Promising Roles of Circular RNAs as Biomarkers and Targets for Potential Diagnosis and Therapy of Tuberculosis
Abstract
:1. Introduction
2. The Biogenesis of circRNAs
3. The Classification of circRNAs
4. The Detection Technology of circRNAs
5. Biological Roles of circRNAs
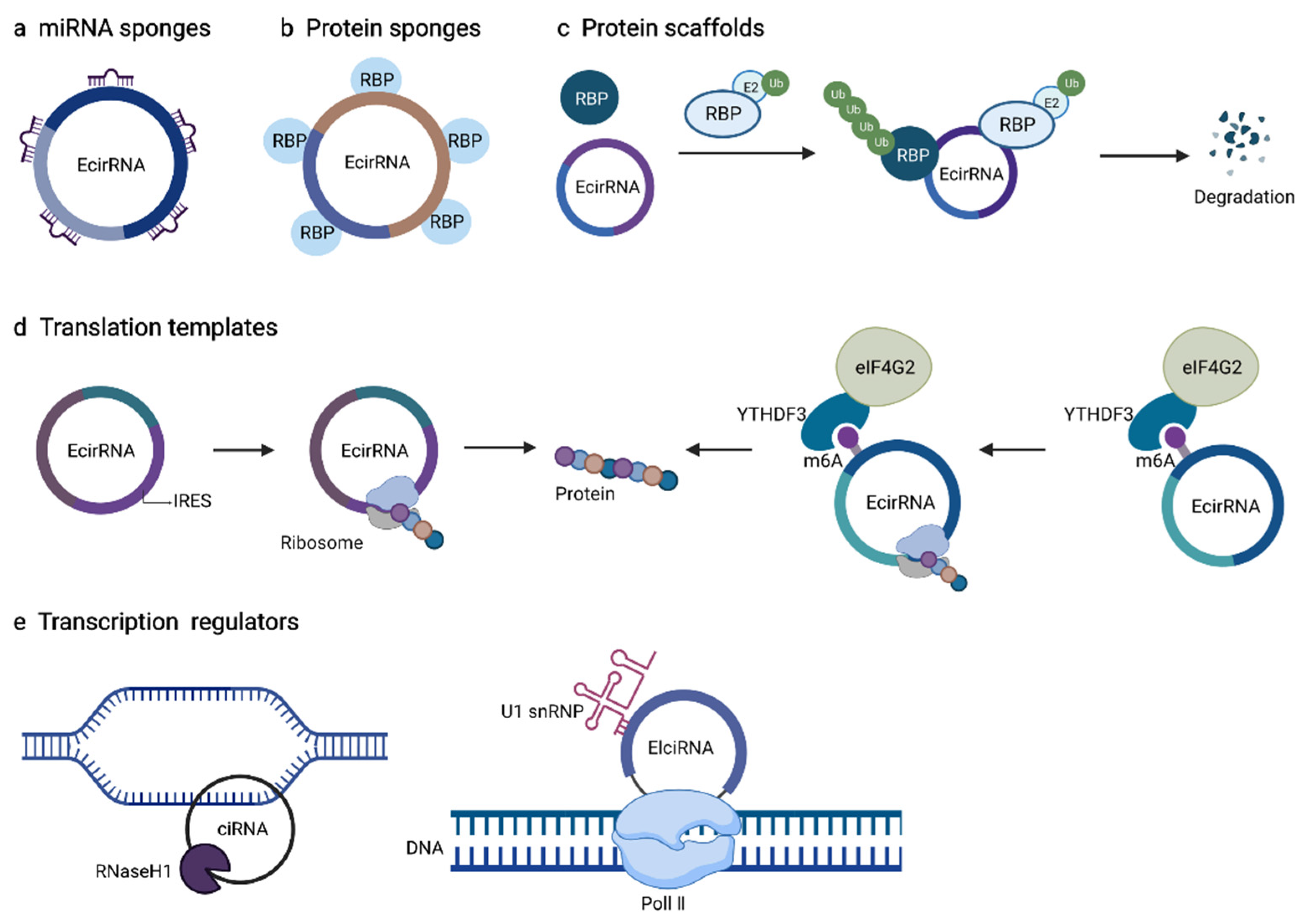
5.1. Role of circRNAs as miRNAs Sponges or Decoys
5.2. Role of circRNAs Contacting with RNA Binding Protein (RBPs)
5.3. Role of circRNAs as Protein Sponges or Decoys
5.4. Role of circRNAs as Protein Scaffolds
5.5. Role of circRNAs as Templates for Translation
5.6. Role of circRNAs Function in Transcription
6. Potentials of circRNAs as Biomarkers in TB
7. CircRNAs Regulate Anti-TB Defense as Potential Therapeutic Targets
8. Conclusions and Prospective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Global Tuberculosis Report. Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 14 October 2021).
- Collaborators, A.R. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Iradukunda, A.; Ndayishimiye, G.P.; Sinarinzi, D.; Odjidja, E.N.; Ntakaburimvo, N.; Nshimirimana, I.; Izere, C. Key factors influencing multidrug-resistant tuberculosis in patients under anti-tuberculosis treatment in two centres in Burundi: A mixed effect modelling study. BMC Public Health 2021, 21, 2142. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.A.; Kim, S.Y.; Jo, K.W.; Kim, H.J.; Park, S.K.; Kim, T.H.; Kim, E.K.; Lee, K.M.; Lee, S.S.; Park, J.S.; et al. Impact of diabetes on treatment outcomes and long-term survival in multidrug-resistant tuberculosis. Respiration 2013, 86, 472–478. [Google Scholar] [CrossRef]
- Brennan, P.J. Bacterial evolution: Emergence of virulence in TB. Nat. Microbiol. 2016, 1, 15031. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Song, H.; Li, G.; Li, Y.; Li, Y.; Zhu, L.; Lu, W.; Chen, C. Relapse or Re-Infection, the Situation of Recurrent Tuberculosis in Eastern China. Front. Cell. Infect. Microbiol. 2021, 11, 638990. [Google Scholar] [CrossRef] [PubMed]
- Vega, V.; Rodríguez, S.; Van der Stuyft, P.; Seas, C.; Otero, L. Recurrent TB: A systematic review and meta-analysis of the incidence rates and the proportions of relapses and reinfections. Thorax 2021, 76, 494–502. [Google Scholar] [CrossRef]
- Khader, S.A.; Divangahi, M.; Hanekom, W.; Hill, P.C.; Maeurer, M.; Makar, K.W.; Mayer-Barber, K.D.; Mhlanga, M.M.; Nemes, E.; Schlesinger, L.S.; et al. Targeting innate immunity for tuberculosis vaccination. J. Clin. Investig. 2019, 129, 3482–3491. [Google Scholar] [CrossRef]
- du Preez, K.; Seddon, J.A.; Schaaf, H.S.; Hesseling, A.C.; Starke, J.R.; Osman, M.; Lombard, C.J.; Solomons, R. Global shortages of BCG vaccine and tuberculous meningitis in children. Lancet Glob. Health 2019, 7, e28–e29. [Google Scholar] [CrossRef]
- Hmama, Z.; Peña-Díaz, S.; Joseph, S.; Av-Gay, Y. Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunol. Rev. 2015, 264, 220–232. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Gross, H.J.; Domdey, H.; Lossow, C.; Jank, P.; Raba, M.; Alberty, H.; Sänger, H.L. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 1978, 273, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Daubersies, P.; Majérus, M.A.; Kerckaert, J.P.; Bailleul, B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992, 11, 1095–1098. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Suzuki, H.; Tsukahara, T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 2014, 15, 9331–9342. [Google Scholar] [CrossRef]
- Wu, D.; Chen, T.; Zhao, X.; Huang, D.; Huang, J.; Huang, Y.; Huang, Q.; Liang, Z.; Chen, C.; Chen, M.; et al. HIF1α-SP1 interaction disrupts the circ-0001875/miR-31-5p/SP1 regulatory loop under a hypoxic microenvironment and promotes non-small cell lung cancer progression. J. Exp. Clin. Cancer Res. 2022, 41, 156. [Google Scholar] [CrossRef]
- Shen, H.; An, O.; Ren, X.; Song, Y.; Tang, S.J.; Ke, X.Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Molias, F.B.; et al. ADARs act as potent regulators of circular transcriptome in cancer. Nat. Commun. 2022, 13, 1508. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef]
- Chen, J.; Wu, Y.; Luo, X.; Jin, D.; Zhou, W.; Ju, Z.; Wang, D.; Meng, Q.; Wang, H.; Fu, X.; et al. Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics 2021, 11, 7507–7526. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.; Du, R.; Chen, T.; Zhang, M.; Zhu, Y.; Luo, X.; Ding, Y. Circular RNA circSLC26A4 Accelerates Cervical Cancer Progression via miR-1287-5p/HOXA7 Axis. Mol. Ther. Nucleic Acids 2020, 19, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Chioccarelli, T.; Falco, G.; Cappetta, D.; De Angelis, A.; Roberto, L.; Addeo, M.; Ragusa, M.; Barbagallo, D.; Berrino, L.; Purrello, M.; et al. FUS driven circCNOT6L biogenesis in mouse and human spermatozoa supports zygote development. Cell. Mol. Life Sci. 2021, 79, 50. [Google Scholar] [CrossRef]
- Yang, T.; Shen, P.; Chen, Q.; Wu, P.; Yuan, H.; Ge, W.; Meng, L.; Huang, X.; Fu, Y.; Zhang, Y.; et al. FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 261. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Yan, P.; Liang, Y.; Sun, Y.; Shen, J.; Zhou, S.; Lin, H.; Liang, X.; Cai, X. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017, 8, e3171. [Google Scholar] [CrossRef]
- Lu, J.; Zhong, C.; Luo, J.; Shu, F.; Lv, D.; Liu, Z.; Tan, X.; Wang, S.; Wu, K.; Yang, T.; et al. HnRNP-L-regulated circCSPP1/miR-520h/EGR1 axis modulates autophagy and promotes progression in prostate cancer. Mol. Ther. Nucleic Acids 2021, 26, 927–944. [Google Scholar] [CrossRef]
- Cao, D. Reverse complementary matches simultaneously promote both back-splicing and exon-skipping. BMC Genom. 2021, 22, 586. [Google Scholar] [CrossRef]
- Shen, S.; Yang, Y.; Shen, P.; Ma, J.; Fang, B.; Wang, Q.; Wang, K.; Shi, P.; Fan, S.; Fang, X. circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann. Rheum. Dis. 2021, 80, 1209–1219. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell. Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, J.; Tian, Y.; Gao, Y.; Dong, X.; Chen, W.; Yuan, X.; Yin, W.; Xu, J.; Chen, K.; et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 2020, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, X.; Liu, M.; Xu, F.; Zhang, Q.; Zhang, Y.; Weng, X.; Liu, S.; Du, Y.; Zhou, X. Direct detection of circRNA in real samples using reverse transcription-rolling circle amplification. Anal. Chim. Acta 2020, 1101, 169–175. [Google Scholar] [CrossRef]
- Li, S.; Teng, S.; Xu, J.; Su, G.; Zhang, Y.; Zhao, J.; Zhang, S.; Wang, H.; Qin, W.; Lu, Z.J.; et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019, 20, 1420–1433. [Google Scholar] [CrossRef]
- Wang, C.; Tan, S.; Liu, W.R.; Lei, Q.; Qiao, W.; Wu, Y.; Liu, X.; Cheng, W.; Wei, Y.Q.; Peng, Y.; et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol. Cancer 2019, 18, 134. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, H.; Xi, F.; Wang, H.; Han, X.; Wei, W.; Zhang, H.; Zhang, Q.; Zheng, Y.; Zhu, Q.; et al. Profiling of circular RNA N(6) -methyladenosine in moso bamboo (Phyllostachys edulis) using nanopore-based direct RNA sequencing. J. Integr. Plant. Biol. 2020, 62, 1823–1838. [Google Scholar] [CrossRef]
- Gaffo, E.; Buratin, A.; Dal Molin, A.; Bortoluzzi, S. Sensitive, reliable and robust circRNA detection from RNA-seq with CirComPara2. Brief Bioinform. 2022, 23, 1–12. [Google Scholar] [CrossRef]
- Cai, Z.; Xue, H.; Xu, Y.; Köhler, J.; Cheng, X.; Dai, Y.; Zheng, J.; Wang, H. Fcirc: A comprehensive pipeline for the exploration of fusion linear and circular RNAs. Gigascience 2020, 9, giaa054. [Google Scholar] [CrossRef]
- Ma, X.K.; Wang, M.R.; Liu, C.X.; Dong, R.; Carmichael, G.G.; Chen, L.L.; Yang, L. CIRCexplorer3: A CLEAR Pipeline for Direct Comparison of Circular and Linear RNA Expression. Genom. Proteom. Bioinform. 2019, 17, 511–521. [Google Scholar] [CrossRef]
- Rabin, A.; Zaffagni, M.; Ashwal-Fluss, R.; Patop, I.L.; Jajoo, A.; Shenzis, S.; Carmel, L.; Kadener, S. SRCP: A comprehensive pipeline for accurate annotation and quantification of circRNAs. Genome Biol. 2021, 22, 277. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Tang, D.; Liao, Y.; Li, P.; Zhang, Y.; Wang, M.; Liang, F.; Wang, X.; Gao, Y.; Wen, L.; et al. Single-cell RNA-seq analysis of mouse preimplantation embryos by third-generation sequencing. PLoS Biol. 2020, 18, e3001017. [Google Scholar] [CrossRef] [PubMed]
- Xin, R.; Gao, Y.; Gao, Y.; Wang, R.; Kadash-Edmondson, K.E.; Liu, B.; Wang, Y.; Lin, L.; Xing, Y. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 2021, 12, 266. [Google Scholar] [CrossRef]
- Zhang, J.; Hou, L.; Zuo, Z.; Ji, P.; Zhang, X.; Xue, Y.; Zhao, F. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat. Biotechnol. 2021, 39, 836–845. [Google Scholar] [CrossRef]
- Rahimi, K.; Venø, M.T.; Dupont, D.M.; Kjems, J. Nanopore sequencing of brain-derived full-length circRNAs reveals circRNA-specific exon usage, intron retention and microexons. Nat. Commun. 2021, 12, 4825. [Google Scholar] [CrossRef]
- Liu, Z.; Tao, C.; Li, S.; Du, M.; Bai, Y.; Hu, X.; Li, Y.; Chen, J.; Yang, E. circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing. Elife 2021, 10, e69457. [Google Scholar] [CrossRef]
- Wei, H.; Liu, B. iCircDA-MF: Identification of circRNA-disease associations based on matrix factorization. Brief Bioinform. 2020, 21, 1356–1367. [Google Scholar] [CrossRef]
- Zheng, K.; You, Z.H.; Li, J.Q.; Wang, L.; Guo, Z.H.; Huang, Y.A. iCDA-CGR: Identification of circRNA-disease associations based on Chaos Game Representation. PLoS Comput. Biol. 2020, 16, e1007872. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhong, J.; Tang, X.; Luo, J. iCDA-CMG: Identifying circRNA-disease associations by federating multi-similarity fusion and collective matrix completion. Mol. Genet. Genom. 2021, 296, 223–233. [Google Scholar] [CrossRef]
- Ji, C.; Liu, Z.; Wang, Y.; Ni, J.; Zheng, C. GATNNCDA: A Method Based on Graph Attention Network and Multi-Layer Neural Network for Predicting circRNA-Disease Associations. Int. J. Mol. Sci. 2021, 22, 8505. [Google Scholar] [CrossRef]
- Deng, L.; Zhang, W.; Shi, Y.; Tang, Y. Fusion of multiple heterogeneous networks for predicting circRNA-disease associations. Sci. Rep. 2019, 9, 9605. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Zhang, Y.; Huang, W.; Shi, J.; Huang, Q.; Jiang, M.; Qiu, Y.; Wang, T.; Chen, H.; Wang, H. Circular RNA Cwc27 contributes to Alzheimer’s disease pathogenesis by repressing Pur-α activity. Cell Death Differ. 2022, 29, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Palermo, C.I.; Barbagallo, C.; Battaglia, R.; Caponnetto, A.; Spina, V.; Ragusa, M.; Di Pietro, C.; Scalia, G.; Purrello, M. Competing endogenous RNA network mediated by circ_3205 in SARS-CoV-2 infected cells. Cell Mol. Life Sci. 2022, 79, 75. [Google Scholar] [CrossRef]
- Hanan, M.; Simchovitz, A.; Yayon, N.; Vaknine, S.; Cohen-Fultheim, R.; Karmon, M.; Madrer, N.; Rohrlich, T.M.; Maman, M.; Bennett, E.R.; et al. A Parkinson’s disease CircRNAs Resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol. Med. 2020, 12, e11942. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Zang, J.; Zhang, T.; Li, Y.; Tan, Z.; Ma, D.; Zhang, T.; Wang, S.; Zhang, Y.; et al. CircOGDH Is a Penumbra Biomarker and Therapeutic Target in Acute Ischemic Stroke. Circ. Res. 2022, 130, 907–924. [Google Scholar] [CrossRef] [PubMed]
- Sang, M.; Meng, L.; Sang, Y.; Liu, S.; Ding, P.; Ju, Y.; Liu, F.; Gu, L.; Lian, Y.; Li, J.; et al. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 2018, 426, 37–46. [Google Scholar] [CrossRef]
- Okholm, T.L.H.; Sathe, S.; Park, S.S.; Kamstrup, A.B.; Rasmussen, A.M.; Shankar, A.; Chua, Z.M.; Fristrup, N.; Nielsen, M.M.; Vang, S.; et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 2020, 12, 112. [Google Scholar] [CrossRef]
- Liu, L.; Xiao, S.; Wang, Y.; Zhu, Z.; Cao, Y.; Yang, S.; Mai, R.; Zheng, Y. Identification of a novel circular RNA circZNF652/miR-486-5p/SERPINE1 signaling cascade that regulates cancer aggressiveness in glioblastoma (GBM). Bioengineered 2022, 13, 1411–1423. [Google Scholar] [CrossRef]
- Shen, P.; Yang, T.; Chen, Q.; Yuan, H.; Wu, P.; Cai, B.; Meng, L.; Huang, X.; Liu, J.; Zhang, Y.; et al. CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing. Mol. Cancer 2021, 20, 51. [Google Scholar] [CrossRef]
- Yu, T.; Ran, L.; Zhao, H.; Yin, P.; Li, W.; Lin, J.; Mao, H.; Cai, D.; Ma, Q.; Pan, X.; et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol. Ther. Nucleic Acids 2021, 26, 649–664. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.X.; Liu, H.S.; Xiong, L.; Yang, X.; Wang, F.W.; Zeng, Z.W.; He, X.W.; Wu, X.R.; Lan, P. A novel NF-κB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol. Cancer 2021, 20, 103. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Latallo, M.J.; Zhang, Z.; Huang, B.; Bobrovnikov, D.G.; Dong, D.; Livingston, N.M.; Tjoeng, W.; Hayes, L.R.; Rothstein, J.D.; et al. Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72-ALS/FTD. Nat. Commun. 2021, 12, 4908. [Google Scholar] [CrossRef]
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef]
- Kosik, K.S. Molecular biology: Circles reshape the RNA world. Nature 2013, 495, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Du, W.W.; Zhang, C.; Yang, W.; Yong, T.; Awan, F.M.; Yang, B.B. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017, 7, 4183–4191. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef] [Green Version]
- Mao, W.; Wang, K.; Xu, B.; Zhang, H.; Sun, S.; Hu, Q.; Zhang, L.; Liu, C.; Chen, S.; Wu, J.; et al. ciRS-7 is a prognostic biomarker and potential gene therapy target for renal cell carcinoma. Mol. Cancer 2021, 20, 142. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wang, H.; Chen, B.; Mao, Q.; Xia, W.; Zhang, T.; Song, X.; Zhang, Z.; Xu, L.; Dong, G.; et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol. Ther. Nucleic Acids 2021, 23, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Hu, Y.; Wang, L.; Li, J. Circ_0003611 acts as a miR-885-5p sponge to aggravate the amyloid-β-induced neuronal injury in Alzheimer’s disease. Metab. Brain Dis. 2022, 37, 961–971. [Google Scholar] [CrossRef]
- Liang, G.; Ling, Y.; Mehrpour, M.; Saw, P.E.; Liu, Z.; Tan, W.; Tian, Z.; Zhong, W.; Lin, W.; Luo, Q.; et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol. Cancer 2020, 19, 65. [Google Scholar] [CrossRef]
- Pan, Z.; Zhao, R.; Li, B.; Qi, Y.; Qiu, W.; Guo, Q.; Zhang, S.; Zhao, S.; Xu, H.; Li, M.; et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer 2022, 21, 16. [Google Scholar] [CrossRef]
- Yu, Y.Z.; Lv, D.J.; Wang, C.; Song, X.L.; Xie, T.; Wang, T.; Li, Z.M.; Guo, J.D.; Fu, D.J.; Li, K.J.; et al. Hsa_circ_0003258 promotes prostate cancer metastasis by complexing with IGF2BP3 and sponging miR-653-5p. Mol. Cancer 2022, 21, 12. [Google Scholar] [CrossRef]
- Lv, X.; Huang, H.; Feng, H.; Wei, Z. Circ-MMP2 (circ-0039411) induced by FOXM1 promotes the proliferation and migration of lung adenocarcinoma cells in vitro and in vivo. Cell Death Dis. 2020, 11, 426. [Google Scholar] [CrossRef]
- Chen, G.; Long, C.; Wang, S.; Wang, Z.; Chen, X.; Tang, W.; He, X.; Bao, Z.; Tan, B.; Zhao, J.; et al. Circular RNA circStag1 promotes bone regeneration by interacting with HuR. Bone Res. 2022, 10, 32. [Google Scholar] [CrossRef]
- Zheng, R.; Zhang, K.; Tan, S.; Gao, F.; Zhang, Y.; Xu, W.; Wang, H.; Gu, D.; Zhu, L.; Li, S.; et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol. Cancer 2022, 21, 49. [Google Scholar] [CrossRef]
- Wang, X.; Chen, M.; Fang, L. hsa_circ_0068631 promotes breast cancer progression through c-Myc by binding to EIF4A3. Mol. Ther. Nucleic Acids 2021, 26, 122–134. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhu, L.; Lu, C.; Wang, C.; Wang, H.; Jin, H.; Ma, X.; Cheng, Z.; Yu, C.; Wang, S.; et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 2021, 12, 295. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 2019, 11, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, L.; Zhou, Y.; Wang, Q.; Zheng, Z.; Xu, B.; Wu, C.; Zhou, Q.; Hu, W.; Wu, C.; et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 2019, 18, 47. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Su, P.; Liang, Y.; Li, Z.; Zhang, H.; Song, X.; Han, D.; Wang, X.; Liu, Y.; et al. circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/beta-catenin pathway. Mol. Ther. 2022, 30, 415–430. [Google Scholar] [CrossRef]
- Lei, M.; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 2020, 19, 30. [Google Scholar] [CrossRef]
- Wethmar, K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip. Rev. RNA 2014, 5, 765–778. [Google Scholar] [CrossRef]
- Chen, C.K.; Cheng, R.; Demeter, J.; Chen, J.; Weingarten-Gabbay, S.; Jiang, L.; Snyder, M.P.; Weissman, J.S.; Segal, E.; Jackson, P.K.; et al. Structured elements drive extensive circular RNA translation. Mol. Cell 2021, 81, 4300–4318.e4313. [Google Scholar] [CrossRef]
- Leppek, K.; Das, R.; Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Z.; Zhang, M.; Zhou, H.; Wu, X.; Zhong, J.; Xiao, F.; Huang, N.; Yang, X.; Zeng, R.; et al. Rolling-translated EGFR variants sustain EGFR signaling and promote glioblastoma tumorigenicity. Neuro Oncol. 2021, 23, 743–756. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018, 110, 304–315. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Bose, R.; Ain, R. Regulation of Transcription by Circular RNAs. Adv. Exp. Med. Biol. 2018, 1087, 81–94. [Google Scholar] [CrossRef]
- Guarnerio, J.; Zhang, Y.; Cheloni, G.; Panella, R.; Mae Katon, J.; Simpson, M.; Matsumoto, A.; Papa, A.; Loretelli, C.; Petri, A.; et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 2019, 29, 628–640. [Google Scholar] [CrossRef]
- Jie, M.; Wu, Y.; Gao, M.; Li, X.; Liu, C.; Ouyang, Q.; Tang, Q.; Shan, C.; Lv, Y.; Zhang, K.; et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol. Cancer 2020, 19, 56. [Google Scholar] [CrossRef]
- Ma, J.; Du, W.W.; Zeng, K.; Wu, N.; Fang, L.; Lyu, J.; Yee, A.J.; Yang, B.B. An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol. Ther. 2021, 29, 2754–2768. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.L.; Lei, Y.N.; Liu, X.Q.; Xue, W.; Zhang, Y.; Nan, F.; Gao, X.; Zhang, J.; Wei, J.; et al. Linking circular intronic RNA degradation and function in transcription by RNase H1. Sci. China Life Sci. 2021, 64, 1795–1809. [Google Scholar] [CrossRef]
- Parsons, L.M.; Somoskövi, A.; Gutierrez, C.; Lee, E.; Paramasivan, C.N.; Abimiku, A.; Spector, S.; Roscigno, G.; Nkengasong, J. Laboratory diagnosis of tuberculosis in resource-poor countries: Challenges and opportunities. Clin. Microbiol. Rev. 2011, 24, 314–350. [Google Scholar] [CrossRef] [Green Version]
- Reis-das-Mercês, L.; Vinasco-Sandoval, T.; Pompeu, R.; Ramos, A.C.; Anaissi, A.K.M.; Demachki, S.; de Assumpção, P.P.; Vidal, A.F.; Ribeiro-Dos-Santos, Â.; Magalhães, L. CircRNAs as Potential Blood Biomarkers and Key Elements in Regulatory Networks in Gastric Cancer. Int. J. Mol. Sci. 2022, 23, 650. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e813. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Zhuang, Z.G.; Zhang, J.A.; Luo, H.L.; Liu, G.B.; Lu, Y.B.; Ge, N.H.; Zheng, B.Y.; Li, R.X.; Chen, C.; Wang, X.; et al. The circular RNA of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol. Immunol. 2017, 90, 264–272. [Google Scholar] [CrossRef]
- Yi, Z.; Gao, K.; Li, R.; Fu, Y. Dysregulated circRNAs in plasma from active tuberculosis patients. J. Cell Mol. Med. 2018, 22, 4076–4084. [Google Scholar] [CrossRef]
- Huang, Z.; Su, R.; Deng, Z.; Xu, J.; Peng, Y.; Luo, Q.; Li, J. Identification of differentially expressed circular RNAs in human monocyte derived macrophages response to Mycobacterium tuberculosis infection. Sci. Rep. 2017, 7, 13673. [Google Scholar] [CrossRef]
- Liu, H.; Lu, G.; Wang, W.; Jiang, X.; Gu, S.; Wang, J.; Yan, X.; He, F.; Wang, J. A Panel of CircRNAs in the Serum Serves as Biomarkers for Mycobacterium tuberculosis Infection. Front. Microbiol. 2020, 11, 1215. [Google Scholar] [CrossRef]
- Huang, Z.; Su, R.; Yao, F.; Peng, Y.; Luo, Q.; Li, J. Circulating circular RNAs hsa_circ_0001204 and hsa_circ_0001747 act as diagnostic biomarkers for active tuberculosis detection. Int. J. Clin. Exp. Pathol. 2018, 11, 586–594. [Google Scholar]
- Huang, Z.; Su, R.; Qing, C.; Peng, Y.; Luo, Q.; Li, J. Plasma Circular RNAs hsa_circ_0001953 and hsa_circ_0009024 as Diagnostic Biomarkers for Active Tuberculosis. Front. Microbiol. 2018, 9, 2010. [Google Scholar] [CrossRef]
- Huang, Z.K.; Yao, F.Y.; Xu, J.Q.; Deng, Z.; Su, R.G.; Peng, Y.P.; Luo, Q.; Li, J.M. Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Active Tuberculosis Patients. Cell Physiol. Biochem. 2018, 45, 1230–1240. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, J.; Qiao, J.; Yi, Z. Signature of circular RNAs in peripheral blood mononuclear cells from patients with active tuberculosis. J. Cell Mol. Med. 2019, 23, 1917–1925. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhang, Q.; Wu, Q.; Tang, H.; Ye, L.; Zhang, Q.; Hua, D.; Zhang, Y.; Li, F. Integrated analyses reveal hsa_circ_0028883 as a diagnostic biomarker in active tuberculosis. Infect. Genet. Evol. 2020, 83, 104323. [Google Scholar] [CrossRef]
- Luo, H.L.; Peng, Y.; Luo, H.; Zhang, J.A.; Liu, G.B.; Xu, H.; Huang, G.X.; Sun, Y.F.; Huang, J.; Zheng, B.Y.; et al. Circular RNA hsa_circ_0001380 in peripheral blood as a potential diagnostic biomarker for active pulmonary tuberculosis. Mol. Med. Rep. 2020, 21, 1890–1896. [Google Scholar] [CrossRef]
- Yi, X.H.; Zhang, B.; Fu, Y.R.; Yi, Z.J. STAT1 and its related molecules as potential biomarkers in Mycobacterium tuberculosis infection. J. Cell Mol. Med. 2020, 24, 2866–2878. [Google Scholar] [CrossRef]
- Luo, H.L.; Pi, J.; Zhang, J.A.; Yang, E.Z.; Xu, H.; Luo, H.; Shen, L.; Peng, Y.; Liu, G.B.; Song, C.M.; et al. Circular RNA TRAPPC6B inhibits intracellular Mycobacterium tuberculosis growth while inducing autophagy in macrophages by targeting microRNA-874-3p. Clin. Transl. Immunol. 2021, 10, e1254. [Google Scholar] [CrossRef]
- Huang, Z.; Yao, F.; Liu, J.; Xu, J.; Guo, Y.; Su, R.; Luo, Q.; Li, J. Up-regulation of circRNA-0003528 promotes mycobacterium tuberculosis associated macrophage polarization via down-regulating miR-224-5p, miR-324-5p and miR-488-5p and up-regulating CTLA4. Aging 2020, 12, 25658–25672. [Google Scholar] [CrossRef]
- Wu, M.; Liu, Z.; Zhang, S. Down-regulation of hsa_circ_0045474 induces macrophage autophagy in tuberculosis via miR-582-5p/TNKS2 axis. Innate Immun. 2022, 28, 11–18. [Google Scholar] [CrossRef]
- Shi, Q.; Wang, J.; Yang, Z.; Liu, Y. CircAGFG1modulates autophagy and apoptosis of macrophages infected by Mycobacterium tuberculosis via the Notch signaling pathway. Ann. Transl. Med. 2020, 8, 645. [Google Scholar] [CrossRef]
- Deng, Q.; Huang, J.; Yan, J.; Mao, E.; Chen, H.; Wang, C. Circ_0001490/miR-579-3p/FSTL1 axis modulates the survival of mycobacteria and the viability, apoptosis and inflammatory response in Mycobacterium tuberculosis-infected macrophages. Tuberculosis 2021, 131, 102123. [Google Scholar] [CrossRef]
- Ma, J.; Chen, X.L.; Sun, Q. microRNA-579 upregulation mediates death of human macrophages with mycobacterium tuberculosis infection. Biochem. Biophys. Res. Commun. 2019, 518, 219–226. [Google Scholar] [CrossRef]
- Kaushik, A.C.; Wu, Q.; Lin, L.; Li, H.; Zhao, L.; Wen, Z.; Song, Y.; Wu, Q.; Wang, J.; Guo, X.; et al. Exosomal ncRNAs profiling of mycobacterial infection identified miRNA-185-5p as a novel biomarker for tuberculosis. Brief. Bioinform. 2021, 22, bbab210. [Google Scholar] [CrossRef]
- Li, B.; Ren, Q.; Li, Y.; Tian, S.; Chong, Y.; Sun, S.; Feng, F. Screening differential circular RNA expression profiles reveals the regulatory role of circMARS in anti-tuberculosis drug-induced liver injury. J. Cell Mol. Med. 2022, 26, 1050–1059. [Google Scholar] [CrossRef]
- Lyu, M.; Cheng, Y.; Zhou, J.; Chong, W.; Wang, Y.; Xu, W.; Ying, B. Systematic evaluation, verification and comparison of tuberculosis-related non-coding RNA diagnostic panels. J. Cell Mol. Med. 2021, 25, 184–202. [Google Scholar] [CrossRef]
- Xu, G.; Wang, J.; Gao, G.F.; Liu, C.H. Insights into battles between Mycobacterium tuberculosis and macrophages. Protein Cell 2014, 5, 728–736. [Google Scholar] [CrossRef]
- Paik, S.; Kim, J.K.; Chung, C.; Jo, E.K. Autophagy: A new strategy for host-directed therapy of tuberculosis. Virulence 2019, 10, 448–459. [Google Scholar] [CrossRef]
- Stutz, M.D.; Allison, C.C.; Ojaimi, S.; Preston, S.P.; Doerflinger, M.; Arandjelovic, P.; Whitehead, L.; Bader, S.M.; Batey, D.; Asselin-Labat, M.L.; et al. Macrophage and neutrophil death programs differentially confer resistance to tuberculosis. Immunity 2021, 54, 1758–1771.e1757. [Google Scholar] [CrossRef]
- Gutierrez, M.G.; Master, S.S.; Singh, S.B.; Taylor, G.A.; Colombo, M.I.; Deretic, V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004, 119, 753–766. [Google Scholar] [CrossRef]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. 2020, 15, 123–147. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef]
- Huang, Z.; Luo, Q.; Guo, Y.; Chen, J.; Xiong, G.; Peng, Y.; Ye, J.; Li, J. Mycobacterium tuberculosis-Induced Polarization of Human Macrophage Orchestrates the Formation and Development of Tuberculous Granulomas In Vitro. PLoS ONE 2015, 10, e0129744. [Google Scholar] [CrossRef]
- Yuan, Q.; Wen, Z.; Yang, K.; Zhang, S.; Zhang, N.; Song, Y.; Chen, F. Identification of Key CircRNAs Related to Pulmonary Tuberculosis Based on Bioinformatics Analysis. Biomed. Res. Int. 2022, 2022, 1717784. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744.e1716. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Chatterjee, S.; Xiao, K.; Riedel, I.; Huang, C.K.; Costa, A.; Cushman, S.; Neufeldt, D.; Rode, L.; Schmidt, A.; et al. A circular RNA derived from the insulin receptor locus protects against doxorubicin-induced cardiotoxicity. Eur. Heart J. 2022, ehac337. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Guo, S.K.; Nan, F.; Xu, Y.F.; Yang, L.; Chen, L.L. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 2022, 82, 420–434.e426. [Google Scholar] [CrossRef]
| Circular RNA | Function | Expression | Derived From | Targets/Signaling Pathways | AUC | Number of TB Patients/Controls | Ref. |
|---|---|---|---|---|---|---|---|
| hsa_circ_0001204 | biomarker | down | plasma | 0.871 | 145/120 | [110] | |
| hsa_circ_0001747 | biomarker | down | plasma | 0.830 | 145/120 | [110] | |
| hsa_circ_0001204; hsa_circ_0001747 | biomarker | down | plasma | 0.928 * | 145/120 | [110] | |
| hsa_circ_0001953 | biomarker | up | plasma | 0.826 | 120/100 | [109] | |
| hsa_circ_0009024 | biomarker | up | plasma | 0.777 | 120/100 | [109] | |
| hsa_circ_0001953; hsa_circ_0009024 | biomarker | up | plasma | 0.915 * | 120/100 | [109] | |
| hsa_circ_001937 | biomarker | up | PBMCs | 0.873 | 115/90 | [111] | |
| hsa_circ_0043497 | biomarker | up | Mtb-infected MDMs | 0.860 | 96/85 | [107] | |
| hsa_circ_0001204 | biomarker | down | Mtb-infected MDMs | 0.848 | 96/85 | [107] | |
| hsa_circ_103017 | biomarker | up | PBMCs | 0.870 | 31/30 | [112] | |
| hsa_circ_059914 | biomarker | up | PBMCs | 0.821 | 31/30 | [112] | |
| hsa_circ_0028883 | biomarker | up | PBMCs | miR-409-5p | 0.773 | 20/20 | [113] |
| hsa_circ_0005836 | biomarker | down | PBMCs | no mention | 49/45 | [105] | |
| hsa_circ_0001380 | biomarker | down | PBMCs | 0.9502 | 32/31 | [114] | |
| hsa_circ_103571 | biomarker | down | plasma | 0.838 | 32/29 | [106] | |
| circ_051239 | biomarker | up | serum | 0.9738 | 72/30 | [108] | |
| circ_029965 | biomarker | up | serum | 0.9443 | 72/30 | [108] | |
| circ_404022 | biomarker | up | serum | 0.9682 | 72/30 | [108] | |
| SAMD8_ hsa_circRNA994 | no mention | no mention | whole blood | no mention | 45/61 | [115] | |
| TWF1_ hsa_circRNA9897 | no mention | no mention | whole blood | no mention | 45/61 | [115] | |
| circTRAPPC6B | miRNA sponge | down | PBMCs | miR-874-3p ATG16L1 autophagy | 0.8609 | 32/31 | [116] |
| hsa_circ_0003528 | miRNA sponge | up | plasma | miR-224-5p miR-324-5p miR-488-5p CTLA4 polarization | no mention | 50/50 | [117] |
| hsa_circ_101128 | biomarker; miRNA sponge | up | PBMCs | let-7a MAPK/P13K-Akt pathway | 0.817 | 31/30 | [112] |
| hsa_circ_0045474 | miRNA sponge | down | PBMCs | miR-582-5p TNKS2 autophagy | no mention | 15/15 | [118] |
| circAGFG1 | miRNA sponge | up | alveolar macrophages in ATB patients | Notch miR-1257 apoptosis autophagy | no mention | no mention | [119] |
| circ_0001490 | miRNA sponge | down | Mtb-infected THP-1 macrophages; serum | miR-579-3p FSTL1 inflammatory response | no mention | 40/23 | [120] |
| cPWWP2A | miRNA sponge | down | primary human MDMs | miR-579 | no mention | no mention | [121] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Li, Y.; Lin, W.; Fan, S.; Chen, H.; Xia, J.; Pi, J.; Xu, J.-F. Promising Roles of Circular RNAs as Biomarkers and Targets for Potential Diagnosis and Therapy of Tuberculosis. Biomolecules 2022, 12, 1235. https://doi.org/10.3390/biom12091235
Huang Y, Li Y, Lin W, Fan S, Chen H, Xia J, Pi J, Xu J-F. Promising Roles of Circular RNAs as Biomarkers and Targets for Potential Diagnosis and Therapy of Tuberculosis. Biomolecules. 2022; 12(9):1235. https://doi.org/10.3390/biom12091235
Chicago/Turabian StyleHuang, Yifan, Ying Li, Wensen Lin, Shuhao Fan, Haorong Chen, Jiaojiao Xia, Jiang Pi, and Jun-Fa Xu. 2022. "Promising Roles of Circular RNAs as Biomarkers and Targets for Potential Diagnosis and Therapy of Tuberculosis" Biomolecules 12, no. 9: 1235. https://doi.org/10.3390/biom12091235
APA StyleHuang, Y., Li, Y., Lin, W., Fan, S., Chen, H., Xia, J., Pi, J., & Xu, J.-F. (2022). Promising Roles of Circular RNAs as Biomarkers and Targets for Potential Diagnosis and Therapy of Tuberculosis. Biomolecules, 12(9), 1235. https://doi.org/10.3390/biom12091235







