LongBondEliminator: A Molecular Simulation Tool to Remove Ring Penetrations in Biomolecular Simulation Systems
Abstract
1. Introduction
2. Materials and Methods
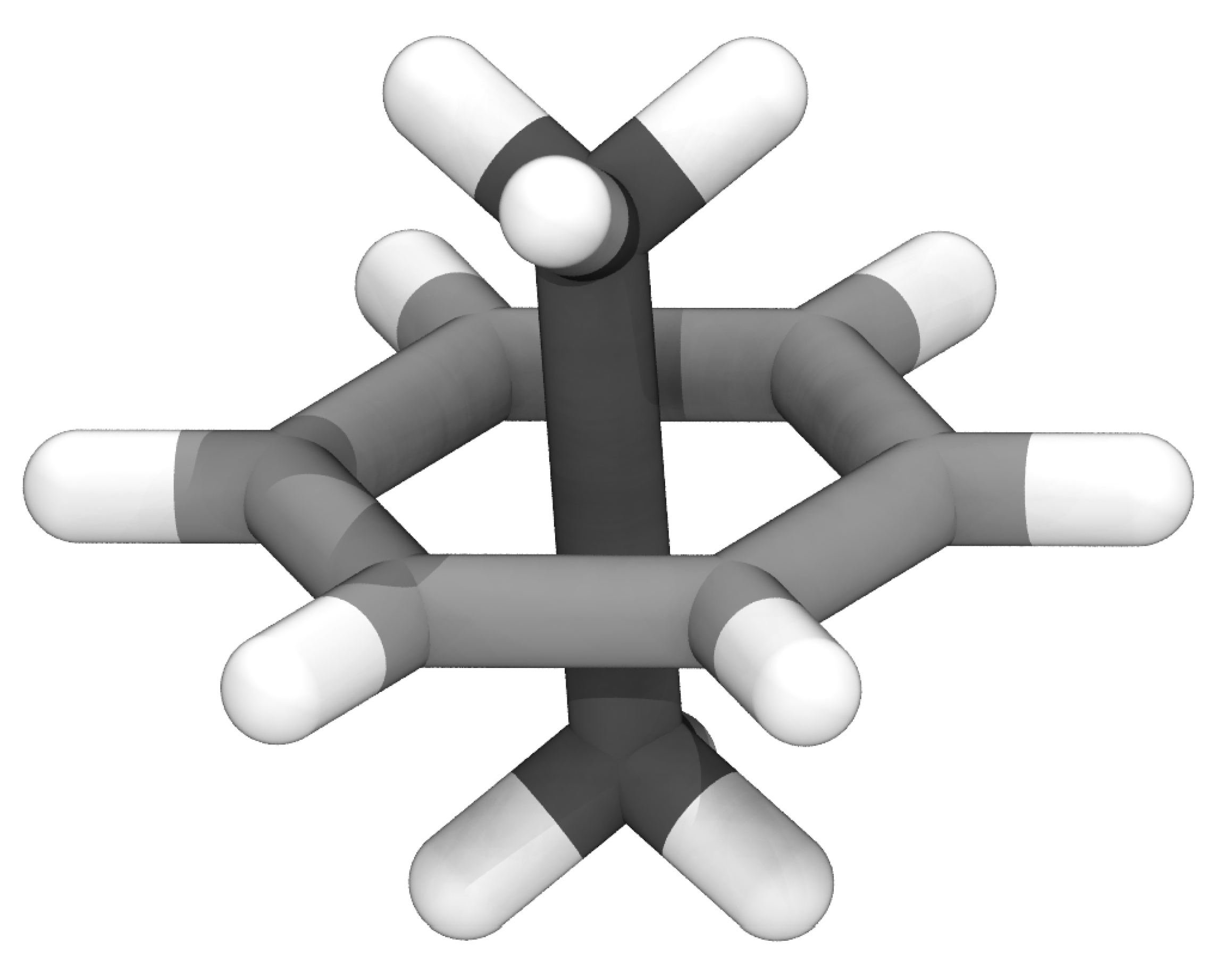
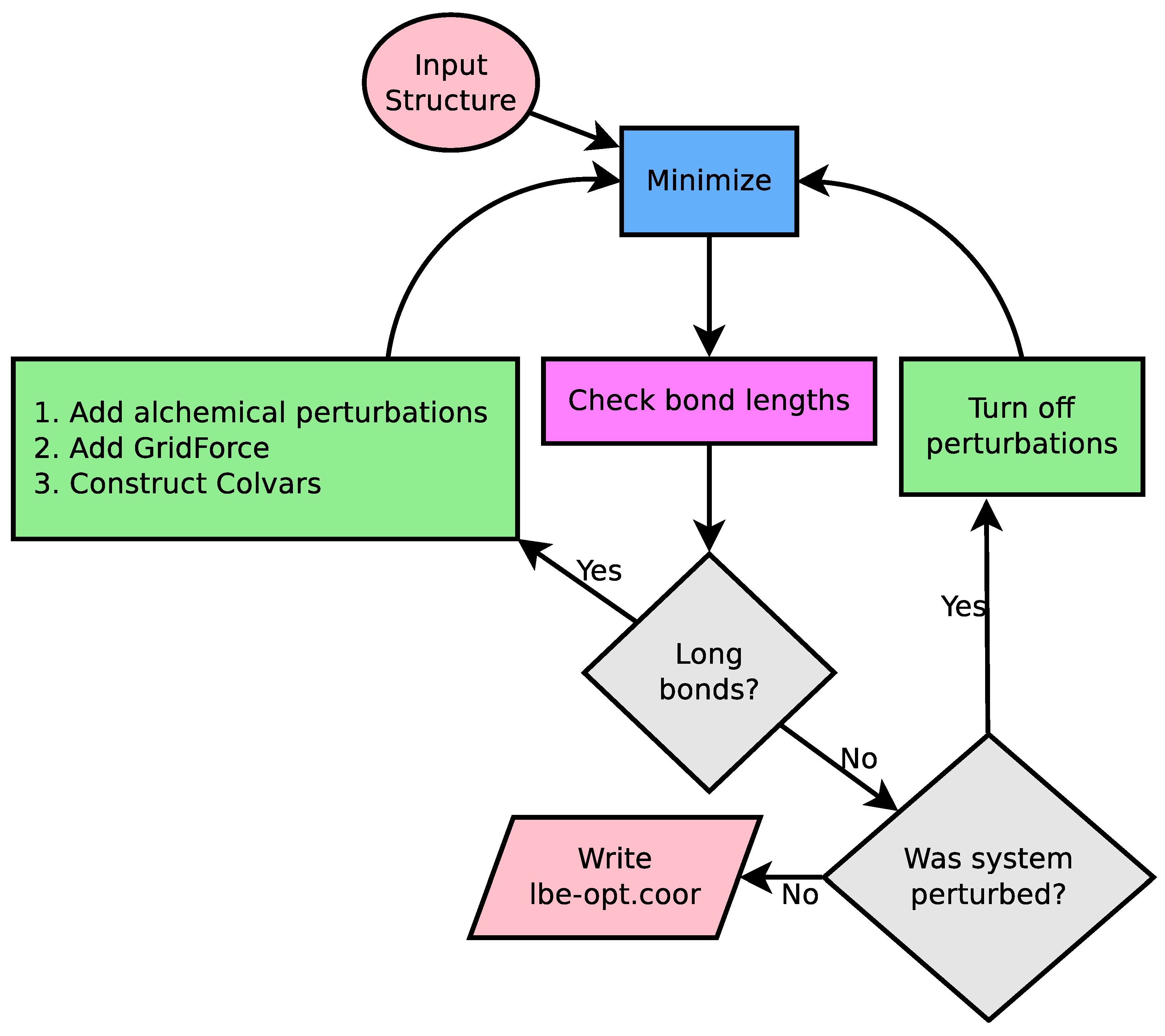
2.1. LongBondEliminator Minimization Algorithm
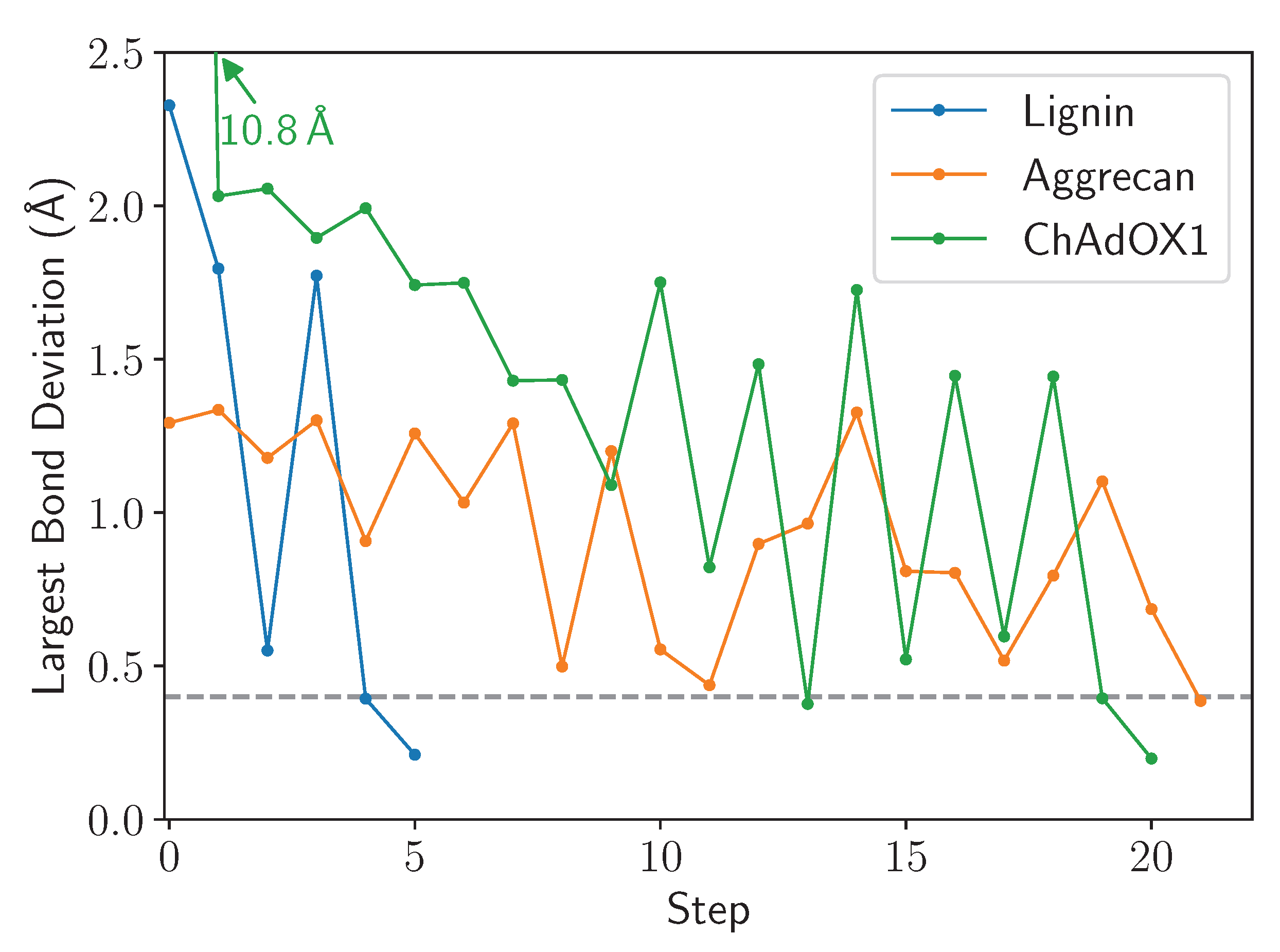
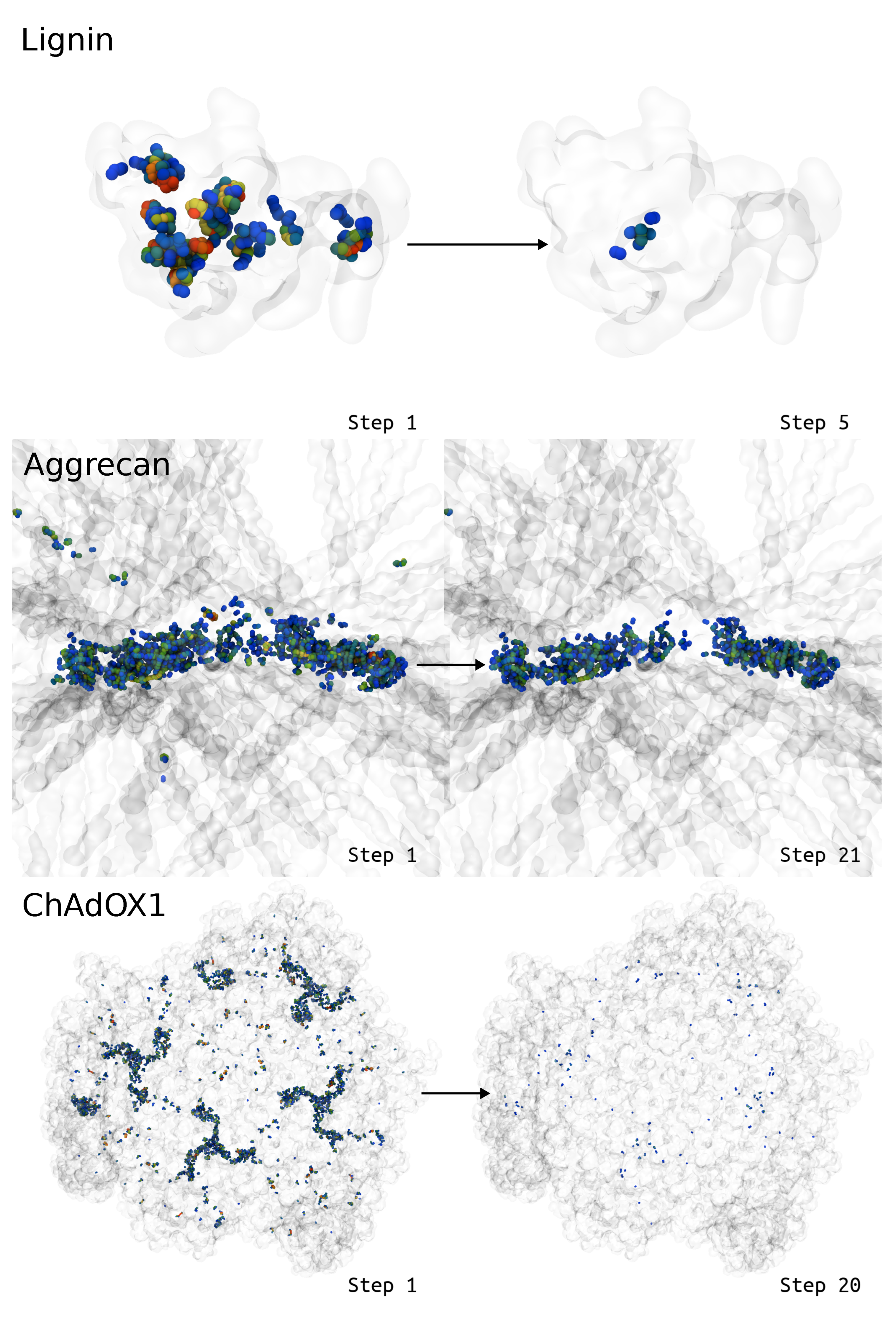
2.2. Test Cases
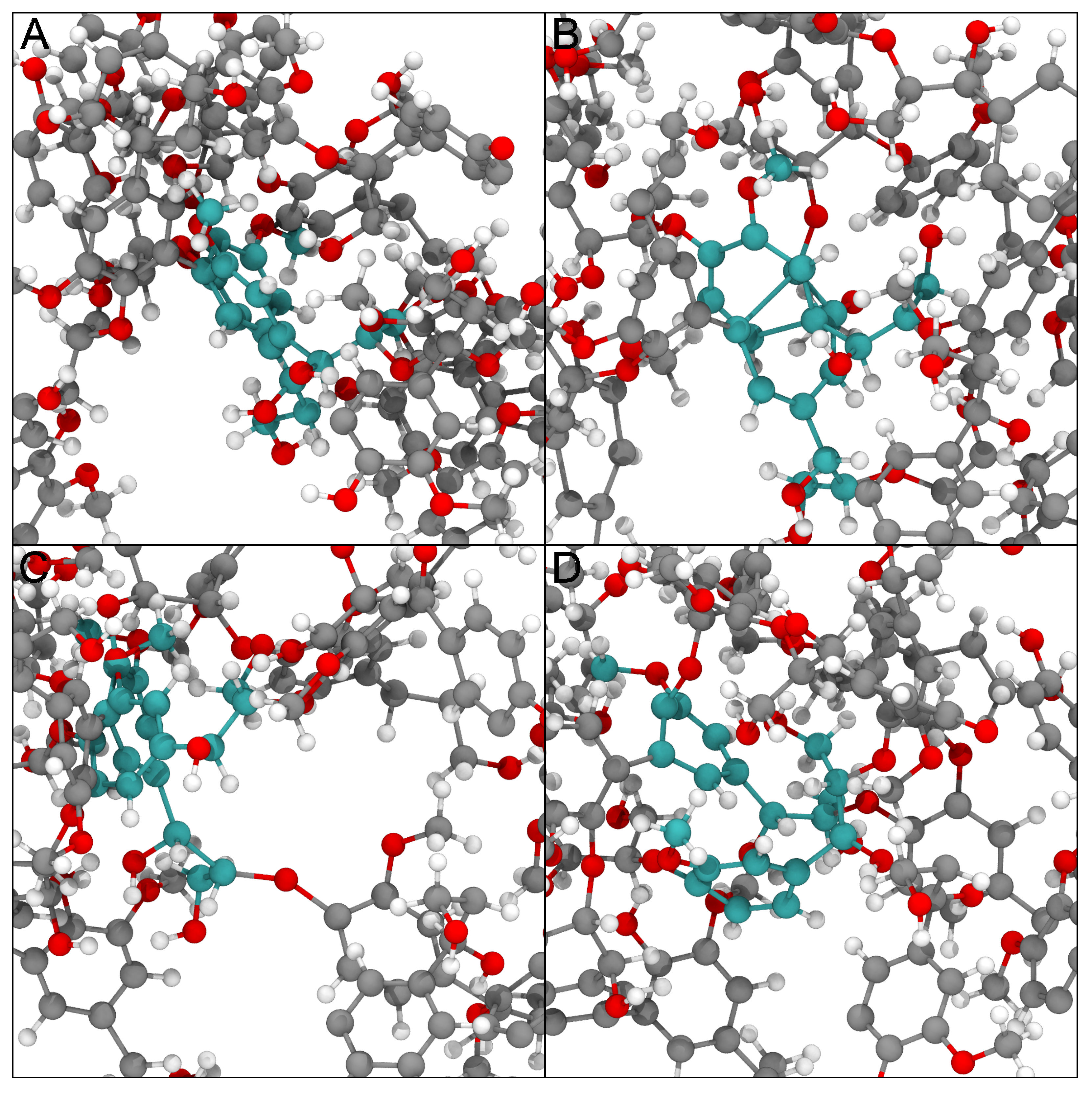
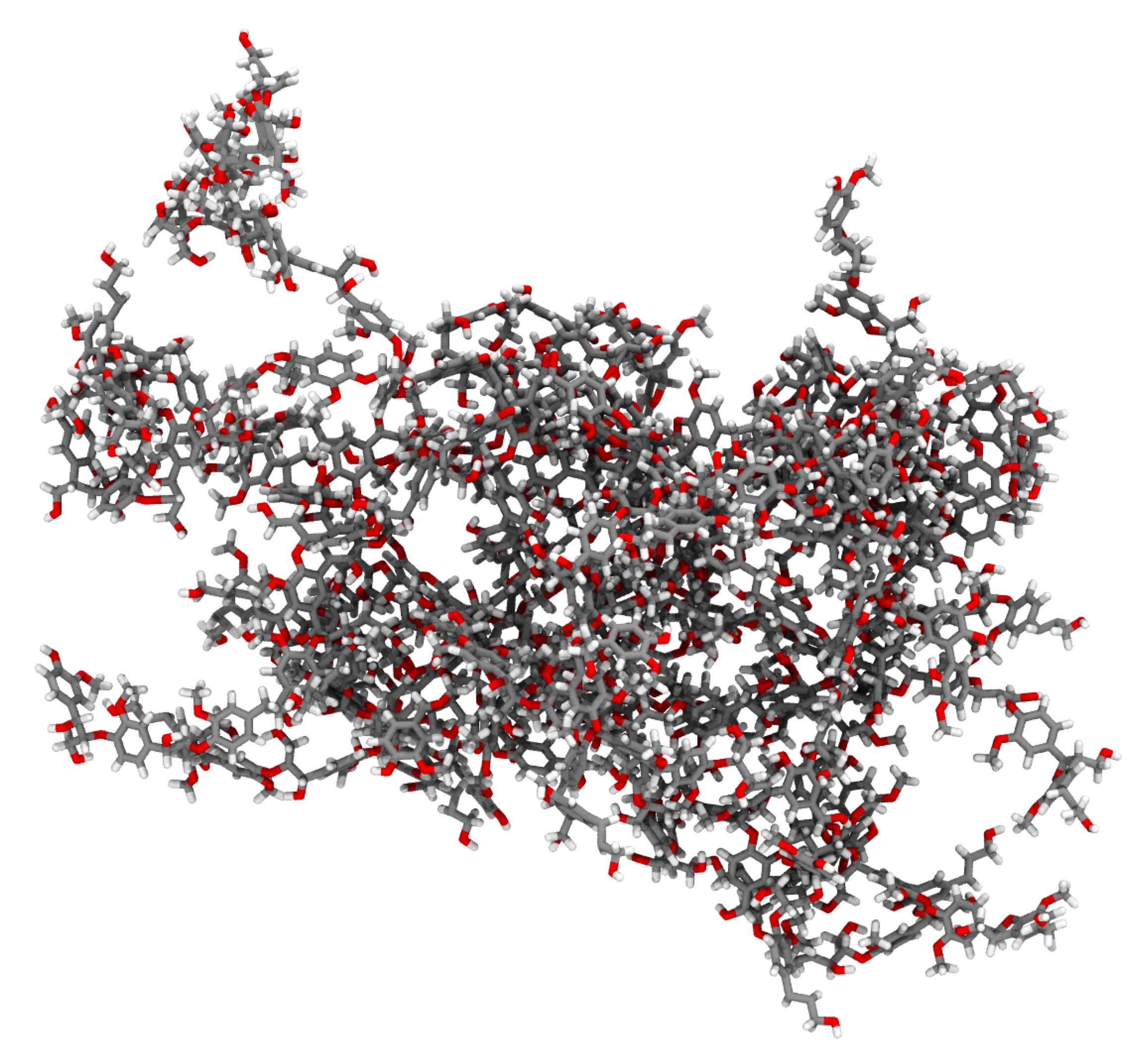
2.2.1. Lignin Polymer
2.2.2. Virus
2.2.3. Glycosylated Protein
2.3. User Guide
- minimize <namdconfigurationfile> <deletelines> <namdrunargs> <cutoff> <genchirality> <writeintermediates>, which goes through the logic to eliminate long bonds. This function is responsible for parsing the NAMD configuration file, and loading in the molecular simulation system that the NAMD configuration file refers to.
- namdconfigurationfile refers to a NAMD configuration file that minimizes the system, but reveals ring penetrations when executed. The existing simulation file will be retained during the subsequent minimization procedures, with the exception of the last few lines.
- deletelines refers to the number of lines that are deleted from the end of the NAMD configuration file. The assumption is that the last lines in the NAMD configuration file will minimize and run a simulation, which LBE will replace. 3 lines are the default.
- namdrunargs shows how you might run a NAMD simulation on your system, e.g., “namd2 +p16” would tell NAMD to use 16 processors when running the simulation. The default is “namd2”.
- cutoff is the metric used to determine when to terminate. Concretely, when the most stretched bond is less than the cutoff larger than the equilibrium bond distance for the interaction found within the force field, we assume the system to be minimized. The default is “0.4”, with units in Å.
- genchirality is either a 0 or a 1. If 1, LBE will generate improper dihedral constraints that are meant to maintain the chirality of potential chiral centers within the molecule. The default option is to generate the constraints.
- writeintermediates is also either a 0 or a 1. If 1, LBE will write a binary coordinate file for the end state at each step. The default option is not to write these intermediate files.
- getlargestdeviation <molid> <parameterlist> <filename>, which measures the most stretched bond in a molecular simulation system prepared in VMD.
- molid is the molecule identification number for a molecule loaded into VMD that you would like to measure.
- parameterlist is a Tcl list that contains all the parameter files necessary to simulate this molecular model.
- filename is the name of the file where the output should be written.
- tagdeviation <molid> <parameterlist> will store the sum of large deviations (>0.1 Å) into the user field for an atom at a particular frame from within a VMD trajectory.
- runloop is the function that implements the central runloop shown in Figure 2, and is intended only to be called from the minimize function.
- forcelongestbond is the function that creates a target location 10 Å away from the center of the bond vector between a stretched bond, and does so perfectly perpendicular to the bond vector.
- writenamdconf writes a new NAMD configuration file based on the existing NAMD configuration file.
- genchiralityextrabondsfile generates the extrabonds file that maintains chirality of the initial model system.
- minimizationscriptname determines what the new NAMD configuration file should be called.
- buildbondtable builds a table of bond parameters from a list of filenames.
- loadsystem loads in the structure and coordinates listed in the original NAMD configuration file.
- parseinputfile parses the NAMD configuration file for key parameters needed to implement the LBE algorithm.
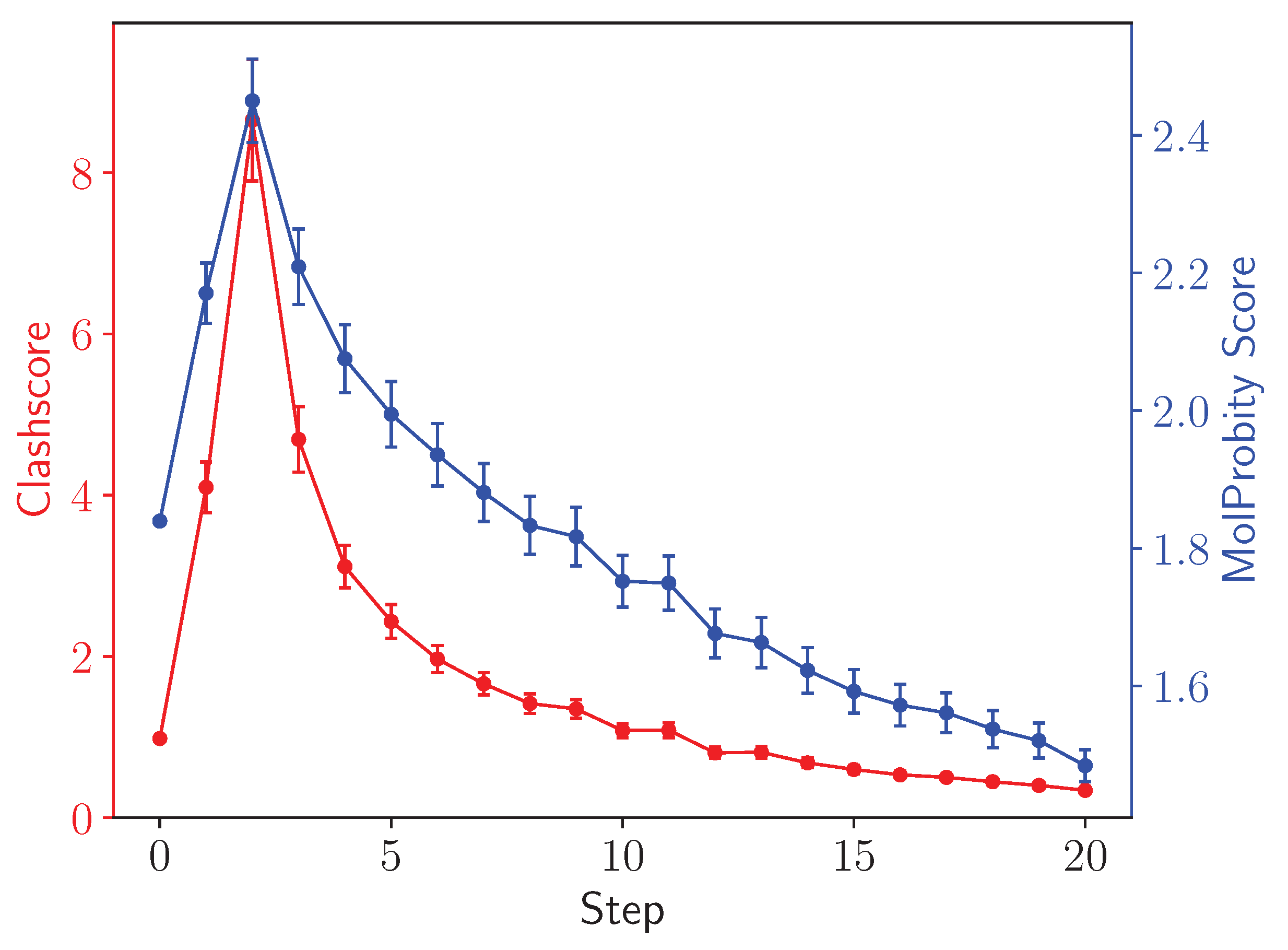
2.4. Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MD | Molecular dynamics |
| LBE | LongBondEliminator |
| FEP | Free Energy Perturbation |
| IGD | Interglobular domain |
| KSD | Keratan sulfate-rich domain |
| ChAd-Y25 | Chimpanzee adenovirus Y25 |
References
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Páll, S.; Zhmurov, A.; Bauer, P.; Abraham, M.; Lundborg, M.; Gray, A.; Hess, B.; Lindahl, E. Heterogeneous Parallelization and Acceleration of Molecular Dynamics Simulations in GROMACS. J. Chem. Phys. 2020, 153, 134110. [Google Scholar] [CrossRef] [PubMed]
- Iftimie, R.; Minary, P.; Tuckerman, M.E. Ab Initio Molecular Dynamics: Concepts, Recent Developments, and Future Trends. Proc. Natl. Acad. Sci. USA 2005, 102, 6654–6659. [Google Scholar] [CrossRef] [PubMed]
- Seritan, S.; Bannwarth, C.; Fales, B.S.; Hohenstein, E.G.; Isborn, C.M.; Kokkila-Schumacher, S.I.L.; Li, X.; Liu, F.; Luehr, N.; Snyder, J.W.; et al. TERACHEM: A Graphical Processing Unit-ACCELERATED Electronic Structure Package for LARGE-SCALE Ab Initio Molecular Dynamics. WIREs Comput. Mol. Sci. 2021, 11, e1494. [Google Scholar] [CrossRef]
- Chan, H.; Narayanan, B.; Cherukara, M.J.; Sen, F.G.; Sasikumar, K.; Gray, S.K.; Chan, M.K.Y.; Sankaranarayanan, S.K.R.S. Machine Learning Classical Interatomic Potentials for Molecular Dynamics from First-Principles Training Data. J. Phys. Chem. C 2019, 123, 6941–6957. [Google Scholar] [CrossRef]
- Doerr, S.; Majewski, M.; Pérez, A.; Krämer, A.; Clementi, C.; Noe, F.; Giorgino, T.; De Fabritiis, G. TorchMD: A Deep Learning Framework for Molecular Simulations. J. Chem. Theory Comput. 2021, 17, 2355–2363. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Stadmiller, S.S.; Pielak, G.J. Protein-Complex Stability in Cells and in Vitro under Crowded Conditions. Curr. Opin. Struct. Biol. 2021, 66, 183–192. [Google Scholar] [CrossRef]
- Feig, M.; Yu, I.; Wang, P.H.; Nawrocki, G.; Sugita, Y. Crowding in Cellular Environments at an Atomistic Level from Computer Simulations. J. Phys. Chem. B 2017, 121, 8009–8025. [Google Scholar] [CrossRef]
- Li, Y.; Liu, D.; Wang, Y.; Su, W.; Liu, G.; Dong, W. The Importance of Glycans of Viral and Host Proteins in Enveloped Virus Infection. Front. Immunol. 2021, 12, 638573. [Google Scholar] [CrossRef]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef]
- Prestegard, J.H. A Perspective on the PDB’s Impact on the Field of Glycobiology. J. Biol. Chem. 2021, 296, 100556. [Google Scholar] [CrossRef]
- Vant, J.W.; Sarkar, D.; Nguyen, J.; Baker, A.T.; Vermaas, J.V.; Singharoy, A. Exploring Cryo-Electron Microscopy with Molecular Dynamics. Biochem. Soc. Trans. 2022, 50, 569–581. [Google Scholar] [CrossRef]
- Gupta, C.; Sarkar, D.; Tieleman, D.P.; Singharoy, A. The Ugly, Bad, and Good Sories of Large-Scale Biomolecular Simulations. Curr. Opin. Struct. Biol. 2022, 73, 102338. [Google Scholar] [CrossRef]
- Maritan, M.; Autin, L.; Karr, J.; Covert, M.W.; Olson, A.J.; Goodsell, D.S. Building Structural Models of a Whole Mycoplasma Cell. J. Mol. Biol. 2022, 434, 167351. [Google Scholar] [CrossRef]
- Jung, J.; Nishima, W.; Daniels, M.; Bascom, G.; Kobayashi, C.; Adedoyin, A.; Wall, M.; Lappala, A.; Phillips, D.; Fischer, W.; et al. Scaling Molecular Dynamics beyond 100,000 Processor Cores for Large-scale Biophysical Simulations. J. Comput. Chem. 2019, 40, 1919–1930. [Google Scholar] [CrossRef]
- Ingólfsson, H.I.; Bhatia, H.; Zeppelin, T.; Bennett, W.F.D.; Carpenter, K.A.; Hsu, P.C.; Dharuman, G.; Bremer, P.T.; Schiøtt, B.; Lightstone, F.C.; et al. Capturing Biologically Complex Tissue-Specific Membranes at Different Levels of Compositional Complexity. J. Phys. Chem. B 2020, 124, 7819–7829. [Google Scholar] [CrossRef]
- Whitmore, E.K.; Vesenka, G.; Sihler, H.; Guvench, O. Efficient Construction of Atomic-Resolution Models of Non-Sulfated Chondroitin Glycosaminoglycan Using Molecular Dynamics Data. Biomolecules 2020, 10, 537. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Mayne, C.G.; Shinn, E.; Tajkhorshid, E. Assembly and Analysis of Cell-Scale Membrane Envelopes. J. Chem. Inf. Model. 2022, 62, 602–617. [Google Scholar] [CrossRef] [PubMed]
- Vermaas, J.V.; Dellon, L.D.; Broadbelt, L.J.; Beckham, G.T.; Crowley, M.F. Automated Transformation of Lignin Topologies into Atomic Structures with LigninBuilder. ACS Sustain. Chem. Eng. 2019, 7, 3443–3453. [Google Scholar] [CrossRef]
- Hagita, K.; Murashima, T. Molecular Dynamics Simulations of Ring Shapes on a Ring Fraction in Ring–Linear Polymer Blends. Macromolecules 2021, 54, 8043–8051. [Google Scholar] [CrossRef]
- Yasuda, Y.; Masumoto, T.; Mayumi, K.; Toda, M.; Yokoyama, H.; Morita, H.; Ito, K. Molecular Dynamics Simulation and Theoretical Model of Elasticity in Slide-Ring Gels. ACS Macro Lett. 2020, 9, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Licari, G.; Dehghani-Ghahnaviyeh, S.; Tajkhorshid, E. Membrane Mixer: A Toolkit for Efficient Shuffling of Lipids in Heterogeneous Biological Membranes. J. Chem. Inf. Model. 2022, 62, 986–996. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009, 97, 50–58. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Sarkar, D.; Kulke, M.; Vermaas, J.V. LongBondEliminator: A Molecular Simulation Tool to Remove Ring Penetrations in Biomolecular Simulation Systems. Zenodo 2023. [Google Scholar] [CrossRef]
- Fiorin, G.; Klein, M.L.; Hénin, J. Using Collective Variables to Drive Molecular Dynamics Simulations. Mol. Phys. 2013, 111, 3345–3362. [Google Scholar] [CrossRef]
- Trabuco, L.G.; Villa, E.; Mitra, K.; Frank, J.; Schulten, K. Flexible Fitting of Atomic Structures into Electron Microscopy Maps Using Molecular Dynamics. Structure 2008, 16, 673–683. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin Biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Wells, D.B.; Abramkina, V.; Aksimentiev, A. Exploring Transmembrane Transport through α-Hemolysin with Grid-Steered Molecular Dynamics. J. Chem. Phys. 2007, 127, 125101. [Google Scholar] [CrossRef]
- Baker, A.T.; Boyd, R.J.; Sarkar, D.; Teijeira-Crespo, A.; Chan, C.K.; Bates, E.; Waraich, K.; Vant, J.; Wilson, E.; Truong, C.D.; et al. ChAdOx1 Interacts with CAR and PF4 with Implications for Thrombosis with Thrombocytopenia Syndrome. Sci. Adv. 2021, 7, eabl8213. [Google Scholar] [CrossRef]
- Dellon, L.D.; Yanez, A.J.; Li, W.; Mabon, R.; Broadbelt, L.J. Computational Generation of Lignin Libraries from Diverse Biomass Sources. Energy Fuels 2017, 31, 8263–8274. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Petridis, L.; Ralph, J.; Crowley, M.F.; Beckham, G.T. Systematic Parameterization of Lignin for the CHARMM Force Field. Green Chem. 2019, 21, 109–122. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; Mackerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone φ, ψ and Side-Chain χ(1) and χ(2) Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting Modern Challenges in Visualization and Analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A Hub for Protein Information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef] [PubMed]
- Guvench, O.; Greenr, S.N.; Kamath, G.; Brady, J.W.; Venable, R.M.; Pastor, R.W.; Mackerell, A.D. Additive Empirical Force Field for Hexopyranose Monosaccharides. J. Comput. Chem. 2008, 29, 2543–2564. [Google Scholar] [CrossRef] [PubMed]
- Guvench, O.; Hatcher, E.; Venable, R.M.; Pastor, R.W.; MacKerell, A.D. CHARMM Additive All-Atom Force Field for Glycosidic Linkages between Hexopyranoses. J. Chem. Theory Comput. 2009, 5, 2353–2370. [Google Scholar] [CrossRef] [PubMed]
- Guvench, O.; Mallajosyula, S.S.; Raman, E.P.; Hatcher, E.; Vanommeslaeghe, K.; Foster, T.J.; Jamison, F.W.; MacKerell, A.D. CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and Its Utility in Polysaccharide and Carbohydrate-Protein Modeling. J. Chem. Theory Comput. 2011, 7, 3162–3180. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and Better Reference Data for Improved All-Atom Structure Validation: PROTEIN SCIENCE.ORG. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A Comprehensive Python-based System for Macromolecular Structure Solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Trabuco, L.G.; Villa, E.; Schreiner, E.; Harrison, C.B.; Schulten, K. Molecular Dynamics Flexible Fitting: A Practical Guide to Combine Cryo-Electron Microscopy and X-ray Crystallography. Methods 2009, 49, 174–180. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Vant, J.W.; Sarkar, D.; Streitwieser, E.; Fiorin, G.; Skeel, R.; Vermaas, J.V.; Singharoy, A. Data-Guided Multi-Map Variables for Ensemble Refinement of Molecular Movies. J. Chem. Phys. 2020, 153, 214102. [Google Scholar] [CrossRef]
- Fiorin, G.; Marinelli, F.; Faraldo-Gómez, J.D. Direct Derivation of Free Energies of Membrane Deformation and Other Solvent Density Variations From Enhanced Sampling Molecular Dynamics. J. Comput. Chem. 2020, 41, 449–459. [Google Scholar] [CrossRef]
- Al-Bluwi, I.; Siméon, T.; Cortés, J. Motion Planning Algorithms for Molecular Simulations: A Survey. Comput. Sci. Rev. 2012, 6, 125–143. [Google Scholar] [CrossRef]
- Sarkar, D.; Lee, H.; Vant, J.W.; Turilli, M.; Jha, S.; Singharoy, A. Scalable Adaptive Protein Ensemble Refinement Integrating Flexible Fitting. bioRxiv 2021. [Google Scholar] [CrossRef]
- Chan, K.Y.; Gumbart, J.; McGreevy, R.; Watermeyer, J.M.; Sewell, B.T.; Schulten, K. Symmetry-Restrained Flexible Fitting for Symmetric EM Maps. Structure 2011, 19, 1211–1218. [Google Scholar] [CrossRef]
- Croll, T.I. ISOLDE: A Physically Realistic Environment for Model Building into Low-Resolution Electron-Density Maps. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 519–530. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarkar, D.; Kulke, M.; Vermaas, J.V. LongBondEliminator: A Molecular Simulation Tool to Remove Ring Penetrations in Biomolecular Simulation Systems. Biomolecules 2023, 13, 107. https://doi.org/10.3390/biom13010107
Sarkar D, Kulke M, Vermaas JV. LongBondEliminator: A Molecular Simulation Tool to Remove Ring Penetrations in Biomolecular Simulation Systems. Biomolecules. 2023; 13(1):107. https://doi.org/10.3390/biom13010107
Chicago/Turabian StyleSarkar, Daipayan, Martin Kulke, and Josh V. Vermaas. 2023. "LongBondEliminator: A Molecular Simulation Tool to Remove Ring Penetrations in Biomolecular Simulation Systems" Biomolecules 13, no. 1: 107. https://doi.org/10.3390/biom13010107
APA StyleSarkar, D., Kulke, M., & Vermaas, J. V. (2023). LongBondEliminator: A Molecular Simulation Tool to Remove Ring Penetrations in Biomolecular Simulation Systems. Biomolecules, 13(1), 107. https://doi.org/10.3390/biom13010107







