Plasma Levels of sFas-sFasL and FASL Gene Expression Are Associated with Tuberculosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Characterization
2.2. DNA Extraction
2.3. Genotyping of FAS -670A/G (rs1800682) and FASL -124A/G (rs5030772)
2.4. Plasma Quantification of sFas and sFasL
2.5. RNA Extraction
2.6. Reverse Transcription
2.7. Gene Expression
2.8. Plasma Measurement of IFN-γ and IL-10
2.9. Tuberculin Skin Test (TST)
2.10. Statistical Analysis
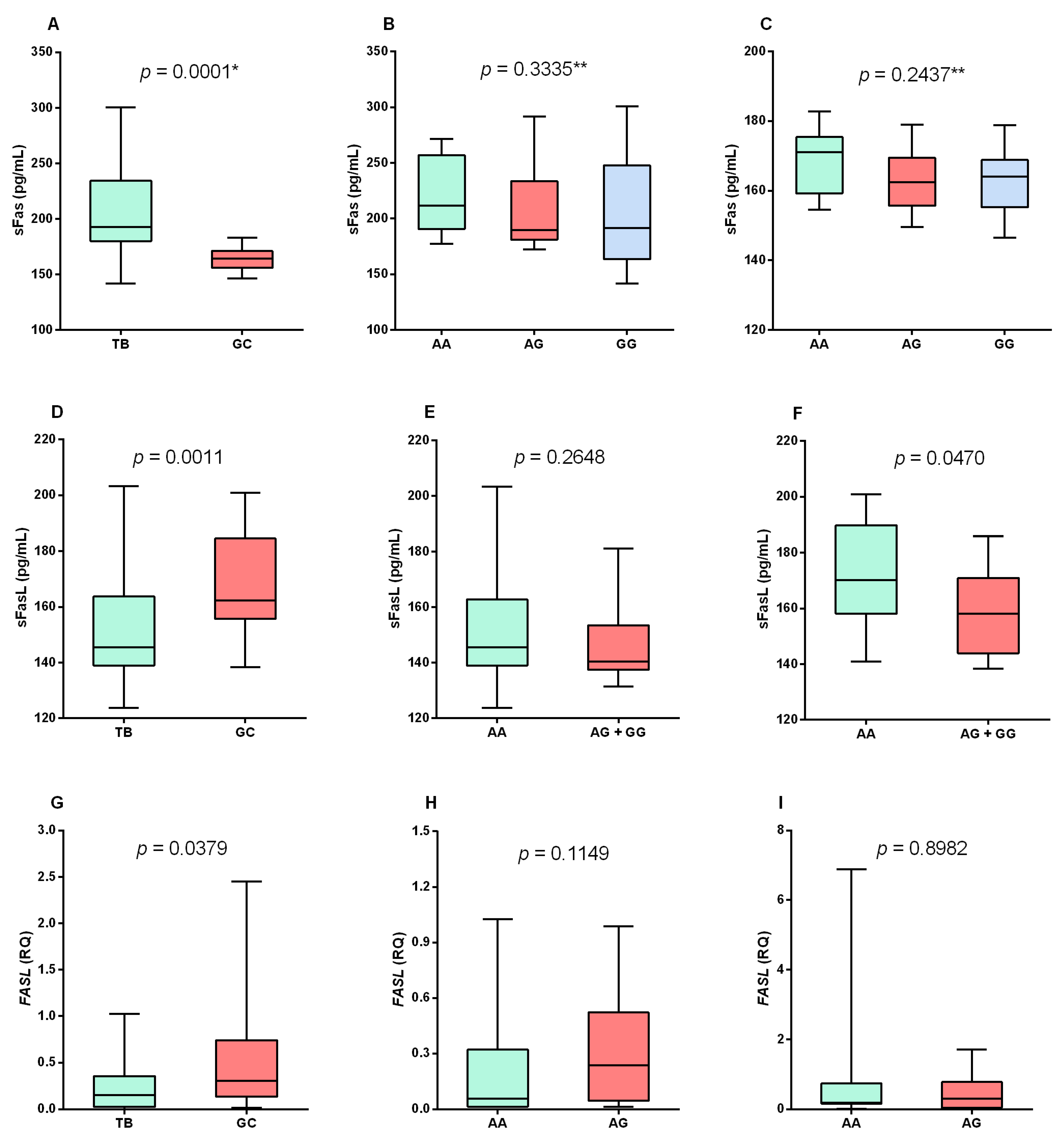
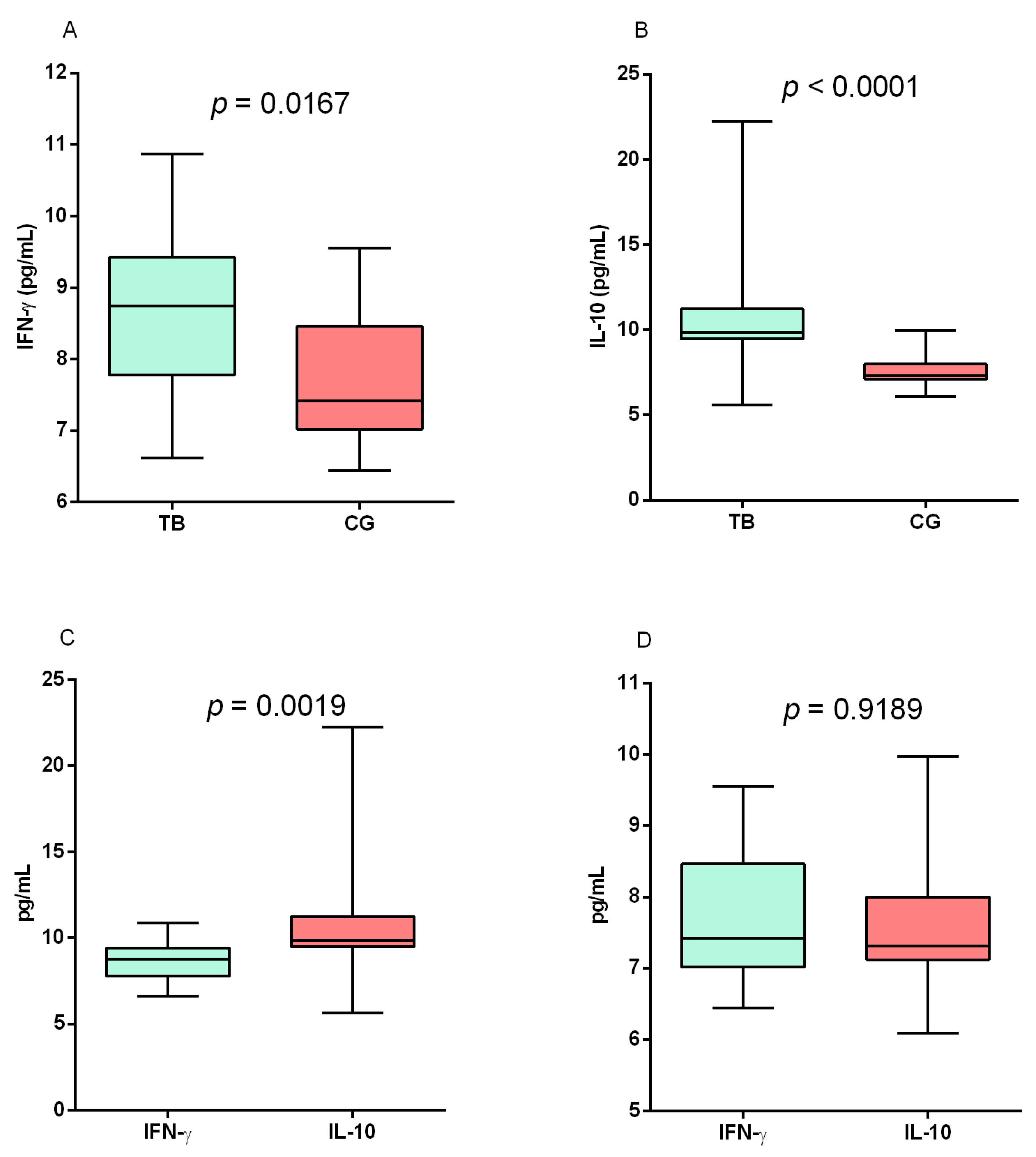
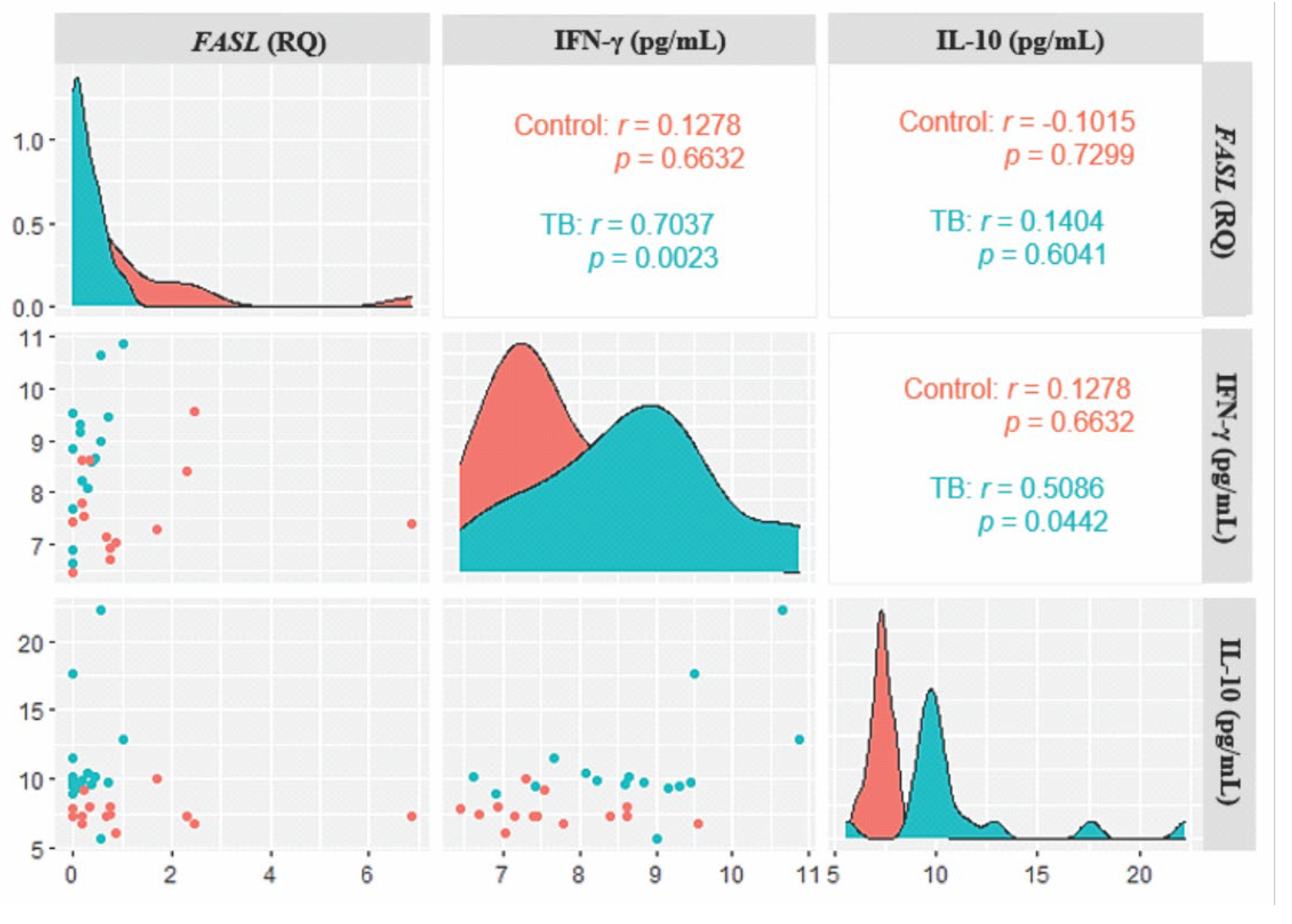
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2020. 2020. Available online: https://www.who.int/publications/i/item/9789240013131 (accessed on 13 January 2022).
- World Health Organization. Global Tuberculosis Report 2018. 2018. Available online: https://apps.who.int/iris/handle/10665/274453 (accessed on 13 January 2022).
- Dheda, K.; Barry, C.E., 3rd; Maartens, G. Tuberculosis. Lancet 2016, 387, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Wu, F.; Zhang, Y.; Fu, Y.; Liu, Z. The Immune Escape Mechanisms of Mycobacterium tuberculosis. Int. J. Mol. Sci. 2019, 20, 340. [Google Scholar] [CrossRef] [PubMed]
- Chai, Q.; Wang, L.; Liu, C.H.; Ge, B. New insights into the evasion of host innate immunity by Mycobacterium tuberculosis. Cell Mol. Immunol. 2020, 17, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Ellinwood, S.; Augenstreich, J.; Mayer-Barber, K.D.; Briken, V. Mycobacterium tuberculosis inhibits the NLRP3 inflammasome activation via its phosphokinase PknF. PLoS Pathog. 2021, 17, e1009712. [Google Scholar] [CrossRef]
- Green, A.M.; Difazio, R.; Flynn, J.L. IFN-γ from CD4 T cells is essential for host survival and enhances CD8 T cell function during Mycobacterium tuberculosis infection. J. Immunol. 2013, 190, 270–277. [Google Scholar] [CrossRef]
- Hamad, A.R.; Arcara, K.; Uddin, S.; Donner, T. The potential of Fas ligand (apoptosis-inducing molecule) as an unconventional therapeutic target in type 1 diabetes. Front. Immunol. 2012, 3, 196. [Google Scholar] [CrossRef]
- Yanti, B.; Mulyadi, M.; Amin, M.; Harapan, H.; Mertaniasih, N.M.; Soetjipto, S. The role of Mycobacterium tuberculosis complex species on apoptosis and necroptosis state of macrophages derived from active pulmonary tuberculosis patients. BMC Res. Notes. 2020, 13, 415. [Google Scholar] [CrossRef]
- Stutz, M.D.; Allison, C.C.; Ojaimi, S.; Preston, S.P.; Doerflinger, M.; Arandjelovic, P.; Whitehead, L.; Bader, S.M.; Batey, D.; Asselin-Labat, M.-L.; et al. Macrophage and neutrophil death programs differentially confer resistance to tuberculosis. Immunity 2021, 54, 1758–1771.e7. [Google Scholar] [CrossRef]
- Papoff, G.; Cascino, I.; Eramo, A.; Starace, G.; Lynch, D.H.; Ruberti, G. An N-terminal domain shared by Fas/Apo-1 (CD95) soluble variants prevents cell death in vitro. J. Immunol. 1996, 156, 4622–4630. [Google Scholar]
- Powell, W.C.; Fingleton, B.; Wilson, C.L.; Boothby, M.; Matrisian, L.M. The metalloproteinase matrilysin proteolytically generates active soluble Fas ligand and potentiates epithelial cell apoptosis. Curr. Biol. 1999, 9, 1441–1447. [Google Scholar] [CrossRef]
- Tanaka, M.; Suda, T.; Takahashi, T.; Nagata, S. Expression of the functional soluble form of human Fas ligand in activated lymphocytes. EMBO J. 1995, 14, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Vargo-Gogola, T.; Crawford, H.C.; Fingleton, B.; Matrisian, L.M. Identification of novel matrix metalloproteinase-7 (matrilysin) cleavage sites in murine and human Fas ligand. Arch. Biochem. Biophys. 2002, 408, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Bajou, K.; Peng, H.; Laug, W.E.; Maillard, C.; Noel, A.; Foidart, J.M.; Martial, J.A.; De Clerck, Y.A. Plasminogen activator inhibitor-1 protects endothelial cells from FasL-mediated apoptosis. Cancer Cell. 2008, 14, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Farre, L.; Bittencourt, A.L.; Silva-Santos, G.; Almeida, A.; Silva, A.C.; Decanine, D.; Soares, G.M.; Alcantara, L.C., Jr.; van Dooren, S.; Galvão-Castro, B.; et al. Fas 670 promoter polymorphism is associated to susceptibility, clinical presentation, and survival in adult T cell leukemia. J. Leukoc. Biol. 2008, 83, 220–222. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Fazaeli, A.; Ghavami, S.; Eskandari-Nasab, E.; Arbabi, F.; Mashhadi, M.A.; Taheri, M.; Chaabane, W.; Jain, M.V.; Łos, M.J. Functional polymorphisms of FAS and FASL gene and risk of breast cancer—Pilot study of 134 cases. PLoS ONE 2013, 8, e53075. [Google Scholar] [CrossRef]
- Yıldır, S.; Sezgin, M.; Barlas, I.Ö.; Türköz, G.; Ankaralı, H.Ç.; Şahin, G.; Erdal, M.E. Relation of the Fas and FasL gene polymorphisms with susceptibility to and severity of rheumatoid arthritis. Rheumatol. Int. 2013, 33, 2637–2645. [Google Scholar] [CrossRef]
- Seleit, I.; Bakry, O.A.; Gayed, E.A.E.; Gawad, A.E.D. Polymorphism of FAS and FAS Ligand Genes in Alopecia Areata: A Case-control Study in Egyptian Population. Indian J. Dermatol. 2018, 63, 220–226. [Google Scholar]
- Wang, T.; Lian, Y. The relationship between Fas and Fas ligand gene polymorphism and preeclampsia risk. Biosci. Rep. 2019, 39, BSR20181901. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Brasil. Ministério da Saúde. Técnicas de Aplicação e Leitura da Prova Tuberculínica. 2014. Available online: https://bvsms.saude.gov.br/bvs/publicacoes/tecnicas_aplicacao_leitura_prova_tuberculinica.pdf (accessed on 27 September 2022).
- Jorgensen, I.; Rayamajhi, M.; Miao, E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017, 17, 151–164. [Google Scholar] [CrossRef]
- Robinson, K.S.; Aw, R. The Commonalities in Bacterial Effector Inhibition of Apoptosis. Trends Microbiol. 2016, 24, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, C.S.; Toossi, Z.; Johnson, J.L.; Luzze, H.; Ntambi, L.; Peters, P.; McHugh, M.; Okwera, A.; Joloba, M.; Mugyenyi, P.; et al. Augmentation of apoptosis and interferon-gamma production at sites of active Mycobacterium tuberculosis infection in human tuberculosis. J. Infect. Dis. 2021, 183, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, T.; Mogga, S.J.; Mfinanga, S.G.; Mørkve, O.; Sviland, L. Significance of Fas and Fas ligand in tuberculous lymphadenitis. Immunology 2005, 114, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Budak, F.; Uzaslan, E.K.; Cangür, S.; Göral, G.; Oral, H.B. Increased pleural soluble fas ligand (sFasL) levels in tuberculosis pleurisy and its relation with T-helper type 1 cytokines. Lung 2008, 186, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.Y.; Zhang, Q.; Su, B.; Li, W.; Tang, S.J. Differential levels of cytokines and soluble Fas ligand between tuberculous and malignant effusions. J. Int. Med. Res. 2010, 38, 2063–2069. [Google Scholar] [CrossRef]
- Wu, S.H.; Li, C.T.; Lin, C.H.; Chu, J.J.; Cheng, M.L.; Lin, K.H. Soluble Fas ligand is another good diagnostic marker for tuberculous pleurisy. Diagn. Microbiol. Infect. Dis. 2010, 68, 395–400. [Google Scholar] [CrossRef]
- Molin, S.; Weiss, E.H.; Ruzicka, T.; Messer, G. The FAS/cd95 promoter single-nucleotide polymorphism -670 A/G and lupus erythematosus. Clin. Exp. Dermatol. 2012, 37, 425–427. [Google Scholar] [CrossRef]
- Mohammadi, A.; Tajik, N.; Shah-Hosseini, A.; Alavian, S.M.; Sharifi, Z.; Jarahi, L. FAS and FAS-Ligand Promoter Polymorphisms in Hepatitis B Virus Infection. Hepat. Mon. 2015, 15, e26490. [Google Scholar] [CrossRef]
- Santos, N.P.; Ribeiro-Rodrigues, E.M.; Ribeiro-Dos-Santos, A.K.; Pereira, R.; Gusmão, L.; Amorim, A.; Guerreiro, J.F.; Zago, M.A.; Matte, C.; Hutz, M.H.; et al. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ances-try-informative marker (AIM) panel. Hum. Mutat. 2010, 31, 184–190. [Google Scholar] [CrossRef]
- Vallinoto, A.C.; Santana, B.B.; dos Santos, E.L.; Santo, R.R.; Hermes, R.B.; Sousa, R.C.; Cayres-Vallinoto, I.; Machado, L.F.; Ishak, M.O.; Ishak, R. FAS −670A/G single nucleotide polymorphism may be associated with human T lymphotropic virus-1 infection and clinical evolution to TSP/HAM. Virus Res. 2012, 163, 178–182. [Google Scholar] [CrossRef]
- Santana, B.B.; Viégas, M.L.; Conde, S.R.; Ishak, M.O.; Ishak, R.; Vallinoto, A.C. FAS and FASL gene polymorphisms are not associated with hepatitis B virus infection based on a case-control study in a Brazilian population. Dis. Markers 2013, 35, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Hermes, R.B.; Santana, B.B.; Lima, S.S.; Neris Martins Feitosa, R.; de Oliveira Guimarães Ishak, M.; Ishak, R.; Vallinoto, A.C. FAS -670 A/G polymorphism may be associated with the depletion of CD4+ T lymphocytes in HIV-1 infection. Hum. Immunol. 2015, 76, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Cascino, I.; Fiucci, G.; Papoff, G.; Ruberti, G. Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J. Immunol. 1995, 154, 2706–2713. [Google Scholar] [PubMed]
- André, S.; Picard, M.; Cezar, R.; Roux-Dalvai, F.; Alleaume-Butaux, A.; Soundaramourty, C.; Cruz, A.S.; Mendes-Frias, A.; Gotti, C.; Leclercq, M.; et al. T cell apoptosis characterizes severe COVID-19 disease. Cell Death Differ. 2018, 22, 1486–1499. [Google Scholar] [CrossRef] [PubMed]
- Suda, T.; Takahashi, T.; Golstein, P.; Nagata, S. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell 1993, 75, 1169–1178. [Google Scholar] [CrossRef]
- Sieg, S.; Smith, D.; Kaplan, D. Differential activity of soluble versus cellular Fas ligand: Regulation by an accessory molecule. Cell. Immunol. 1999, 195, 89–95. [Google Scholar] [CrossRef]
- Nishigaki, K.; Minatoguchi, S.; Seishima, M.; Asano, K.; Noda, T.; Yasuda, N.; Sano, H.; Kumada, H.; Takemura, M.; Noma, A.; et al. Plasma Fas ligand, an inducer of apoptosis, and plasma soluble Fas, an inhibitor of apoptosis, in patients with chronic congestive heart failure. J. Am. Coll. Cardiol. 1997, 29, 1214–1220. [Google Scholar] [CrossRef]
- Yasuda, N.; Gotoh, K.; Minatoguchi, S.; Asano, K.; Nishigaki, K.; Nomura, M.; Ohno, A.; Watanabe, M.; Sano, H.; Kumada, H.; et al. An increase of soluble Fas, an inhibitor of apoptosis, associated with progression of COPD. Respir. Med. 1998, 92, 993–999. [Google Scholar] [CrossRef]
- Mouawad, R.; Khayat, D.; Soubrane, C. Plasma Fas ligand, an inducer of apoptosis, and plasma soluble Fas, an inhibitor of apoptosis, in advanced melanoma. Melanoma Res. 2000, 10, 461–467. [Google Scholar] [CrossRef]
- Oddo, M.; Renno, T.; Attinger, A.; Bakker, T.; MacDonald, H.R.; Meylan, P.R. Fas ligand-induced apoptosis of infected human macrophages reduces the viability of intracellular Mycobacterium tuberculosis. J. Immunol. 1998, 160, 5448–5454. [Google Scholar]
- Lee, J.; Hartman, M.; Kornfeld, H. Macrophage apoptosis in tuberculosis. Yonsei Med. J. 2009, 50, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, S.; Abraham, P.; Daniel, H.D.; Ramakrishna, B.S.; Sridharan, G. Characterization of soluble FAS, FAS ligand and tumour necrosis factor-alpha in patients with chronic HCV infection. J. Clin. Virol. 2005, 34, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Lin, Y.T.; Yan, X.L.; Ding, Y.L.; Wu, Y.L.; Chen, W.N.; Lin, X. Hepatitis B virus core protein inhibits Fas-mediated apoptosis of hepatoma cells via regulation of mFas/FasL and sFas expression. FASEB J. 2015, 29, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, T.A.; Shambat, S.M.; Vulin, C.; Hoeller, S.; Acevedo, C.; Huemer, M.; Gomez-Mejia, A.; Chang, C.; Baum, J.; Hertegonne, S.; et al. Blunted sFasL signalling exacerbates TNF-driven neutrophil necroptosis in critically ill COVID-19 patients. Clin. Transl. Immunol. 2021, 10, e1357. [Google Scholar] [CrossRef] [PubMed]
- Arrigucci, R.; Lakehal, K.; Vir, P.; Handler, D.; Davidow, A.L.; Herrera, R.; Estrada-Guzmán, J.D.; Bushkin, Y.; Tyagi, S.; Lardizabal, A.A.; et al. Active Tuberculosis Is Characterized by Highly Differentiated Effector Memory Th1 Cells. Front. Immunol. 2018, 9, 2127. [Google Scholar] [CrossRef]
- Wong, E.A.; Evans, S.; Kraus, C.R.; Engelman, K.D.; Maiello, P.; Flores, W.J.; Cadena, A.M.; Klein, E.; Thomas, K.; White, A.G.; et al. IL-10 Impairs Local Immune Response in Lung Granulomas and Lymph Nodes during Early Mycobacterium tuberculosis Infection. J. Immunol. 2020, 204, 644–659. [Google Scholar] [CrossRef]
- Stringari, L.L.; Covre, L.P.; da Silva, F.D.C.; de Oliveira, V.L.; Campana, M.C.; Hadad, D.J.; Palaci, M.; Salgame, P.; Dietze, R.; Gomes, D.C.O.; et al. Increase of CD4+CD25highFoxP3+ cells impairs in vitro human microbicidal activity against Mycobacterium tuberculosis during latent and acute pulmonary tuberculosis. PLoS Negl. Trop. Dis. 2021, 15, e0009605. [Google Scholar] [CrossRef]
- Boselli, D.; Losana, G.; Bernabei, P.; Bosisio, D.; Drysdale, P.; Kiessling, R.; Gaston, J.S.; Lammas, D.; Casanova, J.L.; Kumararatne, D.S.; et al. IFN-gamma regulates Fas ligand expression in human CD4+ T lymphocytes and controls their anti-mycobacterial cytotoxic functions. Eur. J. Immunol. 2007, 37, 2196–2204. [Google Scholar] [CrossRef]
- Cope, A.; Le Friec, G.; Cardone, J.; Kemper, C. The Th1 life cycle: Molecular control of IFN-γ to IL-10 switching. Trends Immunol. 2011, 32, 278–286. [Google Scholar] [CrossRef]
- Moreira-Teixeira, L.; Redford, P.S.; Stavropoulos, E.; Ghilardi, N.; Maynard, C.L.; Weaver, C.T.; Rosário, A.P.F.D.; Wu, X.; Langhorne, J.; O’Garra, A. T Cell-Derived IL-10 Impairs Host Resistance to Mycobacterium tuberculosis Infection. J. Immunol. 2017, 199, 613–623. [Google Scholar] [CrossRef]
- Redford, P.S.; Boonstra, A.; Read, S.; Pitt, J.; Graham, C.; Stavropoulos, E.; Bancroft, G.J.; O’Garra, A. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur. J. Immunol. 2010, 40, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Cyktor, J.C.; Carruthers, B.; Kominsky, R.A.; Beamer, G.L.; Stromberg, P.; Turner, J. IL-10 inhibits mature fibrotic granuloma formation during Mycobacterium tuberculosis infection. J. Immunol. 2013, 190, 2778–2790. [Google Scholar] [CrossRef] [PubMed]

| Genotypes and Alleles | TB n = 200 n (%) | CG n = 200 n (%) | p† |
|---|---|---|---|
| FAS -670A/G | |||
| AA | 30 (15.0) | 36 (18.0) | 0.4294 |
| GA | 88 (44.0) | 94 (47.0) | |
| GG | 82 (41.0) | 70 (35.0) | |
| *A | 0.37 | 0.41 | 0.2184 |
| *G | 0.63 | 0.59 | |
| FASL -124A/G | |||
| AA | 168 (84.0) | 162 (81.0) | 0.5106 |
| AG + GG # | 32 (16.0) | 38 (19.0) | |
| *A | 0.91 | 0.9 | 0.5418 |
| *G | 0.09 | 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, I.d.P.; Amoras, E.d.S.G.; Sousa, F.D.M.d.; Sousa, P.V.N.R.d.; Lima, S.S.; Cayres-Vallinoto, I.M.V.; Ishak, R.; Vallinoto, A.C.R.; Queiroz, M.A.F. Plasma Levels of sFas-sFasL and FASL Gene Expression Are Associated with Tuberculosis. Biomolecules 2023, 13, 80. https://doi.org/10.3390/biom13010080
Souza IdP, Amoras EdSG, Sousa FDMd, Sousa PVNRd, Lima SS, Cayres-Vallinoto IMV, Ishak R, Vallinoto ACR, Queiroz MAF. Plasma Levels of sFas-sFasL and FASL Gene Expression Are Associated with Tuberculosis. Biomolecules. 2023; 13(1):80. https://doi.org/10.3390/biom13010080
Chicago/Turabian StyleSouza, Iury de Paula, Ednelza da Silva Graça Amoras, Francisca Dayse Martins de Sousa, Paulo Victor Negrão Raiol de Sousa, Sandra Souza Lima, Izaura Maria Vieira Cayres-Vallinoto, Ricardo Ishak, Antonio Carlos Rosário Vallinoto, and Maria Alice Freitas Queiroz. 2023. "Plasma Levels of sFas-sFasL and FASL Gene Expression Are Associated with Tuberculosis" Biomolecules 13, no. 1: 80. https://doi.org/10.3390/biom13010080
APA StyleSouza, I. d. P., Amoras, E. d. S. G., Sousa, F. D. M. d., Sousa, P. V. N. R. d., Lima, S. S., Cayres-Vallinoto, I. M. V., Ishak, R., Vallinoto, A. C. R., & Queiroz, M. A. F. (2023). Plasma Levels of sFas-sFasL and FASL Gene Expression Are Associated with Tuberculosis. Biomolecules, 13(1), 80. https://doi.org/10.3390/biom13010080









