Nucleolar Architecture Is Modulated by a Small Molecule, the Inositol Pyrophosphate 5-InsP7
Abstract
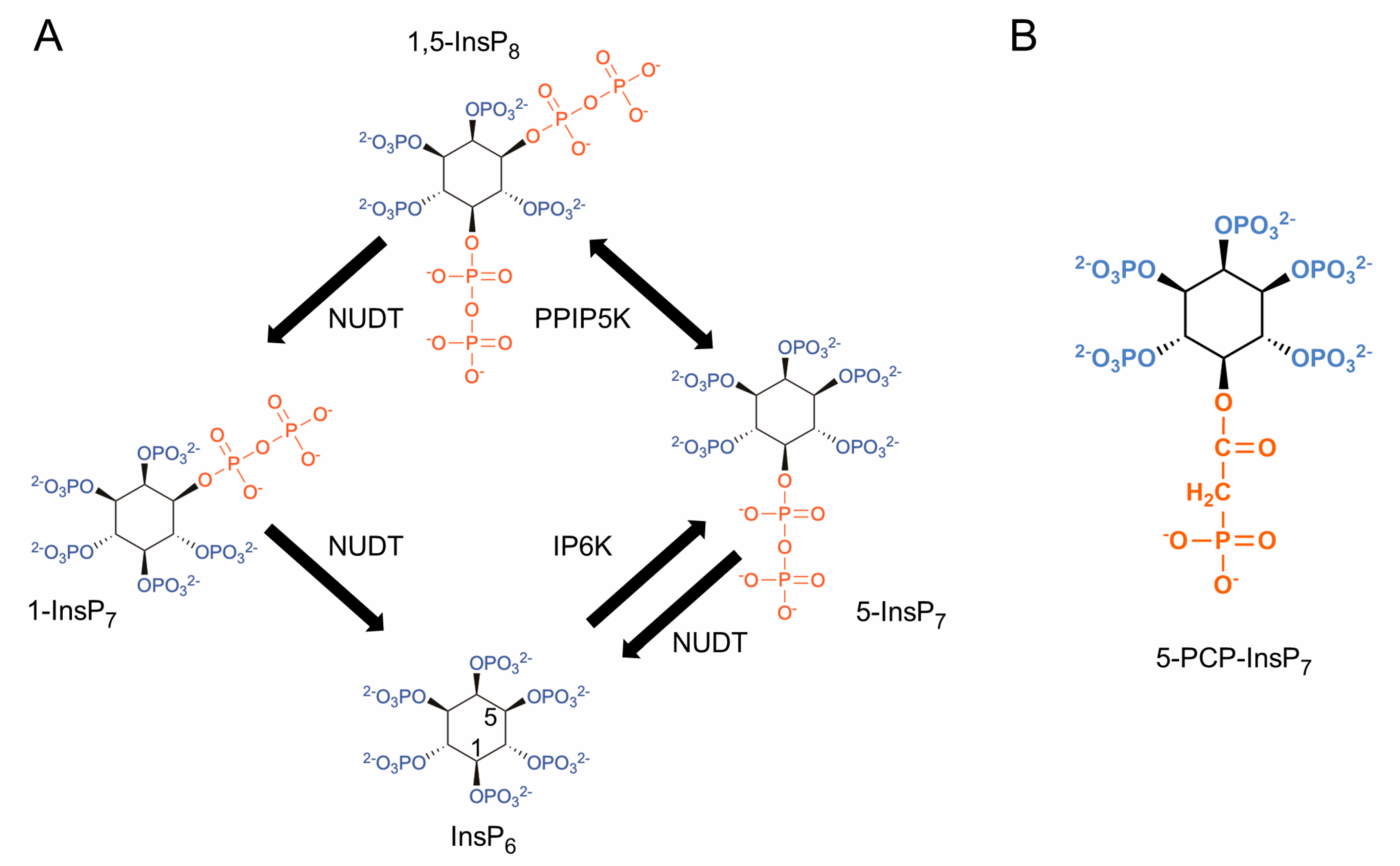
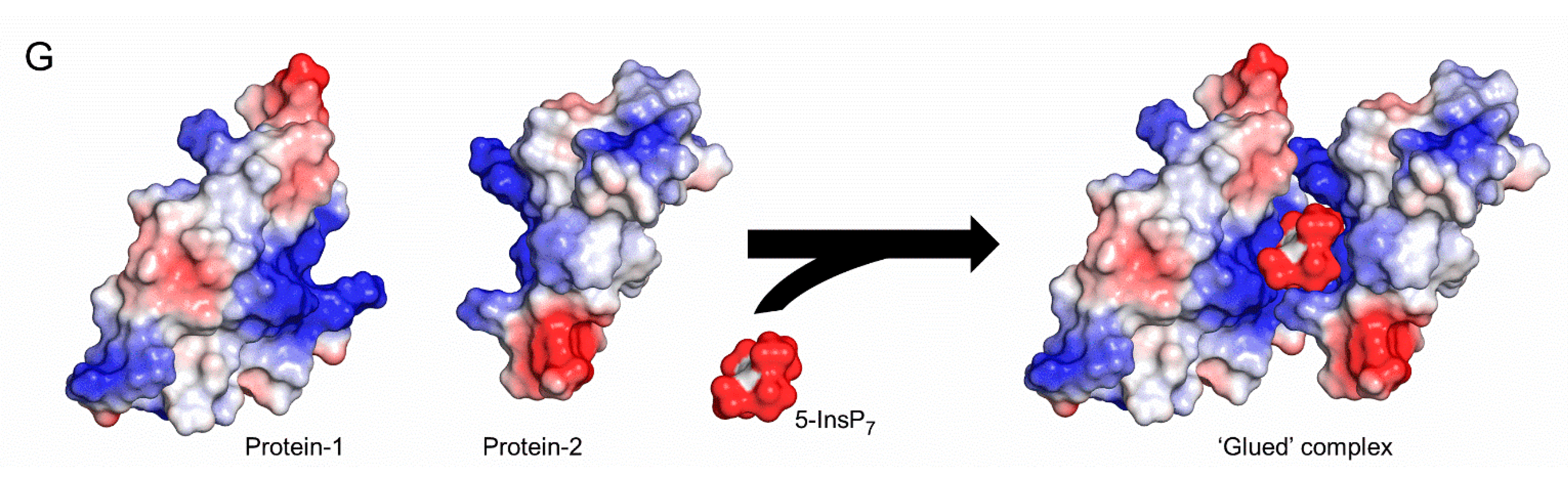
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Loading of 5-PCP-InsP7 into Cells Using Liposomes
2.3. Generation of NUDT3 KO Cells
2.4. Lentiviral Transduction of HCT116 Cells
2.5. Western Blotting and Analysis
2.6. Confocal Microscopy
2.7. Sucrose Density Gradient Fractionation of Ribosome Subunits
2.8. Total RNA Analysis by Automated Electrophoresis
3. Results
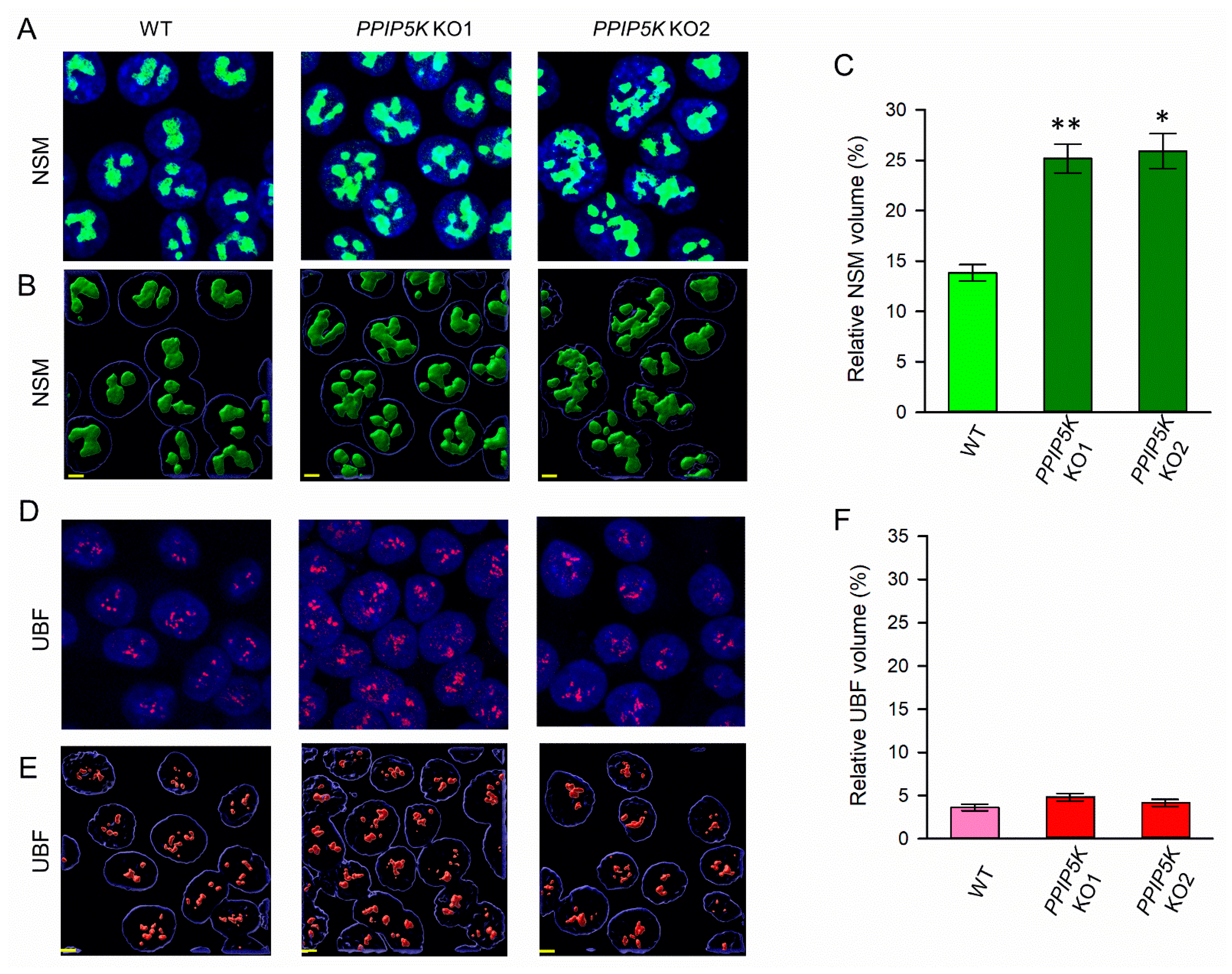
3.1. Nucleolar Granular Volume Is Elevated in PPIP5K KO HCT116 Cells
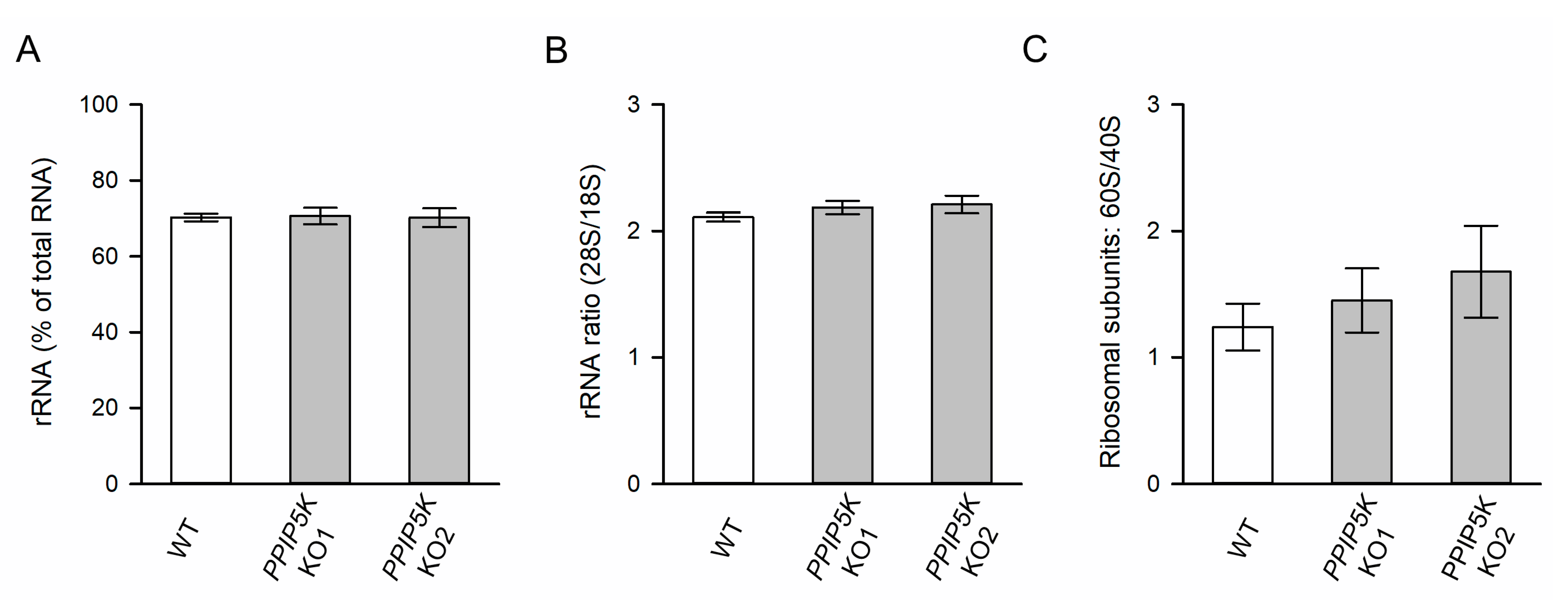
3.2. The KO of PPIP5Ks Does Not Affect rRNA Synthesis
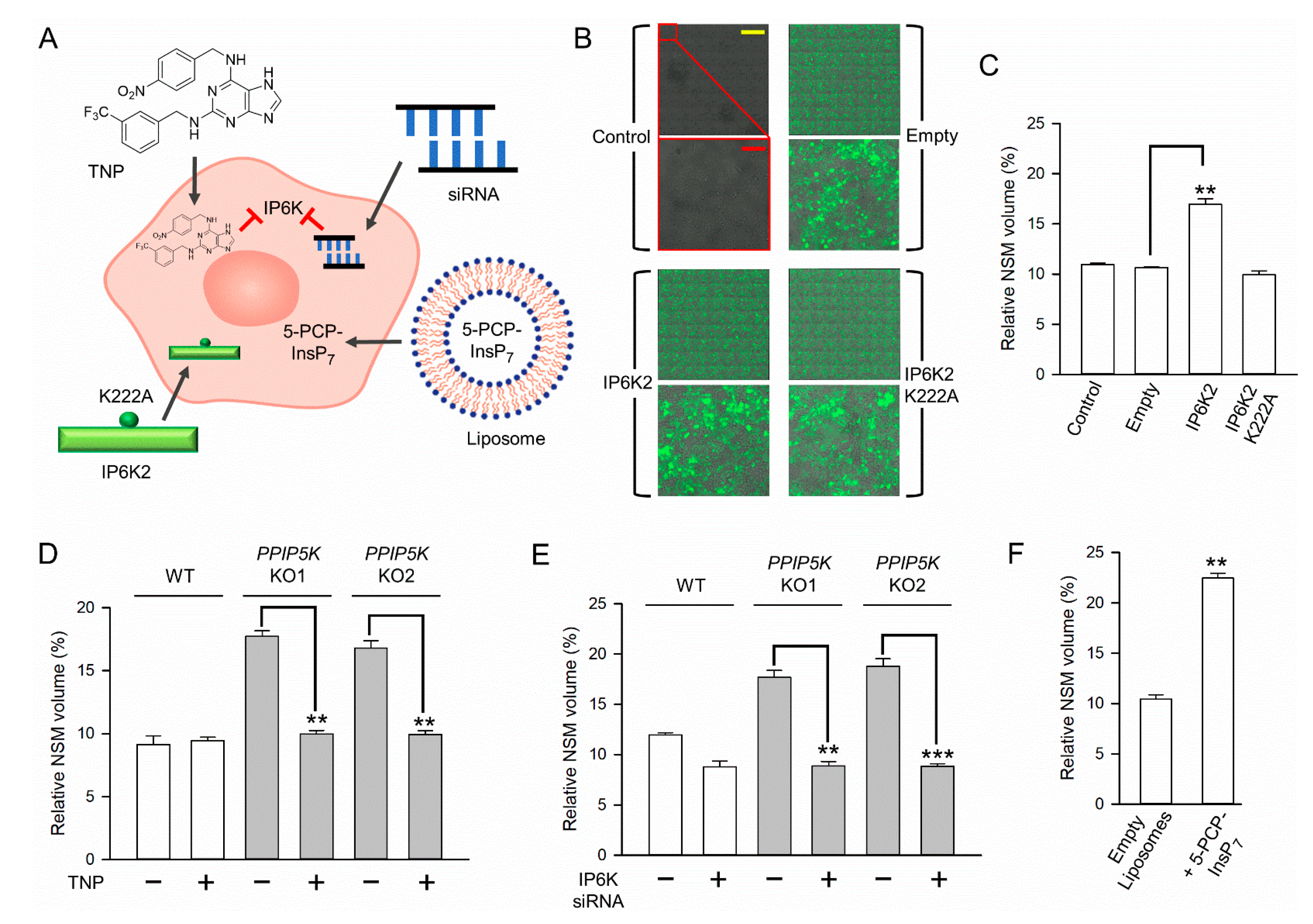
3.3. The Impact of NUDT3 KO upon Nucleolar Granular Volume
3.4. The Impact upon Nucleolar Granular Volume of Changes in Cellular IP6K Activity
3.5. Nucleolar Granular Volume Is Expanded by Liposomal Delivery of Metabolically Stable 5-PCP-InsP7
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, S.; Kim, M.G.; Ahn, H.; Kim, S. Inositol Pyrophosphates: Signaling Molecules with Pleiotropic Actions in Mammals. Molecules 2020, 25, 2208. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Trung, M.; Furkert, D.; Fiedler, D. Versatile signaling mechanisms of inositol pyrophosphates. Curr. Opin. Chem. Biol. 2022, 70, 102177. [Google Scholar] [CrossRef] [PubMed]
- Randall, T.A.; Gu, C.; Li, X.; Wang, H.; Shears, S.B. A two-way switch for inositol pyrophosphate signaling: Evolutionary history and biological significance of a unique, bifunctional kinase/phosphatase. Adv. Biol. Regul. 2020, 75, 100674. [Google Scholar] [CrossRef]
- Gu, C.; Wilson, M.S.C.; Jessen, H.J.; Saiardi, A.; Shears, S.B. Inositol Pyrophosphate Profiling of two HCT116 Cell Lines Uncovers Variation in InsP8 Levels. PLoS ONE 2016, 11, e0165286. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lau, K.; Puschmann, R.; Harmel, R.K.; Zhang, Y.; Pries, V.; Gaugler, P.; Broger, L.; Dutta, A.K.; Jessen, H.J.; et al. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. eLife 2019, 8, e43582. [Google Scholar] [CrossRef]
- Li, X.; Gu, C.; Hostachy, S.; Sahu, S.; Wittwer, C.J.; Jessen, H.J.; Fiedler, D.; Wang, H.; Shears, S.B. Control of XPR1-dependent cellular phosphate efflux by InsP8 is an exemplar for functionally-exclusive inositol pyrophosphate signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3568–3574. [Google Scholar] [CrossRef]
- Alam, M.T.; Olin-Sandoval, V.; Stincone, A.; Keller, M.A.; Zelezniak, A.; Luisi, B.F.; Ralser, M. The self-inhibitory nature of metabolic networks and its alleviation through compartmentalization. Nat. Commun. 2017, 8, 16018. [Google Scholar] [CrossRef]
- Gu, C.; Liu, J.; Liu, X.; Zhang, H.; Luo, J.; Wang, H.; Locasale, J.W.; Shears, S.B. Metabolic supervision by PPIP5K, an inositol pyrophosphate kinase/phosphatase, controls proliferation of the HCT116 tumor cell line. Proc. Natl. Acad. Sci. USA 2021, 118, e2020187118. [Google Scholar] [CrossRef]
- Thota, S.G.; Unnikannan, C.P.; Thampatty, S.R.; Manorama, R.; Bhandari, R. Inositol pyrophosphates regulate RNA polymerase I-mediated rRNA transcription in Saccharomyces cerevisiae. Biochem. J. 2015, 466, 105–114. [Google Scholar] [CrossRef]
- Barker, C.J.; Leibiger, I.B.; Leibiger, B.; Berggren, P.O. Phosphorylated inositol compounds in beta -cell stimulus-response coupling. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E1113–E1122. [Google Scholar] [CrossRef]
- Zhang, X.; Li, N.; Zhang, J.; Zhang, Y.; Yang, X.; Luo, Y.; Zhang, B.; Xu, Z.; Zhu, Z.; Yang, X.; et al. 5-IP7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis. Nat. Metab. 2021, 3, 1400–1414. [Google Scholar] [CrossRef]
- Zhu, Q.; Ghosal, S.; Tyagi, R.; Chakraborty, A. Global IP6K1 deletion enhances temperature modulated energy expenditure which reduces carbohydrate and fat induced weight gain. Mol. Metab. 2016, 6, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.C.; Gao, Z.; Riley, A.M.; Furkert, D.; Wittwer, C.; Dutta, A.; Rojas, T.; Semenza, E.R.; Felder, R.A.; Pluznick, J.L.; et al. The inositol pyrophosphate 5-InsP7 drives sodium-potassium pump degradation by relieving an autoinhibitory domain of PI3K p85alpha. Sci. Adv. 2020, 6, eabb8542. [Google Scholar] [CrossRef]
- Sahu, S.; Wang, Z.; Jiao, X.; Gu, C.; Jork, N.; Wittwer, C.; Li, X.; Hostachy, S.; Fiedler, D.; Wang, H.; et al. InsP7 is a small-molecule regulator of NUDT3-mediated mRNA decapping and processing-body dynamics. Proc. Natl. Acad. Sci. USA 2020, 117, 19245–19253. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Bhandari, R. IP6K1 upregulates the formation of processing bodies by influencing protein-protein interactions on the mRNA cap. J. Cell Sci. 2021, 134, jcs259117. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, R.; Saiardi, A.; Ahmadibeni, Y.; Snowman, A.M.; Resnick, A.C.; Kristiansen, T.Z.; Molina, H.; Pandey, A.; Werner, J.K., Jr.; Juluri, K.R.; et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. USA 2007, 104, 15305–15310. [Google Scholar] [CrossRef]
- Saiardi, A.; Bhandari, A.; Resnick, R.; Cain, A.; Snowman, A.M.; Snyder, S.H. Inositol Pyrophosphate: Physiologic Phosphorylation of Proteins. Science 2004, 306, 2101–2105. [Google Scholar] [CrossRef]
- Ganguli, S.; Shah, A.; Hamid, A.; Singh, A.; Palakurti, R.; Bhandari, R. A high energy phosphate jump—From pyrophospho-inositol to pyrophospho-serine. Adv. Biol. Reg. 2019, 75, 100662. [Google Scholar] [CrossRef]
- Wu, M.; Chong, L.S.; Perlman, D.H.; Resnick, A.C.; Fiedler, D. Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc. Nat. Acad. Sci. USA 2016, 113, E6757–E6765. [Google Scholar] [CrossRef]
- Chakraborty, A.; Koldobskiy, M.A.; Bello, N.T.; Maxwell, M.; Potter, J.J.; Juluri, K.R.; Maag, D.; Kim, S.; Huang, A.S.; Dailey, M.J.; et al. Inositol pyrophosphates inhibit akt signaling, thereby regulating insulin sensitivity and weight gain. Cell 2010, 143, 897–910. [Google Scholar] [CrossRef]
- Pavlovic, I.; Thakor, D.T.; Vargas, J.R.; McKinlay, C.J.; Hauke, S.; Anstaett, P.; Camuna, R.C.; Bigler, L.; Gasser, G.; Schultz, C.; et al. Cellular delivery and photochemical release of a caged inositol-pyrophosphate induces PH-domain translocation in cellulo. Nat. Commun. 2016, 7, 10622. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, N.A.; Zaremba, A.; Janoshazi, A.K.; Weaver, J.D.; Shears, S.B. PPIP5K1 Modulates Ligand Competition Between Diphosphoinositol Polyphosphates and PtdIns(3,4,5)P3 for Polyphosphoinositide-Binding Domains. Biochem. J. 2013, 453, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Wild, R.; Gerasimaite, R.; Jung, J.Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jessen, H.J.; Poirier, Y.; Hothorn, M.; et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef]
- Zhou, J.; Hu, Q.; Xiao, X.; Yao, D.; Ge, S.; Ye, J.; Li, H.; Cai, R.; Liu, R.; Meng, F.; et al. Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure. Nat. Commun. 2021, 12, 7040. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanchez, U.; Tury, S.; Nicolas, G.; Wilson, M.S.; Jurici, S.; Ayrignac, X.; Courgnaud, V.; Saiardi, A.; Sitbon, M.; Battini, J.L. Interplay between PFBC-associated SLC20A2 and XPR1 phosphate transporters requires inositol polyphosphates for control of cellular phosphate homeostasis. J. Biol. Chem. 2020, 295, 9366–9378. [Google Scholar] [CrossRef]
- Furkert, D.; Hostachy, S.; Nadler-Holly, M.; Fiedler, D. Triplexed Affinity Reagents to Sample the Mammalian Inositol Pyrophosphate Interactome. Cell Chem. Biol. 2020, 27, 1097–1108. [Google Scholar] [CrossRef]
- Mantilla, B.S.; Kalesh, K.; Brown, N.W., Jr.; Fiedler, D.; Docampo, R. Affinity-based proteomics reveals novel targets of inositol pyrophosphate (5-IP7 )-dependent phosphorylation and binding in Trypanosoma cruzi replicative stages. Mol. Microbiol. 2021, 115, 986–1004. [Google Scholar] [CrossRef]
- Latonen, L. Phase-to-Phase With Nucleoli—Stress Responses, Protein Aggregation and Novel Roles of RNA. Front. Cell Neurosci. 2019, 13, 151. [Google Scholar] [CrossRef]
- Scott, M.S.; Boisvert, F.M.; McDowall, M.D.; Lamond, A.I.; Barton, G.J. Characterization and prediction of protein nucleolar localization sequences. Nucleic. Acids Res. 2010, 38, 7388–7399. [Google Scholar] [CrossRef]
- Dignon, G.L.; Best, R.B.; Mittal, J. Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef]
- Gu, C.; Nguyen, H.N.; Ganini, D.; Chen, Z.; Jessen, H.J.; Gu, Z.; Wang, H.; Shears, S.B. KO of 5-InsP7 kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc. Natl. Acad. Sci. USA 2017, 114, 11968–11973. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; DeRose, E.F.; London, R.E.; Shears, S.B. IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family. Nat. Commun. 2014, 5, 4178. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-H.; Haam, J.; Walker, M.; Scappini, E.; Naughton, J.; Martin, N.P. Production of Viral Constructs for Neuroanatomy, Calcium Imaging, and Optogenetics. Curr. Protoc. Neurosci. 2019, 87, e66. [Google Scholar] [CrossRef]
- Gordon, J.; Pillon, M.C.; Stanley, R.E. Nol9 Is a Spatial Regulator for the Human ITS2 Pre-rRNA Endonuclease-Kinase Complex. J. Mol. Biol. 2019, 431, 3771–3786. [Google Scholar] [CrossRef]
- Jeucken, K.C.M.; Koning, J.J.; van Hamburg, J.P.; Mebius, R.E.; Tas, S.W. A Straightforward Method for 3D Visualization of B Cell Clusters and High Endothelial Venules in Lymph Nodes Highlights Differential Roles of TNFRI and -II. Front. Immunol. 2021, 12, 699336. [Google Scholar] [CrossRef] [PubMed]
- Pillon, M.C.; Goslen, K.H.; Gordon, J.; Wells, M.L.; Williams, J.G.; Stanley, R.E. It takes two (Las1 HEPN endoribonuclease domains) to cut RNA correctly. J. Biol. Chem. 2020, 295, 5857–5870. [Google Scholar] [CrossRef]
- Harmel, R.K.; Puschmann, R.; Nguyen Trung, M.; Saiardi, A.; Schmieder, P.; Fiedler, D. Harnessing 13C-labeled myo-inositol to interrogate inositol phosphate messengers by NMR. Chem. Sci. 2019, 10, 5267–5274. [Google Scholar] [CrossRef]
- Avitabile, D.; Bailey, B.; Cottage, C.T.; Sundararaman, B.; Joyo, A.; McGregor, M.; Gude, N.; Truffa, S.; Zarrabi, A.; Konstandin, M.; et al. Nucleolar stress is an early response to myocardial damage involving nucleolar proteins nucleostemin and nucleophosmin. Proc. Natl. Acad. Sci. USA 2011, 108, 6145–6150. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339. [Google Scholar] [CrossRef]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Korner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef]
- Hua, L.; Yan, D.; Wan, C.; Hu, B. Nucleolus and Nucleolar Stress: From Cell Fate Decision to Disease Development. Cells 2022, 11, 3017. [Google Scholar] [CrossRef]
- Kilari, R.S.; Weaver, J.D.; Shears, S.B.; Safrany, S.T. Understanding inositol pyrophosphate metabolism and function: Kinetic characterization of the DIPPs. FEBS Lett. 2013, 587, 3464–3470. [Google Scholar] [CrossRef]
- Gu, C.; Nguyen, H.N.; Hofer, A.; Jessen, H.J.; Dai, X.; Wang, H.; Shears, S.B. The Significance of the Bifunctional Kinase/Phosphatase Activities of PPIP5Ks for Coupling Inositol Pyrophosphate Cell-Signaling to Cellular Phosphate Homeostasis. J. Biol. Chem. 2017, 292, 4544–4555. [Google Scholar] [CrossRef]
- Paramasivam, P.; Franke, C.; Stöter, M.; Höijer, A.; Bartesaghi, S.; Sabirsh, A.; Lindfors, L.; Arteta, M.Y.; Dahlén, A.; Bak, A.; et al. Endosomal escape of delivered mRNA from endosomal recycling tubules visualized at the nanoscale. J. Cell Biol. 2021, 221, e202110137. [Google Scholar] [CrossRef]
- Wu, M.; Chong, L.S.; Capolicchio, S.; Jessen, H.J.; Resnick, A.C.; Fiedler, D. Elucidating Diphosphoinositol Polyphosphate Function with Nonhydrolyzable Analogues. Angew. Chem. Int. Ed. 2014, 53, 9508–9511. [Google Scholar] [CrossRef]
- Lorenzo-Orts, L.; Couto, D.; Hothorn, M. Identity and functions of inorganic and inositol polyphosphates in plants. New Phytol. 2020, 225, 637–652. [Google Scholar] [CrossRef]
- Tiku, V.; Antebi, A. Nucleolar Function in Lifespan Regulation. Trends Cell Biol. 2018, 28, 662–672. [Google Scholar] [CrossRef]
- Orti, F.; Navarro, A.M.; Rabinovich, A.; Wodak, S.J.; Marino-Buslje, C. Insight into membraneless organelles and their associated proteins: Drivers, Clients and Regulators. Comput. Struct. Biotechnol. J. 2021, 19, 3964–3977. [Google Scholar] [CrossRef]
- Tiku, V.; Jain, C.; Raz, Y.; Nakamura, S.; Heestand, B.; Liu, W.; Späth, M.; Suchiman, H.E.D.; Müller, R.-U.; Slagboom, P.E.; et al. Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 2017, 8, 16083. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, C.; Liu, B.; Liang, D.; Qin, X.; Li, X.; Zhang, R.; Li, C.; Wang, H.; Sun, D.; et al. Inositol pyrophosphates mediate the effects of aging on bone marrow mesenchymal stem cells by inhibiting Akt signaling. Stem. Cell Res. 2014, 5, 33. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahu, S.; Gordon, J.; Gu, C.; Sobhany, M.; Fiedler, D.; Stanley, R.E.; Shears, S.B. Nucleolar Architecture Is Modulated by a Small Molecule, the Inositol Pyrophosphate 5-InsP7. Biomolecules 2023, 13, 153. https://doi.org/10.3390/biom13010153
Sahu S, Gordon J, Gu C, Sobhany M, Fiedler D, Stanley RE, Shears SB. Nucleolar Architecture Is Modulated by a Small Molecule, the Inositol Pyrophosphate 5-InsP7. Biomolecules. 2023; 13(1):153. https://doi.org/10.3390/biom13010153
Chicago/Turabian StyleSahu, Soumyadip, Jacob Gordon, Chunfang Gu, Mack Sobhany, Dorothea Fiedler, Robin E. Stanley, and Stephen B. Shears. 2023. "Nucleolar Architecture Is Modulated by a Small Molecule, the Inositol Pyrophosphate 5-InsP7" Biomolecules 13, no. 1: 153. https://doi.org/10.3390/biom13010153
APA StyleSahu, S., Gordon, J., Gu, C., Sobhany, M., Fiedler, D., Stanley, R. E., & Shears, S. B. (2023). Nucleolar Architecture Is Modulated by a Small Molecule, the Inositol Pyrophosphate 5-InsP7. Biomolecules, 13(1), 153. https://doi.org/10.3390/biom13010153






