Ippolito, G.; Mfinanga, S.; Ntoumi, F.; Yeboah-Manu, D.; Vilaplana, C.; Zumla, A.; Maeurer, M. Latent TB Infection (LTBI)-Mycobacterium tuberculosis pathogenesis and the dynamics of the granuloma battleground. Int. J. Infect. Dis. 2019, 80, S58–S61. [Google Scholar] [CrossRef]
- Martinot, A.J. Microbial Offense vs Host Defense: Who Controls the TB Granuloma? Vet. Pathol. 2018, 55, 14–26. [Google Scholar] [CrossRef]
- Gibson, S.E.R.; Harrison, J.; Cox, J.A.G. Modelling a Silent Epidemic: A Review of the In Vitro Models of Latent Tuberculosis. Pathogens 2018, 7, 88. [Google Scholar] [CrossRef]
- Natarajan, S.; Ranganathan, M.; Hanna, L.E.; Tripathy, S. Transcriptional Profiling and Deriving a Seven-Gene Signature That Discriminates Active and Latent Tuberculosis: An Integrative Bioinformatics Approach. Genes 2022, 13, 616. [Google Scholar] [CrossRef]
- Chin, K.L.; Anibarro, L.; Sarmiento, M.E.; Acosta, A. Challenges and the Way forward in Diagnosis and Treatment of Tuberculosis Infection. Trop. Med. Infect. Dis. 2023, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Carrère-Kremer, S.; Kolia-Diafouka, P.; Pisoni, A.; Bolloré, K.; Peries, M.; Godreuil, S.; Bourdin, A.; Van de Perre, P.; Tuaillon, E. QuantiFERON-TB Gold Plus Assay in Patients With Latent vs. Active Tuberculosis in a Low Incidence Setting: Level of IFN-γ, CD4/CD8 Responses, and Release of IL-2, IP-10, and MIG. Front. Microbiol. 2022, 13, 825021. [Google Scholar] [CrossRef] [PubMed]
- WHO. Latent Tuberculosis Infection: Updated and Consolidated Guidelines for Programmatic Management; World Health Organization: Geneva, Switzerland, 2018; Available online: https://apps.who.int/iris/handle/10665/260233 (accessed on 2 October 2023).

| TB n = 245 | LTBI n = 80 | HC n = 100 | |
|---|---|---|---|
| Age, mean (SD) | 41.2 (10.1) | 38.4 (9.4) | 35.7 (11.6) |
| Sex, n (%) Female Male | 100 (41.0) 145 (59.0) | 34 (42.5) 46 (57.5) | 42 (42.0) 58 (58.0) |
| Location, n (%) TB, pulmonary TB, extrapulmonary | 201 (82.0) 44 (18.0) | - | - |
| Alleles and Genotypes | TB | LTBI | HC | p | OR |
|---|---|---|---|---|---|
| n = 245 | n = 80 | n = 100 | (95% IC) | ||
| n (%) | n (%) | n (%) | |||
| IFNG +874A/T | |||||
| AA | 139 (56.7) † | 22 (27.5) | 33 (33.0) | 0.0027 a∆ | 3.6104 (1.36–9.60) a |
| p = 0.0168 | |||||
| AT | 92 (37.55) | 50 (62.5) † | 49 (49.0) | <0.0001 b∆ | 5.5156 (2.45–11.9) b |
| p < 0.0001 | |||||
| TT | 14 (5.71) | 8 (10.0) | 18 (18.0) † | 0.1436 c∆ | - |
| * A | 0.755 | 0.588 | 0.571 | <0.0001 a∆ | 2.1649 (1.48–3.15) a |
| p < 0.0001 | |||||
| * T | 0.245 | 0.412 | 0.429 | <0.0001 b∆ | 2.2790 (1.60–3.22) b |
| p < 0.0001 | |||||
| 0.8112 c∆ | - | ||||
| IL6 -174G/C | |||||
| GG | 178 (72.7) | 53 (66.3) | 54 (54.0) | 0.0557 a∞ | - |
| GC | 61 (24.9) | 21 (26.2) | 38 (38.0) | 0.0021 b∞ | 4.3955 (1.46–13.22) b |
| p = 0.0118 | |||||
| CC | 6 (2.4) | 6 (7.6) | 8 (8.0) | 0.2222 c∞ | - |
| * G | 0.851 | 0.794 | 0.73 | 0.9374 | - |
| * C | 0.149 | 0.206 | 0.27 | 0.0003 b∆ | 2.1128 (1.41–3.14) b |
| p < 0.0001 | |||||
| 0.1603 c∆ | - | ||||
| IL4 -560C/T | |||||
| CC | 62 (25.3) | 31 (38.8) † | 39 (39.0) † | 0.0286 a∆ | 2.8000 (1.26–6.22) a |
| p = 0.0165 | |||||
| CT | 127 (51.8) | 39 (48.7) | 48 (48.0) | 0.01655 b∆ | 2.7097 (1.31–5.60) b |
| p = 0.0099 | |||||
| TT | 56 (22.9) † | 10 (12.5) | 13 (13.0) | 0.9927 c∆ | - |
| * C | 0.512 | 0.631 | 0.63 | 0.0029 a∆ | 1.6300 (1.12–2.35) a |
| p = 0.0114 | |||||
| * T | 0.488 | 0.369 | 0.37 | 0.0013 b∆ | 1.6213 (1.58–2.27) b |
| p = 0.0062 | |||||
| 0.9805 c∆ | - | ||||
| IL10 -1082A/G | |||||
| AA | 148 (60.4) † | 38 (47.5) | 50 (50.0) | 0.0361 a∆ | - |
| AG | 72 (29.4) | 36 (45.0) † | 45 (45.0) † | 0.0132 b∆ | - |
| GG | 25 (10.2) | 6 (7.5) | 5 (5.0) | 0.7758 c∆ | - |
| * A | 0.746 | 0.703 | 0.738 | 0.3368 a∆ | - |
| * G | 0.254 | 0.297 | 0.262 | 0.8903 b∆ | - |
| 0.6020 c∆ | - |
| IFNG +874 | IL6 -174 | IL4 -560 | IL10 +1082 | TB | LTBI | HC | TB vs. LTBI OR (95% CI) | TB vs. HC OR (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | A | G | C | A | 163 | 53 | 64 | 0.998 (0.67–1.44) p = 0.9965 | 1.042 (0.73–1.48) p = 0.8900 |
| 2 | A | G | C | G | 27 | 6 | 7 | 1.473 (0.59–3.63) p = 0.5256 | 1.588 (0.68–3.70) p = 0.3764 |
| 3 | A | G | T | A | 104 | 14 | 15 | 2.753 (1.52–4.96) * p = 0.0008 | 3.273 (1.85–5.78) * p < 0.0001 |
| 4 | A | G | T | G | 35 | 8 | 10 | 1.348 (0.61–2.98) p = 0.5506 | 1.356 (0.65–2.81) p = 0.5166 |
| 5 | A | C | C | A | 14 | 6 | 7 | 0.742 (0.28–1.96) p = 0.7361 | 0.817 (0.31–2.01) p = 0.8213 |
| 6 | A | C | C | G | 7 | 2 | 3 | 1.126 (0.23–5.47) p = 0.8064 | 0.941 (0.24–3.67) p = 0.7916 |
| 7 | A | C | T | A | 14 | 2 | 8 | 2.285 (0.51–10.10) p = 0.4120 | 0.698 (0.28–1.69) p = 0.5742 |
| 8 | A | C | T | G | 24 | 2 | 2 | 4.00 (0.93–17.11) p = 0.0743 | 5.038 (1.17–21.52) * p = 0.0280 |
| 9 | T | G | C | A | 21 | 16 | 12 | 0.396 (0.20–0.77) * p = 0.0103 | 0.693 (0.33–1.43) p = 0.4282 |
| 10 | T | G | C | G | 10 | 9 | 7 | 0.344 (0.14–0.86) * p = 0.0352 | 0.568 (0.21–1.51) p = 0.3820 |
| 11 | T | G | T | A | 36 | 9 | 17 | 1.307 (0.62–2.77) p = 0.6036 | 0.843 (0.46–1.54) p = 0.6909 |
| 12 | T | G | T | G | 14 | 12 | 11 | 0.357 (0.16–0.78) * p = 0.0157 | 0.499 (0.22–1.12) p = 0.1362 |
| 13 | T | C | C | A | 5 | 4 | 8 | 0.396 (0.11–1.49) p = 0.3048 | 0.245 (0.08–0.75) * p = 0.0200 |
| 14 | T | C | C | G | 6 | 5 | 8 | 0.378 (0.11–1.26) p = 0.1958 | 0.294 (0.10–0.85) * p = 0.0382 |
| 15 | T | C | T | A | 6 | 7 | 10 | 0.266 (0.09–0.80) * p = 0.0292 | 0.233 (0.08–0.65) * p = 0.0062 |
| 16 | T | C | T | G | 12 | 5 | 12 | 0.815 (0.28–2.34) p = 0.9220 | 0.388 (0.17–0.88) * p = 0.0348 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva Graça Amoras, E.; de Morais, T.G.; do Nascimento Ferreira, R.; Gomes, S.T.M.; de Sousa, F.D.M.; de Paula Souza, I.; Ishak, R.; Vallinoto, A.C.R.; Queiroz, M.A.F. Association of Cytokine Gene Polymorphisms and Their Impact on Active and Latent Tuberculosis in Brazil’s Amazon Region. Biomolecules 2023, 13, 1541. https://doi.org/10.3390/biom13101541
da Silva Graça Amoras E, de Morais TG, do Nascimento Ferreira R, Gomes STM, de Sousa FDM, de Paula Souza I, Ishak R, Vallinoto ACR, Queiroz MAF. Association of Cytokine Gene Polymorphisms and Their Impact on Active and Latent Tuberculosis in Brazil’s Amazon Region. Biomolecules. 2023; 13(10):1541. https://doi.org/10.3390/biom13101541
Chicago/Turabian Styleda Silva Graça Amoras, Ednelza, Thais Gouvea de Morais, Rafaella do Nascimento Ferreira, Samara Tatielle Monteiro Gomes, Francisca Dayse Martins de Sousa, Iury de Paula Souza, Ricardo Ishak, Antonio Carlos Rosário Vallinoto, and Maria Alice Freitas Queiroz. 2023. "Association of Cytokine Gene Polymorphisms and Their Impact on Active and Latent Tuberculosis in Brazil’s Amazon Region" Biomolecules 13, no. 10: 1541. https://doi.org/10.3390/biom13101541
APA Styleda Silva Graça Amoras, E., de Morais, T. G., do Nascimento Ferreira, R., Gomes, S. T. M., de Sousa, F. D. M., de Paula Souza, I., Ishak, R., Vallinoto, A. C. R., & Queiroz, M. A. F. (2023). Association of Cytokine Gene Polymorphisms and Their Impact on Active and Latent Tuberculosis in Brazil’s Amazon Region. Biomolecules, 13(10), 1541. https://doi.org/10.3390/biom13101541










