Newborns with Favourable Outcomes after Perinatal Asphyxia Have Upregulated Glucose Metabolism-Related Proteins in Plasma
Abstract
1. Introduction
2. Materials and Methods
2.1. Recruitment
2.2. Plasma Sample Preparation and TMT10plex™ Labelling
2.3. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
2.4. Proteomics Data Analysis
2.5. Western Blot Analysis
2.6. Statistical Analysis
3. Results
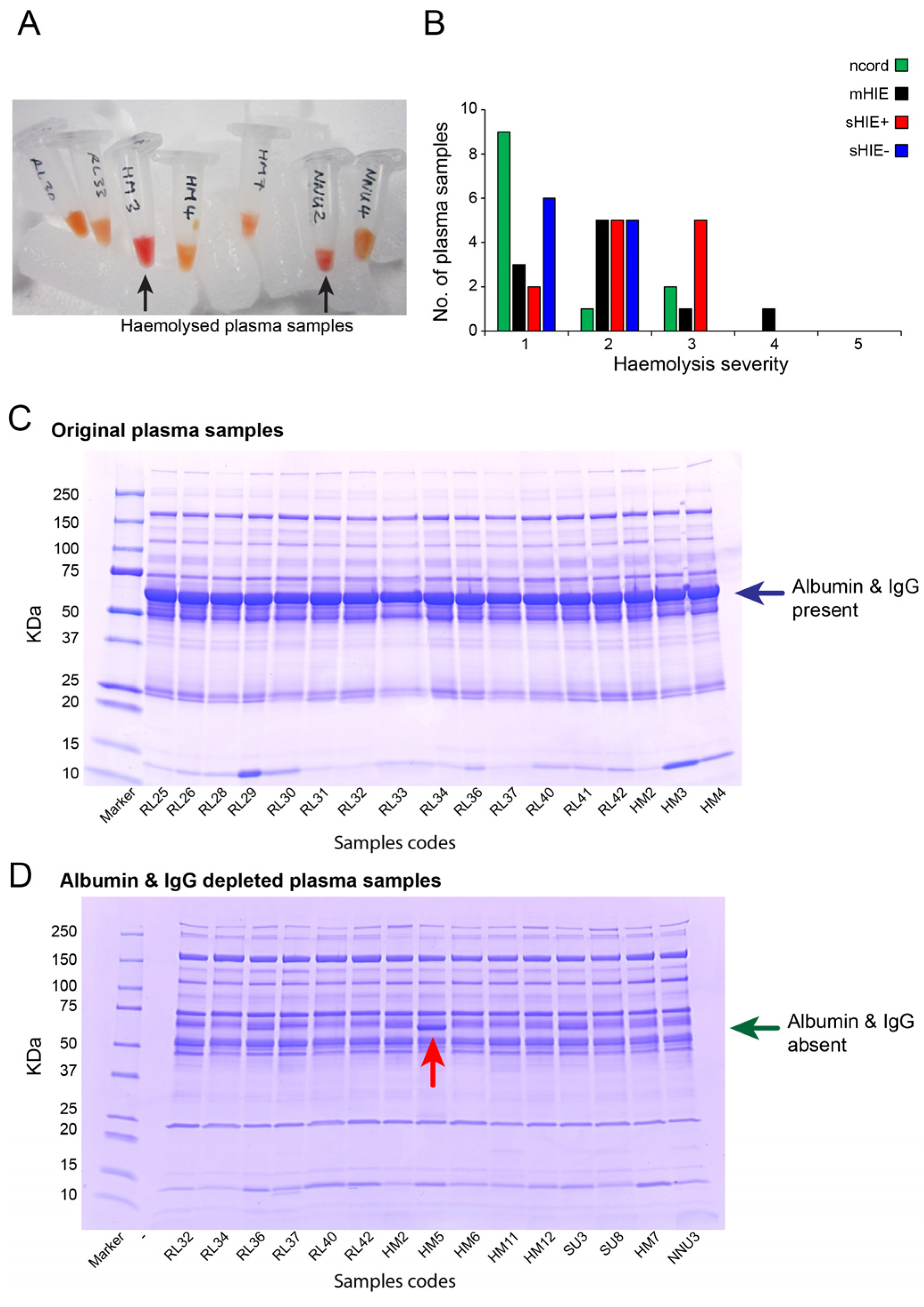
3.1. Plasma Sample Quality
3.2. Demographic Characteristics
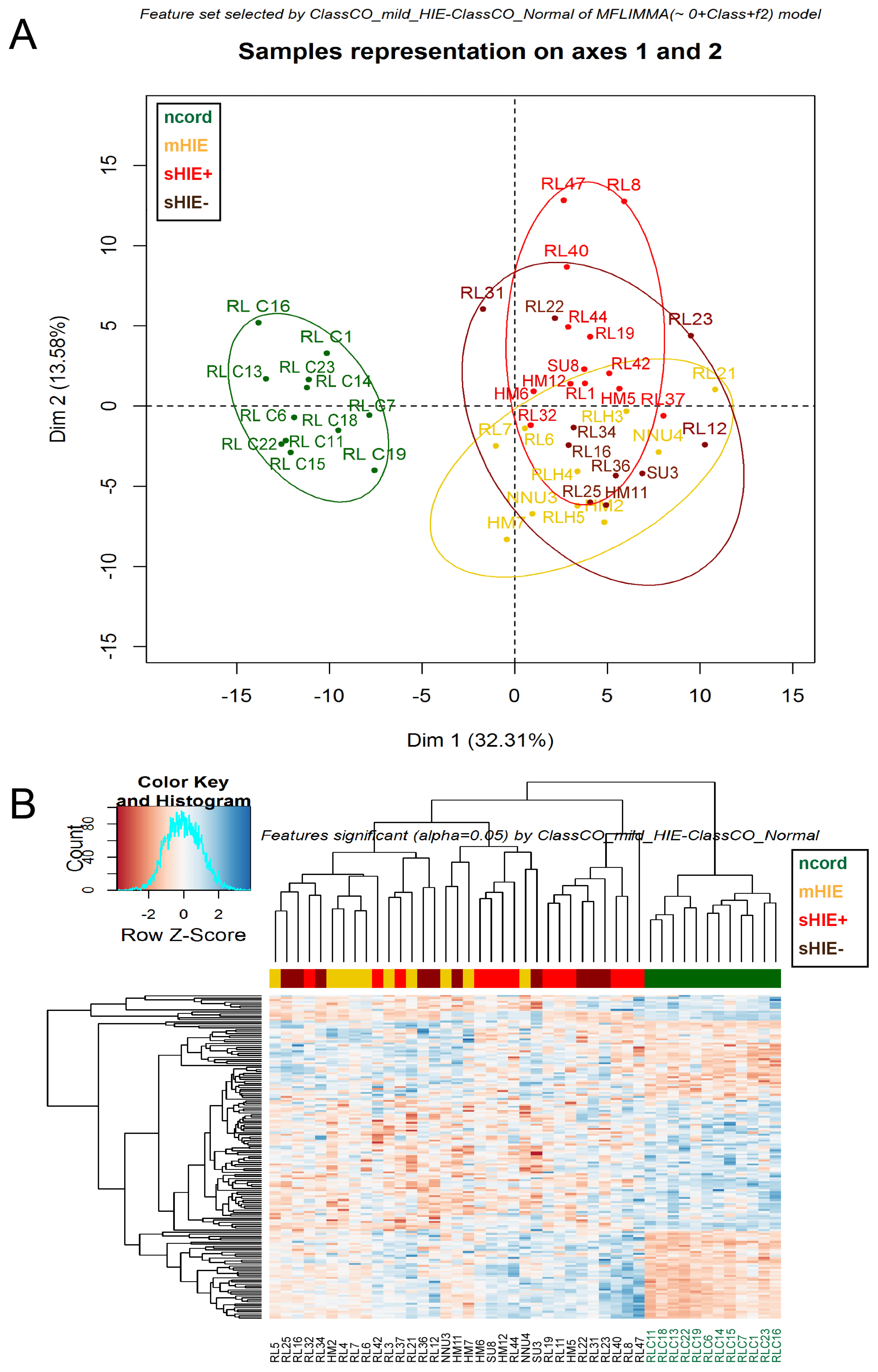
3.3. Cord Blood from Normal Pregnancies Exhibits a Different Protein Profile to the Blood of HIE Newborns
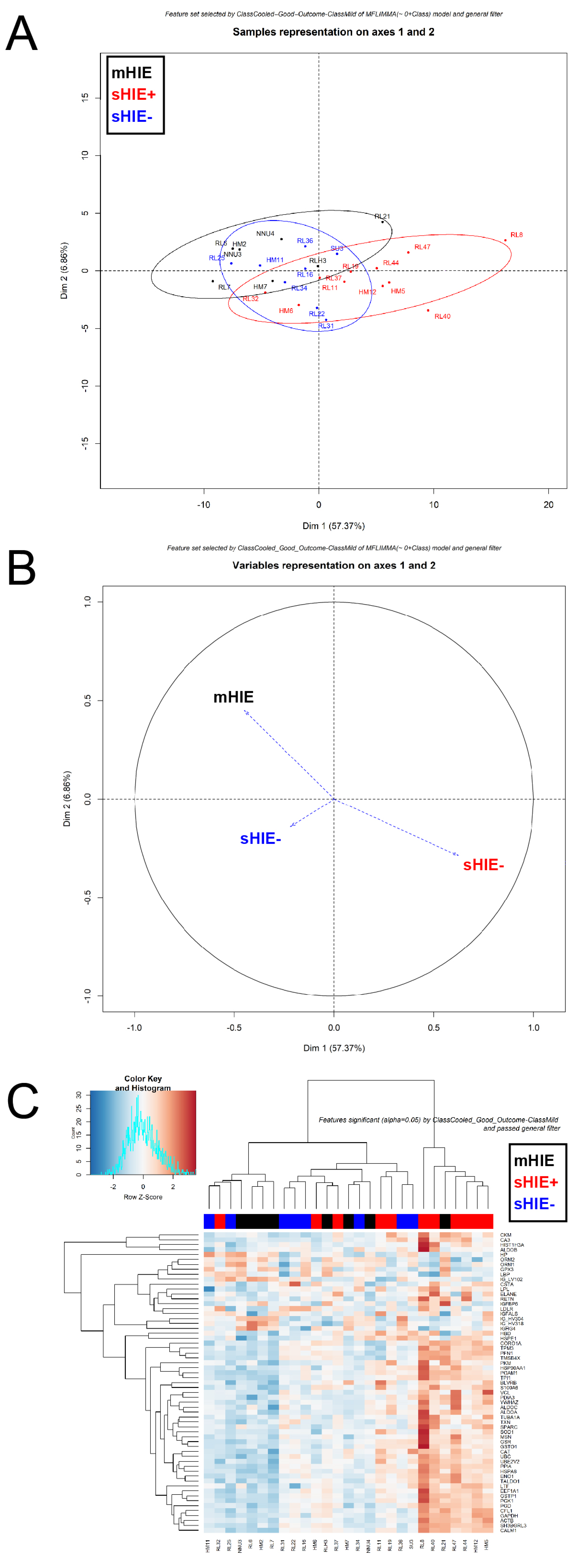
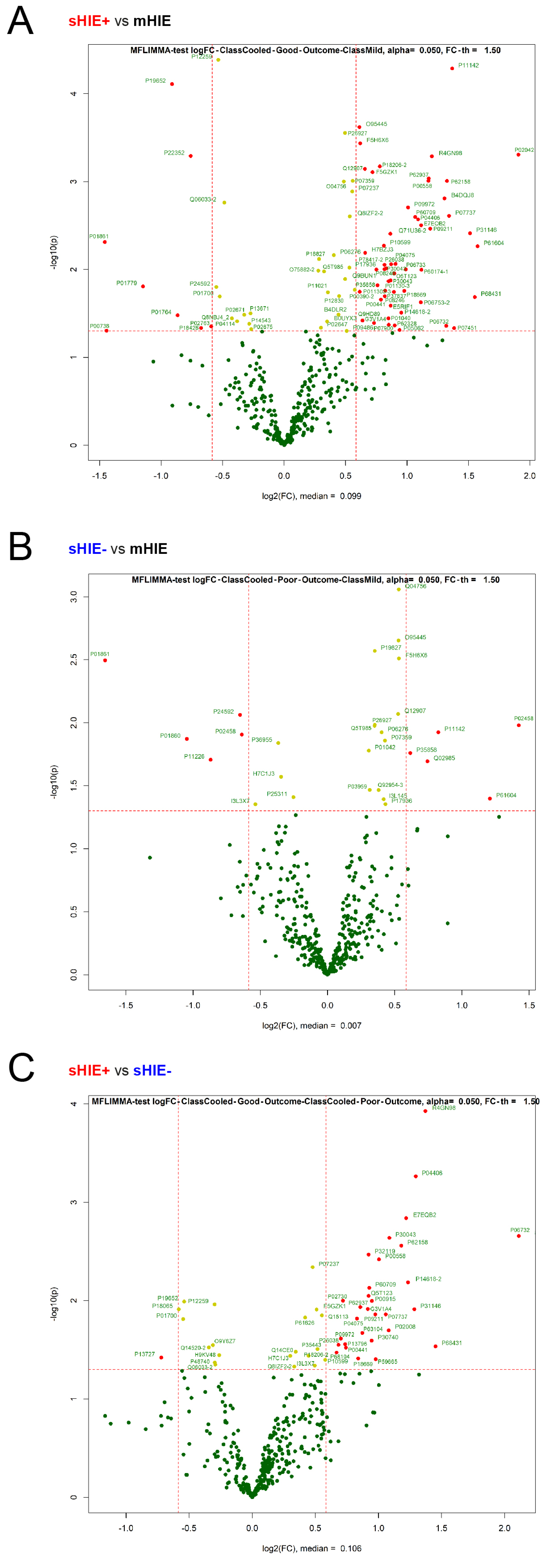
3.4. mHIE Plasma Exhibit a Different Protein Profile to Plasma from sHIE+ and sHIE− Groups
3.5. Comparison between sHIE+ and sHIE− Groups
3.6. Validation of Selected Protein Targets with Western Blotting
3.7. Biological Processes and Pathways
4. Discussion
4.1. Proteomic Study in HIE Newborns
4.2. Glucose Metabolism
4.3. Unsuitability of Cord Blood from Normal Pregnancies as Control for This Study
4.4. Certain Protein Biomarkers Were Not Detected in HIE Plasma
4.5. Suitability of Certain Endogenous Control Proteins
5. Limitations and Strengths
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Badawi, N.; Kurinczuk, J.J.; Keogh, J.M.; Alessandri, L.M.; O’Sullivan, F.; Burton, P.R.; Pemberton, P.J.; Stanley, F.J. Intrapartum risk factors for newborn encephalopathy: The Western Australian case-control study. BMJ 1998, 317, 1554–1558. [Google Scholar] [CrossRef]
- Edwards, A.D.; Brocklehurst, P.; Gunn, A.J.; Halliday, H.; Juszczak, E.; Levene, M.; Strohm, B.; Thoresen, M.; Whitelaw, A.; Azzopardi, D. Neurological outcomes at 18 months of age after moderate hypothermia for perinatal hypoxic ischaemic encephalopathy: Synthesis and meta-analysis of trial data. BMJ 2010, 340, c363. [Google Scholar] [CrossRef]
- Chiesa, C.; Pellegrini, G.; Panero, A.; De Luca, T.; Assumma, M.; Signore, F.; Pacifico, L. Umbilical cord interleukin-6 levels are elevated in term neonates with perinatal asphyxia. Eur. J. Clin. Investig. 2003, 33, 352–358. [Google Scholar] [CrossRef]
- Gazzolo, D.; Frigiola, A.; Bashir, M.; Iskander, I.; Mufeed, H.; Aboulgar, H.; Venturini, P.; Marras, M.; Serra, G.; Frulio, R.; et al. Diagnostic accuracy of S100B urinary testing at birth in full-term asphyxiated newborns to predict neonatal death. PLoS ONE 2009, 4, e4298. [Google Scholar] [CrossRef]
- Lv, H.; Wang, Q.; Wu, S.; Yang, L.; Ren, P.; Yang, Y.; Gao, J.; Li, L. Neonatal hypoxic ischemic encephalopathy-related biomarkers in serum and cerebrospinal fluid. Clin. Chim. Acta 2015, 450, 282–297. [Google Scholar] [CrossRef]
- O’Sullivan, M.P.; Looney, A.M.; Moloney, G.M.; Finder, M.; Hallberg, B.; Clarke, G.; Boylan, G.B.; Murray, D.M. Validation of Altered Umbilical Cord Blood MicroRNA Expression in Neonatal Hypoxic-Ischemic Encephalopathy. JAMA Neurol. 2019, 76, 333–341. [Google Scholar] [CrossRef]
- Celtik, C.; Acunas, B.; Oner, N.; Pala, O. Neuron-specific enolase as a marker of the severity and outcome of hypoxic ischemic encephalopathy. Brain Dev. 2004, 26, 398–402. [Google Scholar] [CrossRef]
- Chalak, L.F.; Sanchez, P.J.; Adams-Huet, B.; Laptook, A.R.; Heyne, R.J.; Rosenfeld, C.R. Biomarkers for severity of neonatal hypoxic-ischemic encephalopathy and outcomes in newborns receiving hypothermia therapy. J. Pediatr. 2014, 164, 468–474.e461. [Google Scholar] [CrossRef]
- Qian, J.; Zhou, D.; Wang, Y.W. Umbilical artery blood S100beta protein: A tool for the early identification of neonatal hypoxic-ischemic encephalopathy. Eur. J. Pediatr. 2009, 168, 71–77. [Google Scholar] [CrossRef]
- Ceccon, M.E. Interleukins in hypoxic-ischemic encephalopathy. J. Pediatr. 2003, 79, 280–281. [Google Scholar] [CrossRef]
- Massaro, A.N.; Wu, Y.W.; Bammler, T.K.; Comstock, B.; Mathur, A.; McKinstry, R.C.; Chang, T.; Mayock, D.E.; Mulkey, S.B.; Van Meurs, K.; et al. Plasma Biomarkers of Brain Injury in Neonatal Hypoxic-Ischemic Encephalopathy. J. Pediatr. 2018, 194, 67–75.e61. [Google Scholar] [CrossRef] [PubMed]
- Douglas-Escobar, M.V.; Heaton, S.C.; Bennett, J.; Young, L.J.; Glushakova, O.; Xu, X.; Barbeau, D.Y.; Rossignol, C.; Miller, C.; Old Crow, A.M.; et al. UCH-L1 and GFAP Serum Levels in Neonates with Hypoxic-Ischemic Encephalopathy: A Single Center Pilot Study. Front. Neurol. 2014, 5, 273. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.K.; Ponnusamy, V.; Evanson, J.; Kapellou, O.; Ekitzidou, G.; Gupta, N.; Clarke, P.; Michael-Titus, A.T.; Yip, P.K. Raised Plasma Neurofilament Light Protein Levels Are Associated with Abnormal MRI Outcomes in Newborns Undergoing Therapeutic Hypothermia. Front. Neurol. 2018, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.K.; Yip, P.K.; Barlas, A.; Tharmapoopathy, P.; Ponnusamy, V.; Michael-Titus, A.T.; Chisholm, P. Raised Plasma Neurofilament Light Protein Levels After Rewarming Are Associated with Adverse Neurodevelopmental Outcomes in Newborns After Therapeutic Hypothermia. Front. Neurol. 2020, 11, 562510. [Google Scholar] [CrossRef]
- Tsai, T.T.; Huang, T.H.; Ho, N.Y.; Chen, Y.P.; Chen, C.A.; Chen, C.F. Development of a multiplex and sensitive lateral flow immunoassay for the diagnosis of periprosthetic joint infection. Sci. Rep. 2019, 9, 15679. [Google Scholar] [CrossRef] [PubMed]
- Pappireddi, N.; Martin, L.; Wuhr, M. A Review on Quantitative Multiplexed Proteomics. ChemBioChem 2019, 20, 1210–1224. [Google Scholar] [CrossRef] [PubMed]
- Bachor, R.; Waliczek, M.; Stefanowicz, P.; Szewczuk, Z. Trends in the Design of New Isobaric Labeling Reagents for Quantitative Proteomics. Molecules 2019, 24, 701. [Google Scholar] [CrossRef]
- Azzopardi, D.; Brocklehurst, P.; Edwards, D.; Halliday, H.; Levene, M.; Thoresen, M.; Whitelaw, A.; Group, T.S. The TOBY Study. Whole body hypothermia for the treatment of perinatal asphyxial encephalopathy: A randomised controlled trial. BMC Pediatr. 2008, 8, 17. [Google Scholar] [CrossRef]
- Tharmapoopathy, P.; Chisholm, P.; Barlas, A.; Varsami, M.; Gupta, N.; Ekitzidou, G.; Ponnusamy, V.; Kappelou, O.; Evanson, J.; Rosser, G.; et al. In clinical practice, cerebral MRI in newborns is highly predictive of neurodevelopmental outcome after therapeutic hypothermia. Eur. J. Paediatr. Neurol. 2020, 25, 127–133. [Google Scholar] [CrossRef]
- Rutherford, M.; Ramenghi, L.A.; Edwards, A.D.; Brocklehurst, P.; Halliday, H.; Levene, M.; Strohm, B.; Thoresen, M.; Whitelaw, A.; Azzopardi, D. Assessment of brain tissue injury after moderate hypothermia in neonates with hypoxic-ischaemic encephalopathy: A nested substudy of a randomised controlled trial. Lancet Neurol. 2010, 9, 39–45. [Google Scholar] [CrossRef]
- Ramont, L.; Thoannes, H.; Volondat, A.; Chastang, F.; Millet, M.C.; Maquart, F.X. Effects of hemolysis and storage condition on neuron-specific enolase (NSE) in cerebrospinal fluid and serum: Implications in clinical practice. Clin. Chem. Lab. Med. 2005, 43, 1215–1217. [Google Scholar] [CrossRef] [PubMed]
- Russell, C.L.; Heslegrave, A.; Mitra, V.; Zetterberg, H.; Pocock, J.M.; Ward, M.A.; Pike, I. Combined tissue and fluid proteomics with Tandem Mass Tags to identify low-abundance protein biomarkers of disease in peripheral body fluid: An Alzheimer’s Disease case study. Rapid Commun. Mass. Spectrom. 2017, 31, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Husson, F.; Josse, J. Handling missing values in multiple factor analysis. Food Qual. Prefer. 2013, 30, 77–85. [Google Scholar] [CrossRef]
- Josse, J.; Husson, F. missMDA: A Package for Handling Missing Values in Multivariate Data Analysis. J. Stat. Softw. 2016, 70, 1–31. [Google Scholar] [CrossRef]
- Yip, P.K.; Bowes, A.L.; Hall, J.C.E.; Burguillos, M.A.; Ip, T.H.R.; Baskerville, T.; Liu, Z.H.; Mohamed, M.; Getachew, F.; Lindsay, A.D.; et al. Docosahexaenoic acid reduces microglia phagocytic activity via miR-124 and induces neuroprotection in rodent models of spinal cord contusion injury. Human Mol. Genet. 2019, 28, 2427–2448. [Google Scholar] [CrossRef]
- Yip, P.K.; Wong, L.F.; Sears, T.A.; Yanez-Munoz, R.J.; McMahon, S.B. Cortical overexpression of neuronal calcium sensor-1 induces functional plasticity in spinal cord following unilateral pyramidal tract injury in rat. PLoS Biol. 2010, 8, e1000399. [Google Scholar] [CrossRef]
- Collins, M.A.; An, J.; Peller, D.; Bowser, R. Total protein is an effective loading control for cerebrospinal fluid western blots. J. Neurosci. Methods 2015, 251, 72–82. [Google Scholar] [CrossRef]
- Zhu, Y.; Yun, Y.; Jin, M.; Li, G.; Li, H.; Miao, P.; Ding, X.; Feng, X.; Xu, L.; Sun, B. Identification of novel biomarkers for neonatal hypoxic-ischemic encephalopathy using iTRAQ. Ital. J. Pediatr. 2020, 46, 67. [Google Scholar] [CrossRef]
- Ehrchen, J.M.; Sunderkotter, C.; Foell, D.; Vogl, T.; Roth, J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J. Leukoc. Biol. 2009, 86, 557–566. [Google Scholar] [CrossRef]
- Li, Y.; Chen, B.; Yang, X.; Zhang, C.; Jiao, Y.; Li, P.; Liu, Y.; Li, Z.; Qiao, B.; Bond Lau, W.; et al. S100a8/a9 Signaling Causes Mitochondrial Dysfunction and Cardiomyocyte Death in Response to Ischemic/Reperfusion Injury. Circulation 2019, 140, 751–764. [Google Scholar] [CrossRef]
- Sun, P.; Li, Q.; Zhang, Q.; Xu, L.; Han, J.Y. Upregulated expression of S100A8 in mice brain after focal cerebral ischemia reperfusion. World J. Emerg. Med. 2013, 4, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Guellec, I.; Ancel, P.Y.; Beck, J.; Loron, G.; Chevallier, M.; Pierrat, V.; Kayem, G.; Vilotitch, A.; Baud, O.; Ego, A.; et al. Glycemia and Neonatal Encephalopathy: Outcomes in the LyTONEPAL (Long-Term Outcome of Neonatal Hypoxic EncePhALopathy in the Era of Neuroprotective Treatment With Hypothermia) Cohort. J. Pediatr. 2023, 257, 113350. [Google Scholar] [CrossRef] [PubMed]
- Kalogeropoulou, M.S.; Thomson, L.; Beardsall, K. Continuous glucose monitoring during therapeutic hypothermia for hypoxic ischaemic encephalopathy: A feasibility study. Arch. Dis. Childhood. Fetal Neonatal Ed. 2023, 108, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Pinchefsky, E.F.; Hahn, C.D.; Kamino, D.; Chau, V.; Brant, R.; Moore, A.M.; Tam, E.W.Y. Hyperglycemia and Glucose Variability Are Associated with Worse Brain Function and Seizures in Neonatal Encephalopathy: A Prospective Cohort Study. J. Pediatr. 2019, 209, 23–32. [Google Scholar] [CrossRef]
- Tam, E.W.Y.; Kamino, D.; Shatil, A.S.; Chau, V.; Moore, A.M.; Brant, R.; Widjaja, E. Hyperglycemia associated with acute brain injury in neonatal encephalopathy. Neuroimage Clin. 2021, 32, 102835. [Google Scholar] [CrossRef]
- Salhab, W.A.; Wyckoff, M.H.; Laptook, A.R.; Perlman, J.M. Initial hypoglycemia and neonatal brain injury in term infants with severe fetal acidemia. Pediatrics 2004, 114, 361–366. [Google Scholar] [CrossRef]
- Kauffman, F.C. The quantitative histochemistry of enzymes of the pentose phosphate pathway in the central nervous system of the rat. J. Neurochem. 1972, 19, 1–9. [Google Scholar] [CrossRef]
- Lu, Z.; Hunter, T. Metabolic Kinases Moonlighting as Protein Kinases. Trends Biochem. Sci. 2018, 43, 301–310. [Google Scholar] [CrossRef]
- Tristan, C.; Shahani, N.; Sedlak, T.W.; Sawa, A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell Signal. 2011, 23, 317–323. [Google Scholar] [CrossRef]
- Chen, D.; Wei, L.; Liu, Z.R.; Yang, J.J.; Gu, X.; Wei, Z.Z.; Liu, L.P.; Yu, S.P. Pyruvate Kinase M2 Increases Angiogenesis, Neurogenesis, and Functional Recovery Mediated by Upregulation of STAT3 and Focal Adhesion Kinase Activities After Ischemic Stroke in Adult Mice. Neurotherapeutics 2018, 15, 770–784. [Google Scholar] [CrossRef]
- Ennen, C.S.; Huisman, T.A.; Savage, W.J.; Northington, F.J.; Jennings, J.M.; Everett, A.D.; Graham, E.M. Glial fibrillary acidic protein as a biomarker for neonatal hypoxic-ischemic encephalopathy treated with whole-body cooling. Am. J. Obstet. Gynecol. 2011, 205, 251.e1–251.e7. [Google Scholar] [CrossRef] [PubMed]
- Imam, S.S.; Gad, G.I.; Atef, S.H.; Shawky, M.A. Cord blood brain derived neurotrophic factor: Diagnostic and prognostic marker in fullterm newborns with perinatal asphyxia. Pak. J. Biol. Sci. 2009, 12, 1498–1504. [Google Scholar] [CrossRef] [PubMed]
- Nagdyman, N.; Komen, W.; Ko, H.K.; Muller, C.; Obladen, M. Early biochemical indicators of hypoxic-ischemic encephalopathy after birth asphyxia. Pediatr. Res. 2001, 49, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Hasegawa, S.; Maeba, S.; Fukunaga, S.; Motoyama, M.; Hamano, H.; Ichiyama, T. Serum tau protein level serves as a predictive factor for neurological prognosis in neonatal asphyxia. Brain Dev. 2014, 36, 670–675. [Google Scholar] [CrossRef]
- Weiss, M.L.; Troyer, D.L. Stem cells in the umbilical cord. Stem Cell Rev. 2006, 2, 155–162. [Google Scholar] [CrossRef]
- Malhotra, A.; Novak, I.; Miller, S.L.; Jenkin, G. Autologous transplantation of umbilical cord blood-derived cells in extreme preterm infants: Protocol for a safety and feasibility study. BMJ Open 2020, 10, e036065. [Google Scholar] [CrossRef]
- Ponnusamy, V.; Ip, R.T.H.; Mohamed, M.; Clarke, P.; Wozniak, E.; Mein, C.; Schwendimann, L.; Barlas, A.; Chisholm, P.; Chakkarapani, E.; et al. Neuronal let-7b-5p acts through the Hippo-YAP pathway in neonatal encephalopathy. Commun. Biol. 2021, 4, 1143. [Google Scholar] [CrossRef]
- Douglas-Escobar, M.; Yang, C.; Bennett, J.; Shuster, J.; Theriaque, D.; Leibovici, A.; Kays, D.; Zheng, T.; Rossignol, C.; Shaw, G.; et al. A pilot study of novel biomarkers in neonates with hypoxic-ischemic encephalopathy. Pediatr. Res. 2010, 68, 531–536. [Google Scholar] [CrossRef]
- Looney, A.M.; Walsh, B.H.; Moloney, G.; Grenham, S.; Fagan, A.; O’Keeffe, G.W.; Clarke, G.; Cryan, J.F.; Dinan, T.G.; Boylan, G.B.; et al. Downregulation of Umbilical Cord Blood Levels of miR-374a in Neonatal Hypoxic Ischemic Encephalopathy. J. Pediatr. 2015, 167, 269–273.e2. [Google Scholar] [CrossRef]
- Van Anh, T.N.; Hao, T.K.; Chi, N.T.D.; Son, N.H. Predictions of Hypoxic-Ischemic Encephalopathy by Umbilical Cord Blood Lactate in Newborns with Birth Asphyxia. Open Access Maced. J. Med. Sci. 2019, 7, 3564–3567. [Google Scholar] [CrossRef]
- Walsh, B.H.; Broadhurst, D.I.; Mandal, R.; Wishart, D.S.; Boylan, G.B.; Kenny, L.C.; Murray, D.M. The metabolomic profile of umbilical cord blood in neonatal hypoxic ischaemic encephalopathy. PLoS ONE 2012, 7, e50520. [Google Scholar] [CrossRef]
- Zaigham, M.; Lundberg, F.; Olofsson, P. Protein S100B in umbilical cord blood as a potential biomarker of hypoxic-ischemic encephalopathy in asphyxiated newborns. Early Hum. Dev. 2017, 112, 48–53. [Google Scholar] [CrossRef]
- Buhimschi, I.A.; Buhimschi, C.S.; Weiner, C.P.; Kimura, T.; Hamar, B.D.; Sfakianaki, A.K.; Norwitz, E.R.; Funai, E.F.; Ratner, E. Proteomic but not enzyme-linked immunosorbent assay technology detects amniotic fluid monomeric calgranulins from their complexed calprotectin form. Clin. Diagn. Lab. Immunol. 2005, 12, 837–844. [Google Scholar] [CrossRef]
- Moritz, C.P. Tubulin or Not Tubulin: Heading Toward Total Protein Staining as Loading Control in Western Blots. Proteomics 2017, 17, 1600189. [Google Scholar] [CrossRef]
- Petrak, J.; Ivanek, R.; Toman, O.; Cmejla, R.; Cmejlova, J.; Vyoral, D.; Zivny, J.; Vulpe, C.D. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics 2008, 8, 1744–1749. [Google Scholar] [CrossRef]
- Nakano, T.; Goto, S.; Takaoka, Y.; Tseng, H.P.; Fujimura, T.; Kawamoto, S.; Ono, K.; Chen, C.L. A novel moonlight function of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for immunomodulation. Biofactors 2018, 44, 597–608. [Google Scholar] [CrossRef]
- Lee, F.H.; Su, P.; Xie, Y.F.; Wang, K.E.; Wan, Q.; Liu, F. Disrupting GluA2-GAPDH Interaction Affects Axon and Dendrite Development. Sci. Rep. 2016, 6, 30458. [Google Scholar] [CrossRef][Green Version]
- Ye, Z.; Ander, B.P.; Sharp, F.R.; Zhan, X. Cleaved beta-Actin May Contribute to DNA Fragmentation Following Very Brief Focal Cerebral Ischemia. J. Neuropathol. Exp. Neurol. 2018, 77, 260–265. [Google Scholar] [CrossRef]
- Wu, X.; Evans, A.T.; Kumar, K. Hypothermia preserves expression of beta-actin mRNA in ischemic brain. Neuroreport 1995, 7, 302–304. [Google Scholar] [CrossRef]
- Arakaki, T.L.; Pezza, J.A.; Cronin, M.A.; Hopkins, C.E.; Zimmer, D.B.; Tolan, D.R.; Allen, K.N. Structure of human brain fructose 1,6-(bis)phosphate aldolase: Linking isozyme structure with function. Protein Sci. 2004, 13, 3077–3084. [Google Scholar] [CrossRef]
- Hassell, K.J.; Ezzati, M.; Alonso-Alconada, D.; Hausenloy, D.J.; Robertson, N.J. New horizons for newborn brain protection: Enhancing endogenous neuroprotection. Arch. Dis. Childhood. Fetal Neonatal Ed. 2015, 100, F541–F552. [Google Scholar] [CrossRef]
- Peeples, E.S.; Genaro-Mattos, T.C. Ferroptosis: A Promising Therapeutic Target for Neonatal Hypoxic-Ischemic Brain Injury. Int. J. Mol. Sci. 2022, 23, 7420. [Google Scholar] [CrossRef]




| Antibody | Species | Dilution | Cat. No | Company |
|---|---|---|---|---|
| Primary antibodies | ||||
| β-Actin | Rabbit | 1:250 | 4970 | New England Biolabs, Ipswich, MA, USA |
| GAPDH | Rabbit | 1:500 | 5174 | New England Biolabs, Ipswich, MA, USA |
| Lactotransferrin | Rabbit | 1:500 | ab15811 | Abcam, Cambridge, UK |
| PGK1 | Rabbit | 1:100 | ab38007 | Abcam, Cambridge, UK |
| Secondary antibodies | ||||
| Mouse immunoglobulins/HRP | Goat | 1:10,000 | P0447 | Dako, Cheshire, UK |
| Rabbit immunoglobulins/HRP | Goat | 1:5000 | P0448 | Dako, Cheshire, UK |
| ncord | mHIE | sHIE+ | sHIE− | p | |
|---|---|---|---|---|---|
| n | 12 | 10 | 12 | 10 | |
| Male | 8 | 4 | 8 | 7 | 0.47 |
| Birth weight (g) | 3340 (2790, 3622) | 3430 (2886, 3905) | 3770 (3172, 4274) | 3685 (3472, 3813) | 0.39 |
| Apgar at 10 min | 10 (10, 10) | 9 (9, 10) | 4 (3, 6) | 4 (3, 5) | <0.001 |
| Post menstrual age (week) | 39.1 (38, 39.2) | 40 (39, 40.8) | 40.6 (39.5, 41.9) | 40.5 (40, 41.1) | 0.08 |
| Sentinel event | 0 | 1 | 4 | 2 | 0.14 |
| Chest compressions | 0 | 1 | 5 | 5 | 0.016 |
| Cord or pH in first hour | not done | 6.97 (6.93, 6.99) | 6.93 (6.87, 6.97) | 6.88 (6.78, 6.98) | 0.33 |
| Cord or base excess in first hour | not done | −12.8 (16.3, −11.4) | −19.1 (−22.8, −16) | −19.5 (−22.0, −18.2) | 0.014 |
| HIE grade (0,1,2,3) | 12,0,0,0 | 1,9,0,0 | 0,0,8,4 | 0,0,5,5 | <0.001 |
| Meconium aspiration (%) | 0 (0) | 1 (10) | 0 (0) | 3 (30) | 0.05 |
| Positive blood cultures (%) | 0 (0) | 0 (0) | 0 (0) | 1 (10) | 0.32 |
| Inotropic support (%) | 0 (0) | 0 (0) | 3 (25) | 8 (80) | <0.001 |
| Seizuures (%) | 0 (0) | 0 (0) | 7 (58) | 9 (90) | <0.001 |
| Age at MRI (d) | not done | not done | 8 (7, 10) | 8 (8, 9) | 0.71 |
| Age at blood sample (hr) | 0.5 | 18.7 ± 7.5 | 18.5 ± 11.5 | 24.0 ± 21.3 |
| Protein ID | Gene Name | Protein Name | Number of Peptides | Mild HIE/ncord | |
|---|---|---|---|---|---|
| Log2FC | p-Value | ||||
| P06276 | CHLE | Cholinesterase | 3 | 0.76 | 2.44 × 10−2 |
| P26927 | HGFL | Hepatocyte growth factor-like protein | 14 | 0.71 | 1.26 × 10−3 |
| P78417-2 | GSTO1 | Isoform 2 of Glutathione S-transferase omega-1 | 3 | 1.58 | 4.60 × 10−3 |
| P37837 | TALDO | Transaldolase | 4 | 1.87 | 5.50 × 10−3 |
| Protein ID | Gene Name | Protein Name | Number of Peptides | sHIE+/ncord | |
|---|---|---|---|---|---|
| Log2FC | p-Value | ||||
| P07737 | PROF1 | Profilin-1 | 4 | 4.08 | 2.09 × 10−8 |
| P26038 | MOES | Moesin | 3 | 3.67 | 8.29 × 10−13 |
| P62328 | TYB4 | Thymosin beta-4 | 3 | 3.31 | 2.05 × 10−6 |
| Q5T123 | SH3BGRL3 | SH3 domain-binding glutamic acid-rich-like protein | 3 | 3.29 | 2.00 × 10−8 |
| P63104 | YWHAZ | 14-3-3 protein zeta/delta | 4 | 3.09 | 9.64 × 10−8 |
| P04075 | ALDOA | Fructose-bisphosphate aldolase A | 13 | 2.94 | 1.21 × 10−8 |
| P00558 | PGK1 | Phosphoglycerate kinase 1 | 7 | 2.83 | 3.44 × 10−8 |
| P13796 | PLS2 | Plastin-2 | 5 | 2.73 | 9.30 × 10−8 |
| P59665 | DEFA1 | Neutrophil defensin 1 | 3 | 2.63 | 1.10 × 10−4 |
| E7EQB2 | LTF | Kaliocin-1 (Fragment) | 4 | 2.45 | 3.28 × 10−5 |
| P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 6 | 2.42 | 1.05 × 10−5 |
| P60709 | ACTB | Actin, cytoplasmic 1 | 5 | 2.40 | 1.15 × 10−6 |
| P02763 | AGP1 | Alpha-1-acid glycoprotein 1 | 10 | 1.52 | 8.80 × 10−3 |
| Protein ID | Gene Name | Protein Name | Number of Peptides | sHIE−/ncord | |
|---|---|---|---|---|---|
| Log2FC | p-Value | ||||
| P11142 | HSPA8 | Heat shock cognate 71 kDa protein | 5 | 2.41 | 9.91 × 10−8 |
| P18065 | IGFBP2 | Insulin-like growth factor-binding protein 2 | 7 | 2.04 | 6.71 × 10−6 |
| Protein ID | Gene Name | Protein Name | Number of Peptides | sHIE+/mHIE | |
|---|---|---|---|---|---|
| Log2FC | p-Value | ||||
| P01861 | IGHG4 | Immunoglobulin heavy constant gamma 4 | 2 | −1.46 | 4.87 × 10−3 |
| P00738 | HP | Haptoglobin | 13 | −1.44 | 4.96 × 10−2 |
| P01779 | IG_HV318 | Immunoglobulin heavy variable 3–23 | 1 | −1.15 | 1.56 × 10−2 |
| P19652 | ORM2 | Alpha−1-acid glycoprotein 2 | 11 | −0.91 | 7.75 × 10−5 |
| P01764 | IG_HV304 | Immunoglobulin heavy variable 3–23 | 2 | −0.86 | 3.32 × 10−2 |
| P22352 | GPX3 | Glutathione peroxidase 3 | 3 | −0.76 | 5.10 × 10−4 |
| P18428 | LBP | Lipopolysaccharide-binding protein | 5 | −0.67 | 4.62 × 10−2 |
| P02763 | ORM1 | Alpha-1-acid glycoprotein 1 | 10 | −0.59 | 4.44 × 10−2 |
| P00390-2 | GSR | Glutathione reductase, mitochondrial | 1 | 0.61 | 1.79 × 10−2 |
| O95445 | APOM | Apolipoprotein M | 5 | 0.61 | 2.41 × 10−4 |
| F5H6 × 6 | GANAB | Neutral alpha-glucosidase AB | 2 | 0.62 | 3.67 × 10−4 |
| Q9HD89 | RETN | Resistin | 1 | 0.64 | 3.81 × 10−2 |
| Q12907 | LMAN2 | Vesicular integral-membrane protein VIP36 | 1 | 0.66 | 7.15 × 10−4 |
| H7BZJ3 | PDIA3 | Protein disulfide-isomerase | 1 | 0.66 | 6.45 × 10−3 |
| F5GZK1 | EXTL2 | Exostosin-like 2 | 1 | 0.72 | 7.82 × 10−4 |
| G3V1A4 | CFL1 | Cofilin-1 | 2 | 0.73 | 4.07 × 10−2 |
| P06858 | LPL | Lipoprotein lipase | 1 | 0.75 | 9.99 × 10−3 |
| P01130-3 | LDLR | Low-density lipoprotein receptor | 3 | 0.76 | 1.52 × 10−2 |
| P18206-2 | VCL | Vinculin | 5 | 0.78 | 6.69 × 10−4 |
| P00441 | SOD1 | Superoxide dismutase [Cu-Zn] | 2 | 0.79 | 2.20 × 10−2 |
| P10599 | TXN | Thioredoxin | 1 | 0.81 | 5.36 × 10−3 |
| P78417-2 | GSTO1 | Glutathione S-transferase omega-1 | 3 | 0.82 | 8.83 × 10−3 |
| P26038 | MSN | Moesin | 3 | 0.82 | 9.88 × 10−3 |
| P08246 | ELANE | Neutrophil elastase | 1 | 0.82 | 2.02 × 10−2 |
| P04040 | CAT | Catalase | 10 | 0.82 | 1.74 × 10−2 |
| P68104 | EEF1A1 | Elongation factor 1-alpha 1 | 1 | 0.83 | 9.68 × 10−3 |
| P01040 | CSTA | Cystatin-A | 2 | 0.85 | 3.60 × 10−2 |
| P30043 | BLVRB | Flavin reductase (NADPH) | 2 | 0.85 | 1.36 × 10−2 |
| P07900 | HSP90AA1 | Heat shock protein HSP 90-alpha | 1 | 0.85 | 4.26 × 10−2 |
| Q71U36-2 | TUBA1A | Tubulin alpha-1A chain | 1 | 0.86 | 3.92 × 10−3 |
| Q5T123 | SH3BGRL3 | SH3 domain-binding glutamic acid-rich-like protein 3 | 3 | 0.87 | 1.33 × 10−2 |
| E5RIF1 | UBE2V2 | Ubiquitin-conjugating enzyme E2 variant 2 | 1 | 0.87 | 2.58 × 10−2 |
| F5H265 | UBC | Polyubiquitin-C | 4 | 0.87 | 8.71 × 10−3 |
| P37837 | TALDO1 | Transaldolase | 4 | 0.89 | 1.80 × 10−2 |
| P06733 | ENO1 | Alpha-enolase | 4 | 0.90 | 1.11 × 10−2 |
| P62328 | TMSB4X | Thymosin beta-4 | 3 | 0.90 | 4.33 × 10−2 |
| P04075 | ALDOA | Fructose-bisphosphate aldolase A | 13 | 0.91 | 8.63 × 10−3 |
| P05062 | ALDOB | Fructose-bisphosphate aldolase B | 3 | 0.94 | 4.88 × 10−2 |
| P14618-2 | PKM | Pyruvate kinase PKM | 1 | 0.95 | 3.09 × 10−2 |
| P18669 | PGAM1 | Phosphoglycerate mutase 1 | 1 | 0.98 | 1.76 × 10−2 |
| P63104 | YWHAZ | 14-3-3 protein zeta/delta | 4 | 0.99 | 9.96 × 10−3 |
| P09972 | ALDOC | Fructose-bisphosphate aldolase C | 1 | 1.01 | 1.97 × 10−3 |
| P60709 | ACTB | Actin, cytoplasmic 1 | 5 | 1.07 | 2.53 × 10−3 |
| P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 6 | 1.09 | 2.69 × 10−3 |
| P06753-2 | TPM3 | Tropomyosin alpha-3 chain | 1 | 1.11 | 2.36 × 10−2 |
| E7EQB2 | LTF | Lactotransferrin | 4 | 1.12 | 3.13 × 10−3 |
| P60174-1 | TPI1 | Triosephosphate isomerase | 4 | 1.12 | 1.01 × 10−2 |
| P00558 | PGK1 | Phosphoglycerate kinase 1 | 7 | 1.17 | 9.79 × 10−4 |
| P62937 | PPIA | Peptidyl-prolyl cis-trans isomerase A | 6 | 1.18 | 9.19 × 10−4 |
| P09211 | GSTP1 | Glutathione S-transferase P | 1 | 1.19 | 3.43 × 10−3 |
| R4GN98 | S100A6 | Protein S100 | 2 | 1.20 | 5.15 × 10−4 |
| B4DQJ8 | PGD | 6-phosphogluconate dehydrogenase, decarboxylating | 1 | 1.31 | 1.55 × 10−3 |
| P06732 | CKM | Creatine kinase M-type | 9 | 1.32 | 4.38 × 10−2 |
| P62158 | CALM1 | Calmodulin-1 | 1 | 1.33 | 9.83 × 10−4 |
| P07737 | PFN1 | Profilin-1 | 4 | 1.34 | 2.45 × 10−3 |
| P11142 | HSPA8 | Heat shock cognate 71 kDa protein | 5 | 1.37 | 5.16 × 10−5 |
| P07451 | CA3 | Carbonic anhydrase 3 | 3 | 1.38 | 4.64 × 10−2 |
| P31146 | CORO1A | Coronin-1A | 1 | 1.51 | 3.86 × 10−3 |
| P68431 | HIST1H3A | Histone H3.1 | 1 | 1.55 | 2.06 × 10−2 |
| P61604 | HSPE1 | 10 kDa heat shock protein, mitochondrial | 2 | 1.57 | 5.44 × 10−3 |
| P02042 | HBD | Hemoglobin subunit delta | 2 | 1.91 | 4.93 × 10−4 |
| Protein ID | Gene Name | Protein Name | Number of Peptides | sHIE−/mHIE | |
|---|---|---|---|---|---|
| Log2FC | p-Value | ||||
| P01861 | IGHG4 | Immunoglobulin heavy constant gamma 4 | 2 | −1.65 | 3.20 × 10−3 |
| P01860 | IGHG3 | Immunoglobulin heavy constant gamma 3 | 5 | −1.05 | 1.35 × 10−2 |
| P11226 | MBL2 | Mannose-binding protein C | 4 | −0.87 | 1.97 × 10−2 |
| P24592 | IGFBP6 | Insulin-like growth factor-binding protein 6 | 2 | −0.65 | 8.67 × 10−3 |
| P02458 | COL2A1 | Collagen alpha-1(II) chain | 2 | −0.64 | 1.24 × 10−2 |
| P35858 | IGFALS | Insulin-like growth factor-binding protein complex acid labile subunit | 12 | 0.62 | 1.74 × 10−2 |
| Q02985 | CFHR3 | Complement factor H-related protein 3 | 1 | 0.74 | 2.02 × 10−2 |
| P11142 | HSPA8 | Heat shock cognate 71 kDa protein | 5 | 0.82 | 1.19 × 10−2 |
| P61604 | HSPE1 | 10 kDa heat shock protein, mitochondrial | 2 | 1.21 | 4.00 × 10−2 |
| P02042 | HBD | Hemoglobin subunit delta | 2 | 1.42 | 1.05 × 10−2 |
| Protein ID | Gene Name | Protein Name | Number of Peptides | sHIE+/sHIE− | |
|---|---|---|---|---|---|
| Log2FC | p-Value | ||||
| P13727 | PRG2 | Bone marrow proteoglycan | 2 | −0.72 | 3.80 × 10−2 |
| P68104 | EEF1A1 | Elongation factor 1-alpha 1 | 1 | 0.67 | 3.37 × 10−2 |
| P26038 | MSN | Moesin | 3 | 0.68 | 2.82 × 10−2 |
| P09972 | ALDOC | Fructose-bisphosphate aldolase C | 1 | 0.70 | 2.43 × 10−2 |
| P02730 | SLC4A1 | Band 3 anion transport protein | 3 | 0.72 | 1.00 × 10−2 |
| P13796 | LCP1 | Plastin-2 | 5 | 0.74 | 2.78 × 10−2 |
| P00441 | SOD1 | Superoxide dismutase [Cu-Zn] | 2 | 0.74 | 3.01 × 10−2 |
| P04075 | ALDOA | Fructose-bisphosphate aldolase A | 13 | 0.83 | 1.52 × 10−2 |
| P18669 | PGAM1 | Phosphoglycerate mutase 1 | 1 | 0.84 | 3.87 × 10−2 |
| P62937 | PPIA | Peptidyl-prolyl cis-trans isomerase A | 6 | 0.86 | 1.17 × 10−2 |
| P63104 | YWHAZ | 14-3-3 protein zeta/delta | 4 | 0.87 | 2.13 × 10−2 |
| G3V1A4 | CFL1 | Cofilin-1 | 2 | 0.92 | 1.22 × 10−2 |
| Q5T123 | SH3BGRL3 | SH3 domain-binding glutamic acid-rich-like protein 3 | 3 | 0.92 | 8.96 × 10−3 |
| P32119 | PRDX2 | Peroxiredoxin-2 | 9 | 0.92 | 3.41 × 10−3 |
| P60709 | ACTB | Actin, cytoplasmic 1 | 5 | 0.93 | 7.41 × 10−3 |
| P30740 | SERPINB1 | Leukocyte elastase inhibitor | 1 | 0.95 | 2.55 × 10−2 |
| P00915 | CA1 | Carbonic anhydrase 1 | 1 | 0.95 | 1.01 × 10−2 |
| P09211 | GSTP1 | Glutathione S-transferase P | 1 | 0.98 | 1.38 × 10−2 |
| P59665 | DEFA1 | Neutrophil defensin 1 | 3 | 0.98 | 3.95 × 10−2 |
| P00558 | PGK1 | Phosphoglycerate kinase 1 | 7 | 1.00 | 3.81 × 10−3 |
| P07737 | PFN1 | Profilin-1 | 4 | 1.06 | 1.39 × 10−2 |
| P02008 | HBZ | Hemoglobin subunit zeta | 3 | 1.08 | 2.00 × 10−2 |
| P30043 | BLVRB | Flavin reductase (NADPH) | 2 | 1.09 | 2.30 × 10−3 |
| P62158 * | CALM1 | Calmodulin-1 | 1 | 1.18 | 2.76 × 10−3 |
| E7EQB2 | LTF | Lactotransferrin | 4 | 1.22 | 1.45 × 10−3 |
| P14618-2 | PKM | Pyruvate kinase PKM | 1 | 1.23 | 6.53 × 10−3 |
| P31146 | CORO1A | Coronin-1A | 1 | 1.28 | 1.23 × 10−2 |
| P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 6 | 1.30 | 5.43 × 10−4 |
| R4GN98 | S100A6 | Protein S100 | 2 | 1.37 | 1.18 × 10−4 |
| P68431 | HIST1H3A | Histone H3.1 | 1 | 1.45 | 2.92 × 10−2 |
| P06732 | CKM | Creatine kinase M-type | 9 | 2.11 | 2.21 × 10−3 |
| Protein ID | Gene Name | Protein Name | Number of Peptides | Log2FC | p-Value |
|---|---|---|---|---|---|
| P00390-2 | GSR | Glutathione reductase, mitochondrial | 1 | 0.61 | 1.79 × 10−2 |
| P37837 | TALDO1 | Transaldolase | 4 | 0.89 | 1.80 × 10−2 |
| P06733 | ENO1 | Alpha-enolase | 4 | 0.9 | 1.11 × 10−2 |
| P04075 | ALDOA | Fructose-bisphosphate aldolase A | 13 | 0.91 | 8.63 × 10−3 |
| P05062 | ALDOB | Fructose-bisphosphate aldolase B | 3 | 0.94 | 4.88 × 10−2 |
| P14618-2 | PKM | Pyruvate kinase PKM | 1 | 0.95 | 3.09 × 10−2 |
| P18669 | PGAM1 | Phosphoglycerate mutase 1 | 1 | 0.98 | 1.76 × 10−2 |
| P09972 | ALDOC | Fructose-bisphosphate aldolase C | 1 | 1.01 | 1.97 × 10−3 |
| P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 6 | 1.09 | 2.69 × 10−3 |
| P60174-1 | TPI1 | Triosephosphate isomerase | 4 | 1.12 | 1.01 × 10−2 |
| P00558 | PGK1 | Phosphoglycerate kinase 1 | 7 | 1.17 | 9.79 × 10−4 |
| B4DQJ8 | PGD | 6-phosphogluconate dehydrogenase, decarboxylating | 1 | 1.31 | 1.55 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yip, P.K.; Bremang, M.; Pike, I.; Ponnusamy, V.; Michael-Titus, A.T.; Shah, D.K. Newborns with Favourable Outcomes after Perinatal Asphyxia Have Upregulated Glucose Metabolism-Related Proteins in Plasma. Biomolecules 2023, 13, 1471. https://doi.org/10.3390/biom13101471
Yip PK, Bremang M, Pike I, Ponnusamy V, Michael-Titus AT, Shah DK. Newborns with Favourable Outcomes after Perinatal Asphyxia Have Upregulated Glucose Metabolism-Related Proteins in Plasma. Biomolecules. 2023; 13(10):1471. https://doi.org/10.3390/biom13101471
Chicago/Turabian StyleYip, Ping K., Michael Bremang, Ian Pike, Vennila Ponnusamy, Adina T. Michael-Titus, and Divyen K. Shah. 2023. "Newborns with Favourable Outcomes after Perinatal Asphyxia Have Upregulated Glucose Metabolism-Related Proteins in Plasma" Biomolecules 13, no. 10: 1471. https://doi.org/10.3390/biom13101471
APA StyleYip, P. K., Bremang, M., Pike, I., Ponnusamy, V., Michael-Titus, A. T., & Shah, D. K. (2023). Newborns with Favourable Outcomes after Perinatal Asphyxia Have Upregulated Glucose Metabolism-Related Proteins in Plasma. Biomolecules, 13(10), 1471. https://doi.org/10.3390/biom13101471







