Heritability of Protein and Metabolite Biomarkers Associated with COVID-19 Severity: A Metabolomics and Proteomics Analysis
Abstract
Key Points
What is already known on this topic
What This Study Adds
1. Introduction
2. Materials and Methods
2.1. Erythrocyte Multi-Omics Twin Study
2.2. Metabolite Pathway Enrichment Analysis
2.3. Gene Ontology Analysis
3. Results
3.1. Serum Biomarkers Associated with COVID-19 Disease States Overlap with Heritable Metabolites and Proteins in Erythrocytes
3.2. Proteins Associated with COVID-19 Disease States Are Highly Correlated in Erythrocytes
3.3. Decreased Levels of Heritable Metabolites Involved in Amino Acid Biosynthesis and Metabolism Pathways Are Associated with Both Non-Severe and Severe COVID-19
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.-Y.; Gu, Y.; Wheeler, B.; Young, H.; Holloway, S.; Sunny, S.-K.; Moore, Z.; Zeng, D. Effectiveness of Covid-19 Vaccines over a 9-Month Period in North Carolina. N. Engl. J. Med. 2022, 386, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Rotshild, V.; Hirsh-Raccah, B.; Miskin, I.; Muszkat, M.; Matok, I. Comparing the Clinical Efficacy of COVID-19 Vaccines: A Systematic Review and Network Meta-Analysis. Sci. Rep. 2021, 11, 22777. [Google Scholar] [CrossRef] [PubMed]
- Sim, B.L.H.; Chidambaram, S.K.; Wong, X.C.; Pathmanathan, M.D.; Peariasamy, K.M.; Hor, C.P.; Chua, H.J.; Goh, P.P. Clinical Characteristics and Risk Factors for Severe COVID-19 Infections in Malaysia: A Nationwide Observational Study. Lancet Reg. Health West. Pac. 2020, 4, 100055. [Google Scholar] [CrossRef]
- Jiang, N.; Liu, Y.-N.; Bao, J.; Li, R.; Ni, W.-T.; Tan, X.-Y.; Xu, Y.; Peng, L.-P.; Wang, X.-R.; Zeng, Y.-M.; et al. Clinical Features and Risk Factors Associated with Severe COVID-19 Patients in China. Chin. Med. J. 2021, 134, 944–953. [Google Scholar] [CrossRef]
- Du, P.; Li, D.; Wang, A.; Shen, S.; Ma, Z.; Li, X. A Systematic Review and Meta-Analysis of Risk Factors Associated with Severity and Death in COVID-19 Patients. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 6660930. [Google Scholar] [CrossRef]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors Associated with COVID-19-Related Death Using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.-S.; Kreidl, M.; et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020, 11, 11–24.e4. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 Infection Alters Kynurenine and Fatty Acid Metabolism, Correlating with IL-6 Levels and Renal Status. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Kimhofer, T.; Lodge, S.; Whiley, L.; Gray, N.; Loo, R.L.; Lawler, N.G.; Nitschke, P.; Bong, S.-H.; Morrison, D.L.; Begum, S.; et al. Integrative Modeling of Quantitative Plasma Lipoprotein, Metabolic, and Amino Acid Data Reveals a Multiorgan Pathological Signature of SARS-CoV-2 Infection. J. Proteome Res. 2020, 19, 4442–4454. [Google Scholar] [CrossRef]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Stefanoni, D.; Dzieciatkowska, M.; Issaian, A.; Nemkov, T.; Hill, R.C.; Francis, R.O.; Hudson, K.E.; Buehler, P.W.; Zimring, J.C.; et al. Evidence of Structural Protein Damage and Membrane Lipid Remodeling in Red Blood Cells from COVID-19 Patients. J. Proteome Res. 2020, 19, 4455–4469. [Google Scholar] [CrossRef]
- Piagnerelli, M.; Vanderelst, J.; Rousseau, A.; Monteyne, D.; Perez-Morga, D.; Biston, P.; Zouaoui Boudjeltia, K. Red Blood Cell Shape and Deformability in Patients with COVID-19 Acute Respiratory Distress Syndrome. Front. Physiol. 2022, 13, 849910. [Google Scholar] [CrossRef] [PubMed]
- Bouchla, A.; Kriebardis, A.G.; Georgatzakou, H.T.; Fortis, S.P.; Thomopoulos, T.P.; Lekkakou, L.; Markakis, K.; Gkotzias, D.; Panagiotou, A.; Papageorgiou, E.G.; et al. Red Blood Cell Abnormalities as the Mirror of SARS-CoV-2 Disease Severity: A Pilot Study. Front. Physiol. 2021, 12, 825055. [Google Scholar] [PubMed]
- Weisenhorn, E.M.M.; van T Erve, T.J.; Riley, N.M.; Hess, J.R.; Raife, T.J.; Coon, J.J. Multi-Omics Evidence for Inheritance of Energy Pathways in Red Blood Cells. Mol. Cell. Proteom. 2016, 15, 3614–3623. [Google Scholar] [CrossRef]
- van ’t Erve, T.J.; Wagner, B.A.; Martin, S.M.; Knudson, C.M.; Blendowski, R.; Keaton, M.; Holt, T.; Hess, J.R.; Buettner, G.R.; Ryckman, K.K.; et al. The Heritability of Metabolite Concentrations in Stored Human Red Blood Cells. Transfusion 2014, 54, 2055–2063. [Google Scholar] [CrossRef]
- Estimating Trait Heritability. Available online: https://www.nature.com/scitable/topicpage/estimating-trait-heritability-46889/ (accessed on 16 December 2022).
- Zhang, J.; Thio, C.H.L.; Gansevoort, R.T.; Snieder, H. Familial Aggregation of CKD and Heritability of Kidney Biomarkers in the General Population: The Lifelines Cohort Study. Am. J. Kidney Dis. 2021, 77, 869–878. [Google Scholar] [CrossRef]
- Rao, F.; Schork, A.J.; Maihofer, A.X.; Nievergelt, C.M.; Marcovina, S.M.; Miller, E.R.; Witztum, J.L.; O’Connor, D.T.; Tsimikas, S. Heritability of Biomarkers of Oxidized Lipoproteins: Twin Pair Study. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1704–1711. [Google Scholar] [CrossRef]
- Loomis, S.J.; Tin, A.; Coresh, J.; Boerwinkle, E.; Pankow, J.S.; Köttgen, A.; Selvin, E.; Duggal, P. Heritability Analysis of Nontraditional Glycemic Biomarkers in the Atherosclerosis Risk in Communities Study. Genet. Epidemiol. 2019, 43, 776–785. [Google Scholar] [CrossRef]
- Povel, C.M.; Boer, J.M.A.; Feskens, E.J.M. Shared Genetic Variance between the Features of the Metabolic Syndrome: Heritability Studies. Mol. Genet. Metab. 2011, 104, 666–669. [Google Scholar] [CrossRef] [PubMed]
- van ‘t Erve, T.J.; Doskey, C.M.; Wagner, B.A.; Hess, J.R.; Darbro, B.W.; Ryckman, K.K.; Murray, J.C.; Raife, T.J.; Buettner, G.R. Heritability of Glutathione and Related Metabolites in Stored Red Blood Cells. Free. Radic. Biol. Med. 2014, 76, 107–113. [Google Scholar] [CrossRef] [PubMed]
- van ‘t Erve, T.J.; Wagner, B.A.; Ryckman, K.K.; Raife, T.J.; Buettner, G.R. The Concentration of Glutathione in Human Erythrocytes Is a Heritable Trait. Free. Radic. Biol. Med. 2013, 65, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Van ’t Erve, T.J.; Wagner, B.A.; Martin, S.M.; Knudson, C.M.; Blendowski, R.; Keaton, M.; Holt, T.; Hess, J.R.; Buettner, G.R.; Ryckman, K.K.; et al. The Heritability of Hemolysis in Stored Human Red Blood Cells. Transfusion 2015, 55, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Chong, J.; Xia, J. Comprehensive Meta-Analysis of COVID-19 Global Metabolomics Datasets. Metabolites 2021, 11, 44. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn Errors of Type I IFN Immunity in Patients with Life-Threatening COVID-19. Science 2020, 370. [Google Scholar] [CrossRef]
- Zhang, Y.; Qin, L.; Zhao, Y.; Zhang, P.; Xu, B.; Li, K.; Liang, L.; Zhang, C.; Dai, Y.; Feng, Y.; et al. Interferon-Induced Transmembrane Protein 3 Genetic Variant Rs12252-C Associated With Disease Severity in Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 34–37. [Google Scholar] [CrossRef]
- Kuo, C.-L.; Pilling, L.C.; Atkins, J.L.; Masoli, J.A.H.; Delgado, J.; Kuchel, G.A.; Melzer, D. APOE E4 Genotype Predicts Severe COVID-19 in the UK Biobank Community Cohort. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 2231–2232. [Google Scholar] [CrossRef]
- Williams, F.M.K.; Freidin, M.B.; Mangino, M.; Couvreur, S.; Visconti, A.; Bowyer, R.C.E.; Le Roy, C.I.; Falchi, M.; Mompeó, O.; Sudre, C.; et al. Self-Reported Symptoms of COVID-19, Including Symptoms Most Predictive of SARS-CoV-2 Infection, Are Heritable. Twin Res. Hum. Genet. 2021, 23, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Forbester, J.L.; Humphreys, I.R. Genetic Influences on Viral-Induced Cytokine Responses in the Lung. Mucosal Immunol. 2021, 14, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Caterino, M.; Fedele, R.; Cevenini, A.; Pontillo, M.; Barra, L.; Ruoppolo, M. COVIDomics: The Proteomic and Metabolomic Signatures of COVID-19. Int. J. Mol. Sci. 2022, 23, 2414. [Google Scholar] [CrossRef]
- Li, T.; Ning, N.; Li, B.; Luo, D.; Qin, E.; Yu, W.; Wang, J.; Yang, G.; Nan, N.; He, Z.; et al. Longitudinal Metabolomics Reveals Ornithine Cycle Dysregulation Correlates with Inflammation and Coagulation in COVID-19 Severe Patients. Front. Microbiol. 2021, 12, 723818. [Google Scholar] [CrossRef] [PubMed]
- Páez-Franco, J.C.; Torres-Ruiz, J.; Sosa-Hernández, V.A.; Cervantes-Díaz, R.; Romero-Ramírez, S.; Pérez-Fragoso, A.; Meza-Sánchez, D.E.; Germán-Acacio, J.M.; Maravillas-Montero, J.L.; Mejía-Domínguez, N.R.; et al. Metabolomics Analysis Reveals a Modified Amino Acid Metabolism That Correlates with Altered Oxygen Homeostasis in COVID-19 Patients. Sci. Rep. 2021, 11, 6350. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Liu, C.; Li, D.; Huang, Q.; Liu, D.; Zhang, Y.; Ye, C.; Zhou, D.; Wang, Y.; Tan, Y.; et al. Metabolomic Analyses Reveal New Stage-Specific Features of COVID-19. Eur. Respir. J. 2022, 59, 2100284. [Google Scholar] [CrossRef] [PubMed]
- Caterino, M.; Costanzo, M.; Fedele, R.; Cevenini, A.; Gelzo, M.; Di Minno, A.; Andolfo, I.; Capasso, M.; Russo, R.; Annunziata, A.; et al. The Serum Metabolome of Moderate and Severe COVID-19 Patients Reflects Possible Liver Alterations Involving Carbon and Nitrogen Metabolism. Int. J. Mol. Sci. 2021, 22, 9548. [Google Scholar] [CrossRef]
- Xiao, N.; Nie, M.; Pang, H.; Wang, B.; Hu, J.; Meng, X.; Li, K.; Ran, X.; Long, Q.; Deng, H.; et al. Integrated Cytokine and Metabolite Analysis Reveals Immunometabolic Reprogramming in COVID-19 Patients with Therapeutic Implications. Nat. Commun. 2021, 12, 1618. [Google Scholar] [CrossRef]
- Kelly, B.; Pearce, E.L. Amino Assets: How Amino Acids Support Immunity. Cell Metab. 2020, 32, 154–175. [Google Scholar] [CrossRef]
- Nie, A.; Sun, B.; Fu, Z.; Yu, D. Roles of Aminoacyl-TRNA Synthetases in Immune Regulation and Immune Diseases. Cell Death Dis. 2019, 10, 901. [Google Scholar] [CrossRef]
- Gilroy, T.E.; Brewer, G.J.; Sing, C.F. Genetic Control of Glycolysis in Human Erythrocytes. Genetics 1980, 94, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Hagenbeek, F.A.; Pool, R.; van Dongen, J.; Draisma, H.H.M.; Jan Hottenga, J.; Willemsen, G.; Abdellaoui, A.; Fedko, I.O.; den Braber, A.; Visser, P.J.; et al. Heritability Estimates for 361 Blood Metabolites across 40 Genome-Wide Association Studies. Nat. Commun. 2020, 11, 39. [Google Scholar] [CrossRef] [PubMed]

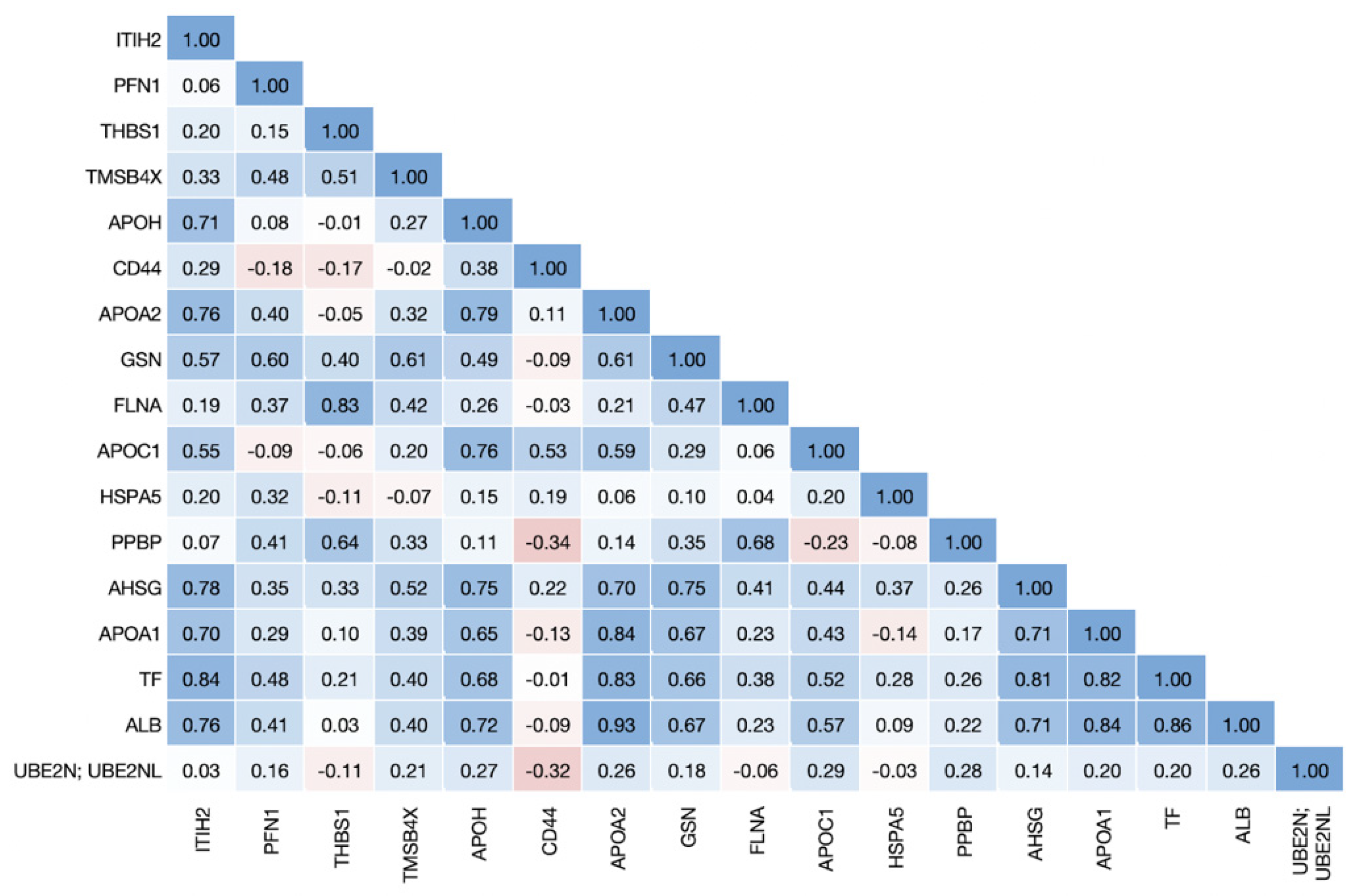
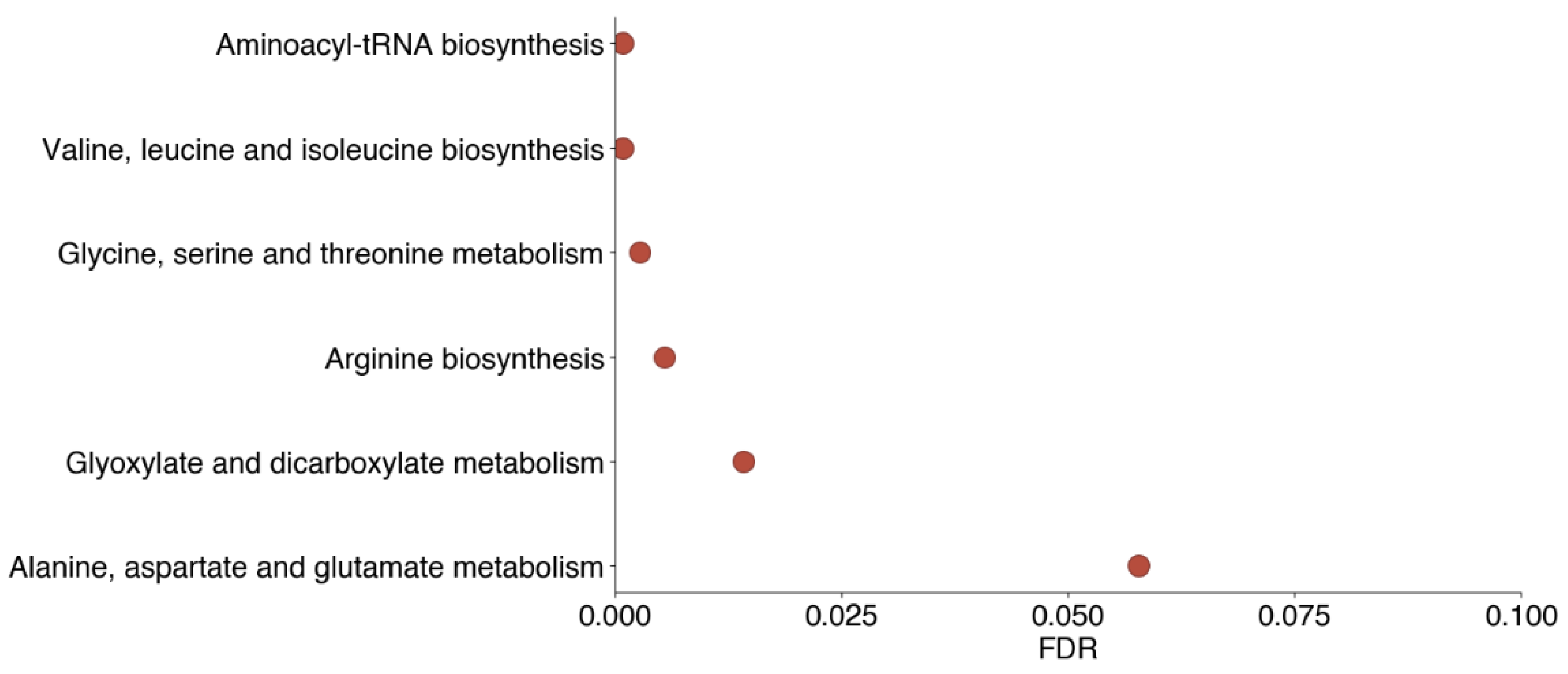

| Metabolite Name | Heritability | Non-COVID vs. Healthy | Non-Severe vs. Healthy | Severe vs. Healthy | Severe vs. Non-Severe |
|---|---|---|---|---|---|
| glutamate | 90.94 | ↓ | ↓ | ||
| thymol sulfate | 90.01 | ↓ | ↓ | ↓ | |
| betaine | 87.62 | ↓ | |||
| cysteinylglycine | 85.05 | ↓ | ↓ | ↓ | |
| arginine | 83.30 | ↓ | ↓ | ||
| spermidine | 82.00 | ↑ | |||
| alpha-tocopherol | 79.38 | ↓ | ↓ | ↓ | |
| isobutyrylcarnitine | 79.20 | ↓ | |||
| 1-stearoylglycerophosphoserine | 78.89 | ↓ | |||
| taurine | 78.70 | ↓ | ↓ | ||
| adenosine | 77.46 | ↓ | |||
| 3-carboxy-4-methyl-5-propyl-2-furanpropanoate (CMPF) | 73.68 | ↓ | |||
| gamma-glutamylglutamate | 72.49 | ↓ | ↓ | ||
| threonate | 72.35 | ↓ | ↓ | ||
| N-acetylmethionine | 72.07 | ↓ | ↓ | ||
| methionine | 71.34 | ↓ | ↓ | ||
| propionylcarnitine | 71.26 | ↓ | |||
| fumarate | 71.16 | ↑ | ↓ | ||
| threonylphenylalanine | 71.08 | ↓ | |||
| 13-HODE + 9-HODE | 67.34 | ↑ | |||
| glycine | 66.05 | ↓ | ↓ | ||
| isovalerylcarnitine | 65.95 | ↑ | |||
| glycodeoxycholate | 65.33 | ↓ | ↓ | ||
| leucylglycine | 64.78 | ↓ | |||
| threonine | 64.40 | ↓ | ↓ | ||
| 1-arachidonoylglycerophosphocholine (20:4n6) | 62.15 | ↓ | |||
| inosine | 62.14 | ↑ | |||
| pyruvate | 60.80 | ↓ | |||
| serine | 60.07 | ↑ | ↓ | ↓ | |
| 4-vinylphenol sulfate | 59.82 | ↓ | ↓ | ↓ | |
| glycerol 3-phosphate (G3P) | 59.03 | ↓ | ↓ | ||
| N-palmitoyl-D-erythro-sphingosine | 58.24 | ↑ | ↑ | ↑ | ↑ |
| choline | 55.80 | ↓ | ↓ | ||
| epiandrosterone sulfate | 55.53 | ↓ | |||
| urea | 54.57 | ↓ | |||
| stearate (18:0) | 54.02 | ↓ | ↓ | ||
| N-delta-acetylornithine | 53.79 | ↓ | ↓ | ||
| docosapentaenoate (n6 DPA; 22:5n6) | 52.87 | ↓ | |||
| 1,5-anhydroglucitol (1,5-AG) | 52.80 | ↓ | |||
| N-acetylmethionine sulfoxide | 52.70 | ↑ | |||
| maltose | 50.65 | ↑ | ↑ | ↑ | |
| alanine | 50.60 | ↓ | ↓ | ||
| 3-methylhistidine | 50.17 | ↓ | |||
| hypotaurine | 49.59 | ↓ | ↓ | ||
| phenylalanine | 48.44 | ↑ | |||
| succinylcarnitine | 48.24 | ↓ | |||
| valine | 47.73 | ↓ | ↓ | ||
| urate | 47.44 | ↓ | |||
| 2-hydroxybutyrate (AHB) | 47.31 | ↑ | ↑ | ||
| 3-methyl-2-oxovalerate | 45.36 | ↓ | ↓ | ||
| 1-linoleoylglycerophosphocholine (18:2n6) | 44.70 | ↓ | ↓ | ↓ | |
| phenol sulfate | 44.41 | ↓ | |||
| 3-methyl-2-oxobutyrate | 44.07 | ↓ | ↓ | ||
| margarate (17:0) | 43.44 | ↓ | |||
| malate | 43.35 | ↑ | ↓ | ||
| stachydrine | 42.36 | ↓ | |||
| orotate | 41.59 | ↑ | |||
| valylglycine | 41.25 | ↓ | |||
| myo-inositol | 38.95 | ↓ | ↑ | ||
| oleate (18:1n9) | 38.62 | ↑ | |||
| nicotinamide | 38.10 | ↓ | |||
| N6-acetyllysine | 37.20 | ↑ | ↓ | ↓ | |
| 3-indoxyl sulfate | 36.66 | ↓ | |||
| cholesterol | 35.88 | ↓ |
| Protein | Heritability | Non-COVID vs. Healthy | Non-Severe vs. Healthy | Severe vs. Healthy | Severe vs. Non-Severe | Upregulated in COVID |
|---|---|---|---|---|---|---|
| CD59 | 75.64 | ↑ | ||||
| ITIH2 | 74.19 | ↓ | ↓ | ↓ | ||
| HP; HPR | 59.44 | ↑ | ↑ | ↑ | yes | |
| CFH | 57.00 | yes | ||||
| PFN1 | 52.35 | ↓ | ||||
| C4B; C4A | 51.68 | ↑ | ||||
| THBS1 | 51.44 | ↓ | ↓ | ↓ | ||
| HIST2H2BF | 51.29 | ↑ | ||||
| SERPINC1 | 41.57 | ↑ | ||||
| TMSB4X | 40.56 | ↓ | ||||
| FGG | 40.46 | yes | ||||
| HIST1H4A | 37.44 | ↑ | ||||
| DEFA3; DEFA1 | 35.25 | ↑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haj, A.K.; Hasan, H.; Raife, T.J. Heritability of Protein and Metabolite Biomarkers Associated with COVID-19 Severity: A Metabolomics and Proteomics Analysis. Biomolecules 2023, 13, 46. https://doi.org/10.3390/biom13010046
Haj AK, Hasan H, Raife TJ. Heritability of Protein and Metabolite Biomarkers Associated with COVID-19 Severity: A Metabolomics and Proteomics Analysis. Biomolecules. 2023; 13(1):46. https://doi.org/10.3390/biom13010046
Chicago/Turabian StyleHaj, Amelia K., Haytham Hasan, and Thomas J. Raife. 2023. "Heritability of Protein and Metabolite Biomarkers Associated with COVID-19 Severity: A Metabolomics and Proteomics Analysis" Biomolecules 13, no. 1: 46. https://doi.org/10.3390/biom13010046
APA StyleHaj, A. K., Hasan, H., & Raife, T. J. (2023). Heritability of Protein and Metabolite Biomarkers Associated with COVID-19 Severity: A Metabolomics and Proteomics Analysis. Biomolecules, 13(1), 46. https://doi.org/10.3390/biom13010046





