Identification of a Multi-Messenger RNA Signature as Type 2 Diabetes Mellitus Candidate Genes Involved in Crosstalk between Inflammation and Insulin Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection of mRNA Set Linked to cGAS/STING, NOD-like Receptor Pathways (NLR) and T2DM
2.2. Study Population and Blood Samples
2.3. Extraction of the mRNA
2.4. Quantitation of the Selected Six-Based mRNA Signature Expression
2.5. Statistics
3. Results
3.1. The Study Groups’ Medical and Anthropometric Characters
3.2. Insulin Sensitivity and the Function of the Pancreatic Islet Cells
3.3. The Expression of ZBP1, HSPA1B, TMEM173, DDX58, NFKB1 and CHUK in the Sera Samples
3.4. Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| T2DM | Type 2 Diabetes Mellitus |
| STING | DNA-sensing stimulator of interferon genes |
| NLR | NOD-like receptor |
| MENA | Middle East and North Africa |
| mRNA | messenger ribonucleic acid |
| NF-κB | nuclear factor-kappa beta |
| cGAS | cyclic GMP AMP synthase |
| NOD | nucleotide-binding oligomerization domain-containing protein |
| TMEM173 | transmembrane protein 173 |
| ZBP1 | Z-DNA-binding protein 1 |
| IRF | interferon regulatory factor |
| RIG-I | retinoic acid inducible gene 1 |
| HSP | Heat shock protein |
| PGN | peptidoglycans |
| DDX58 | DexD/H-Box Helicase 58 |
| MAVs | mitochondrial antiviral signaling |
| MHC | Major histocompatibility complex |
| CHUK | conserved helix–loop–helix ubiquitous kinase |
| IKK α | Inhibitor-κB kinase α |
| mTORC1 | mammalian target of rapamycin complex 1 |
| NLRC3 | NLR family domain-containing protein 3 |
| FFA | free fatty acids |
| LPS | lipopolysachrides |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PBMCs | Peripheral Blood Mononuclear Cells |
| PAMPs | Pathogen-associated molecular pattern |
| DAMPs | Damage-associated molecular pattern |
| HSPA1B | Heat Shock Protein Family A (Hsp70) Member 1B |
| PPI | Protein–protein interaction |
| ADA | The American Diabetes Association |
| BMI | Body Mass Index |
| HOMA-IR | Homeostatic Model Assessment of Insulin Resistance |
| FSI | fasting serum Insulin |
| FSG | fasting serum glucose |
| HOMA-B | Homeostatic Model Assessment of beta cell function |
| TC | total cholesterol |
| TG | triglyceride |
| HDL-c | high density lipoprotein- cholesterol |
| LDL-c | low density lipoprotein- cholesterol |
| GAPDH | glyceraldehyde 3 phosphate dehydrogenase |
| PCR | polymerase chain reaction |
| CT | cycle threshold |
| SPSS | The Statistical Package for the Social Sciences |
| ANOVA | Analysis of Variance |
| ROC curve | Receiver operating characteristic |
| HFD | High-fat diet |
| RIPK3 | Receptor-interacting serine/threonine-protein kinase 3 |
| mtDNA | mitochondrial DNA |
| TNF-α | Tumor necrosis factor alpha |
| IL | interleukin |
| IKK | IκB kinase |
References
- American Diabetes Association. Introduction: Standards of Medical Care in Diabetes—2022. Diabetes Care 2021, 45 (Suppl. S1), S1–S2. [Google Scholar] [CrossRef]
- International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2021. [Google Scholar]
- El-Kebbi, I.M.; Bidikian, N.H.; Hneiny, L.; Nasrallah, M.P. Epidemiology of type 2 diabetes in the Middle East and North Africa: Challenges and call for action. World J. Diabetes 2021, 12, 1401–1425. [Google Scholar] [CrossRef] [PubMed]
- 4American Diabetes Association. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care 2021, 44 (Suppl. S1), S15–S33. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37 (Suppl. S1), S81–S90. [Google Scholar] [CrossRef]
- Fukuda, H.; Mizobe, M. Impact of nonadherence on complication risks and healthcare costs in patients newly-diagnosed with diabetes. Diabetes Res. Clin. Pract. 2016, 123, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Vasu, S.; Kumano, K.; Darden, C.M.; Rahman, I.; Lawrence, M.C.; Naziruddin, B. MicroRNA Signatures as Future Biomarkers for Diagnosis of Diabetes States. Cells 2019, 8, 1533. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Zhang, Z.; Ji, S.; Liu, T.; Zhang, X.; Huang, Y.; Feng, W.; Wang, K.; Wang, J.; Wang, S.; et al. A distinct role of STING in regulating glucose homeostasis through insulin sensitivity and insulin secretion. Proc. Natl. Acad. Sci. USA 2022, 119, e2101848119. [Google Scholar] [CrossRef]
- Yang, Q.; Ouyang, J.; Sun, F.; Yang, J. Short-Chain Fatty Acids: A Soldier Fighting against Inflammation and Protecting from Tumorigenesis in People with Diabetes. Front. Immunol. 2020, 11, 590685. [Google Scholar] [CrossRef]
- Scheithauer, T.P.M.; Rampanelli, E.; Nieuwdorp, M.; Vallance, B.A.; Verchere, C.B.; Van Raalte, D.H.; Herrema, H. Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front. Immunol. 2020, 11, 571731. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Mao, Y.; Luo, W.; Zhang, L.; Wu, W.; Yuan, L.; Xu, H.; Song, J.; Fujiwara, K.; Abe, J.-I.; Lemaire, S.A.; et al. STING–IRF3 Triggers Endothelial Inflammation in Response to Free Fatty Acid-Induced Mitochondrial Damage in Diet-Induced Obesity. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Cervantes, C.; Liu, J.; He, S.; Zhou, H.; Zhang, B.; Cai, H.; Yin, D.; Hu, D.; Li, Z.; et al. DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc. Natl. Acad. Sci. USA 2017, 114, 12196–12201. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Rao, H.; Zhao, J.; Wee, A.; Li, X.; Fei, R.; Huang, R.; Wu, C.; Liu, F.; Wei, L. STING expression in monocyte-derived macrophages is associated with the progression of liver inflammation and fibrosis in patients with nonalcoholic fatty liver disease. Lab. Investig. 2020, 100, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Vanportfliet, J.J.; Chen, Y.F.; Bryant, J.D.; Ragan, K.B.; Tong, C.W.; West, L.C.; Bosenberg, M.W.; Li, P.; Shadel, G.S.; et al. ZBP1 sequesters cGAS in the cytoplasm and sustains type I interferon responses to mitochondrial DNA. bioRxiv 2022. [Google Scholar] [CrossRef]
- Peng, R.; Wang-Kan, X.; Idorn, M.; Zhou, F.Y.; Orozco, S.L.; McCarthy, J.; Leung, C.S.; Lu, X.; Bagola, K.; Rehwinkel, J.; et al. ZBP1 induces inflammatory signaling via RIPK3 and promotes SARS-CoV-2-induced cytokine expression. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rebsamen, M.; Heinz, L.; Meylan, E.; Michallet, M.; Schroder, K.; Hofmann, K.; Vazquez, J.; A Benedict, C.; Tschopp, J. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-κB. EMBO Rep. 2009, 10, 916–922. [Google Scholar] [CrossRef]
- Gautheron, J.; Vucur, M.; Schneider, A.T.; Severi, I.; Roderburg, C.; Roy, S.; Bartneck, M.; Schrammen, P.; Diaz, M.B.; Ehling, J.; et al. The necroptosis-inducing kinase RIPK3 dampens adipose tissue inflammation and glucose intolerance. Nat. Commun. 2016, 7, 11869. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Schertzer, J.D.; Tamrakar, A.K.; Magalhães, J.G.; Pereira, S.; Bilan, P.J.; Fullerton, M.D.; Liu, Z.; Steinberg, G.R.; Giacca, A.; Philpott, D.J.; et al. NOD1 Activators Link Innate Immunity to Insulin Resistance. Diabetes 2011, 60, 2206–2215. [Google Scholar] [CrossRef]
- Rivers, S.L.; Klip, A.; Giacca, A. NOD1: An Interface Between Innate Immunity and Insulin Resistance. Endocrinology 2019, 160, 1021–1030. [Google Scholar] [CrossRef]
- Chan, K.L.; Tam, T.H.; Boroumand, P.; Prescott, D.; Costford, S.R.; Escalante, N.K.; Fine, N.; Tu, Y.; Robertson, S.J.; Prabaharan, D.; et al. Circulating NOD1 activators and hematopoietic NOD1 contribute to metabolic inflammation and insulin resistance. Cell Rep. 2017, 18, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, V.; Grimes, C.L. The Molecular Chaperone HSP70 Binds to and Stabilizes NOD2, an Important Protein Involved in Crohn Disease. J. Biol. Chem. 2014, 289, 18987–18998. [Google Scholar] [CrossRef]
- Brudzynski, K. Insulitis-caused redistribution of heat-shock protein HSP60 inside beta-cells correlates with induction of HSP60 autoantibodies. Diabetes 1993, 42, 908–913. [Google Scholar] [CrossRef] [PubMed]
- Juwono, J.; Martinus, R.D. Does Hsp60 Provide a Link between Mitochondrial Stress and Inflammation in Diabetes Mellitus? J. Diabetes Res. 2016, 2016, 8017571. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, Y. Attribution of NF-κB Activity to CHUK/IKKα-Involved Carcinogenesis. Cancers 2021, 13, 1411. [Google Scholar] [CrossRef]
- Bai, J.; Liu, F. The cGAS-cGAMP-STING Pathway: A Molecular Link Between Immunity and Metabolism. Diabetes 2019, 68, 1099–1108. [Google Scholar] [CrossRef]
- Zhang, L.; Mo, J.; Swanson, K.V.; Wen, H.; Petrucelli, A.; Gregory, S.M.; Zhang, Z.; Schneider, M.; Jiang, Y.; Fitzgerald, K.A.; et al. NLRC3, a Member of the NLR Family of Proteins, Is a Negative Regulator of Innate Immune Signaling Induced by the DNA Sensor STING. Immunity 2014, 40, 329–341. [Google Scholar] [CrossRef]
- Prajapati, B.; Jena, P.K.; Rajput, P.; Purandhar, K.; Seshadri, S. Understanding and Modulating the Toll Like Receptors (TLRs) and NOD Like Receptors (NLRs) Cross Talk in Type 2 Diabetes. Curr. Diabetes Rev. 2014, 10, 190–200. [Google Scholar] [CrossRef]
- American Diabetes Association. Standards of Medical Care in Diabetes—2010. Diabetes Care 2010, 33 (Suppl. S1), 2010. [Google Scholar]
- Chobanian, A.V.; Bakris, G.L.; Black, H.R.; Cushman, W.C.; Green, L.A.; Izzo, J.L., Jr.; Jones, D.W.; Materson, B.J.; Oparil, S.; Wright, J.T., Jr.; et al. The seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure: The JNC 7 report. JAMA 2003, 289, 2560–2571. [Google Scholar] [CrossRef]
- WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet 2004, 363, 157–163. [Google Scholar] [CrossRef]
- Gonen, B.; Rubenstien, A.H. Determination of glycohemoglobin. Diabetologia 1978, 15, 5. [Google Scholar]
- Świderska, E.; Strycharz, J.; Wróblewski, A.; Szemraj, J.; Drzewoski, J.; Śliwińska, A. Role of PI3K/AKT Pathway in Insulin-Mediated Glucose Uptake. In Blood Glucose Levels; Szablewski, L., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chu, J.; Hao, W.; Zhang, J.; Li, H.; Yang, C.; Yang, J.; Chen, X.; Wang, H. Gut Microbiota and Type 2 Diabetes Mellitus: Association, Mechanism, and Translational Applications. Mediat. Inflamm. 2021, 2021, 5110276. [Google Scholar] [CrossRef]
- Eleftheriadou, A.; Williams, S.; Nevitt, S.; Brown, E.; Roylance, R.; Wilding, J.P.H. The prevalence of cardiac autonomic neuropathy in prediabetes: A systematic review. Diabetologia 2021, 64, 288–303. [Google Scholar] [CrossRef]
- Menini, S.; Iacobini, C.; Vitale, M.; Pugliese, G. The Inflammasome in Chronic Complications of Diabetes and Related Metabolic Disorders. Cells 2020, 9, 1812. [Google Scholar] [CrossRef]
- Jiao, H.; Wachsmuth, L.; Kumari, S.; Schwarzer, R.; Lin, J.; Eren, R.O.; Fisher, A.; Lane, R.; Young, G.R.; Kassiotis, G.; et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 2020, 580, 391–395. [Google Scholar] [CrossRef]
- Blum, S.I.; Tse, H.M. Innate Viral Sensor MDA5 and Coxsackievirus Interplay in Type 1 Diabetes Development. Microorganisms 2020, 8, 993. [Google Scholar] [CrossRef]
- An, T.; Zhang, J.; Liu, Y.; Wu, Y.; Lian, J.; Wang, T.; Hu, Y.; Zhu, J.; Huang, J.; Zhao, D.; et al. Combined analysis of whole-exon sequencing and lncRNA sequencing in type 2 diabetes mellitus patients with obesity. J. Cell. Mol. Med. 2020, 24, 2451–2463. [Google Scholar] [CrossRef]
- Yang, G.; Lee, H.E.; Seok, J.K.; Kang, H.C.; Cho, Y.-Y.; Lee, H.S.; Lee, J.Y. RIG-I Deficiency Promotes Obesity-Induced Insulin Resistance. Pharmaceuticals 2021, 14, 1178. [Google Scholar] [CrossRef] [PubMed]
- Uppal, T.; Sarkar, R.; Dhelaria, R.; Verma, S.C. Role of Pattern Recognition Receptors in KSHV Infection. Cancers 2018, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Fujimoto, Y.; Lucas, P.C.; Nakano, H.; Fukase, K.; Nunez, G.; Inohara, N. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008, 27, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mir, K.A.; Pugazhendhi, S.; Paul, M.J.; Nair, A.; Ramakrishna, B.S. Heat-shock protein 70 gene polymorphism is associated with the severity of diabetic foot ulcer and the outcome of surgical treatment. Br. J. Surg. 2009, 96, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Schröder, O.; Schulte, K.-M.; Ostermann, P.; Röher, H.-D.; Ekkernkamp, A.; Laun, R.A. Heat shock protein 70 genotypes HSPA1B and HSPA1L influence cytokine concentrations and interfere with outcome after major injury. Crit. Care Med. 2003, 31, 73–79. [Google Scholar] [CrossRef]
- Haäcker, H.; Karin, M. Regulation and Function of IKK and IKK-Related Kinases. Sci. STKE 2006, 2006, re13. [Google Scholar] [CrossRef]
- Llona-Minguez, S.; Baiget, J.; Mackay, S.P. Small-molecule inhibitors of IκB kinase (IKK) and IKK-related kinases. Pharm. Pat. Anal. 2013, 2, 481–498. [Google Scholar] [CrossRef]
- Arkan, M.C.; Hevener, A.L.; Greten, F.R.; Maeda, S.; Li, Z.W.; Long, J.M.; Wynshaw-Boris, A.; Poli, G.; Olefsky, J.; Karin, M. IKK-beta links inflammation to obesity-induced insulin resistance. Nat. Med. 2005, 11, 191–198. [Google Scholar] [CrossRef]
- Tantiwong, P.; Shanmugasundaram, K.; Monroy, A.; Ghosh, S.; Li, M.; DeFronzo, R.A.; Cersosimo, E.; Sriwijitkamol, A.; Mohan, S.; Musi, N. NF-κB activity in muscle from obese and type 2 diabetic subjects under basal and exercise-stimulated conditions. Am. J. Physiol. Metab. 2010, 299, E794–E801. [Google Scholar] [CrossRef]
- Evans, J.L.; Goldfine, I.D.; Maddux, B.A.; Grodsky, G.M. Oxidative Stress and Stress-Activated Signaling Pathways: A Unifying Hypothesis of Type 2 Diabetes. Endocr. Rev. 2002, 23, 599–622. [Google Scholar] [CrossRef]
- Kim, J.K.; Kim, Y.J.; Fillmore, J.J.; Chen, Y.; Moore, I.; Lee, J.; Yuan, M.; Li, Z.W.; Karin, M.; Perret, P.; et al. Prevention of fat-induced insulin resistance by salicylate. J. Clin. Investig. 2001, 108, 437–446. [Google Scholar] [CrossRef]
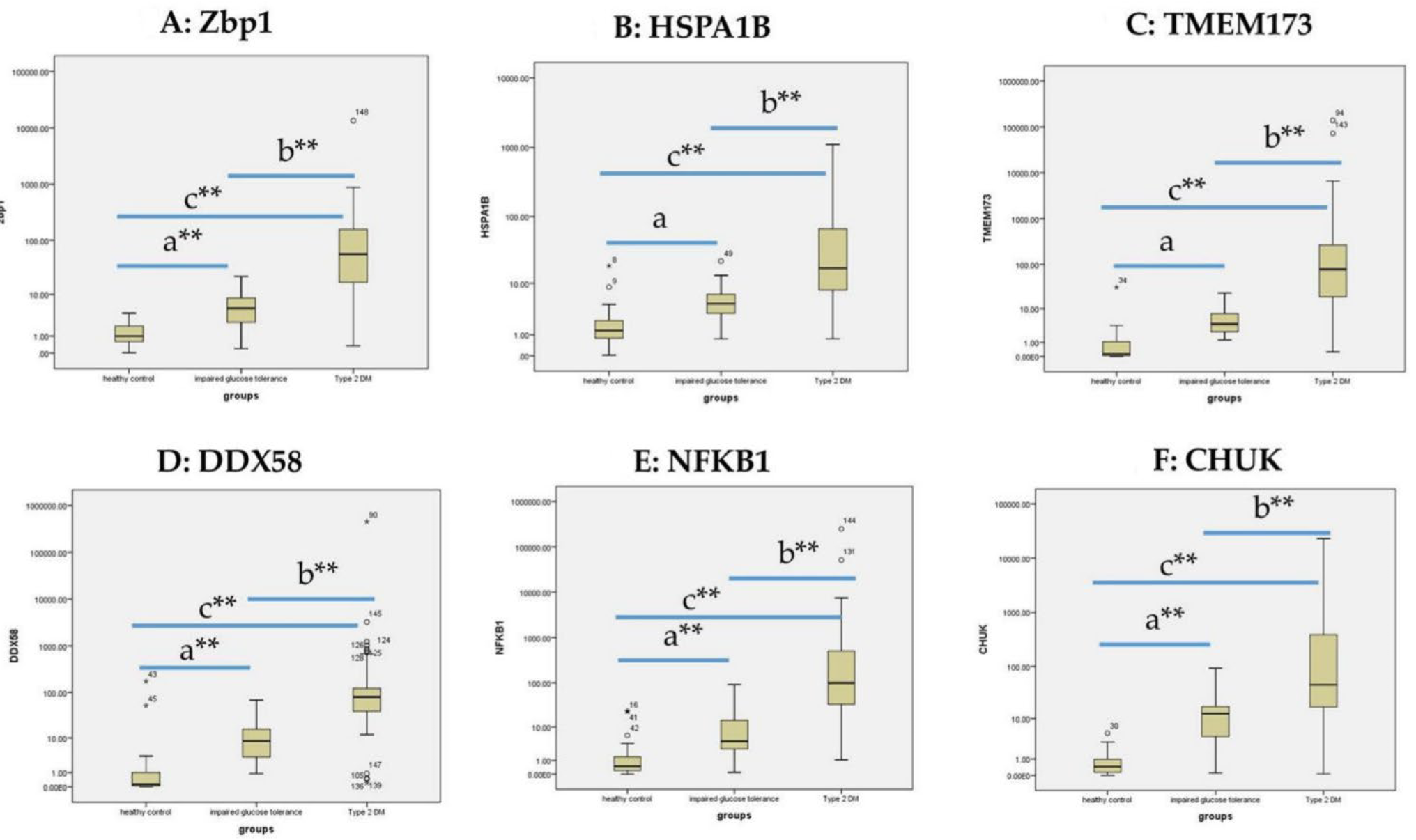
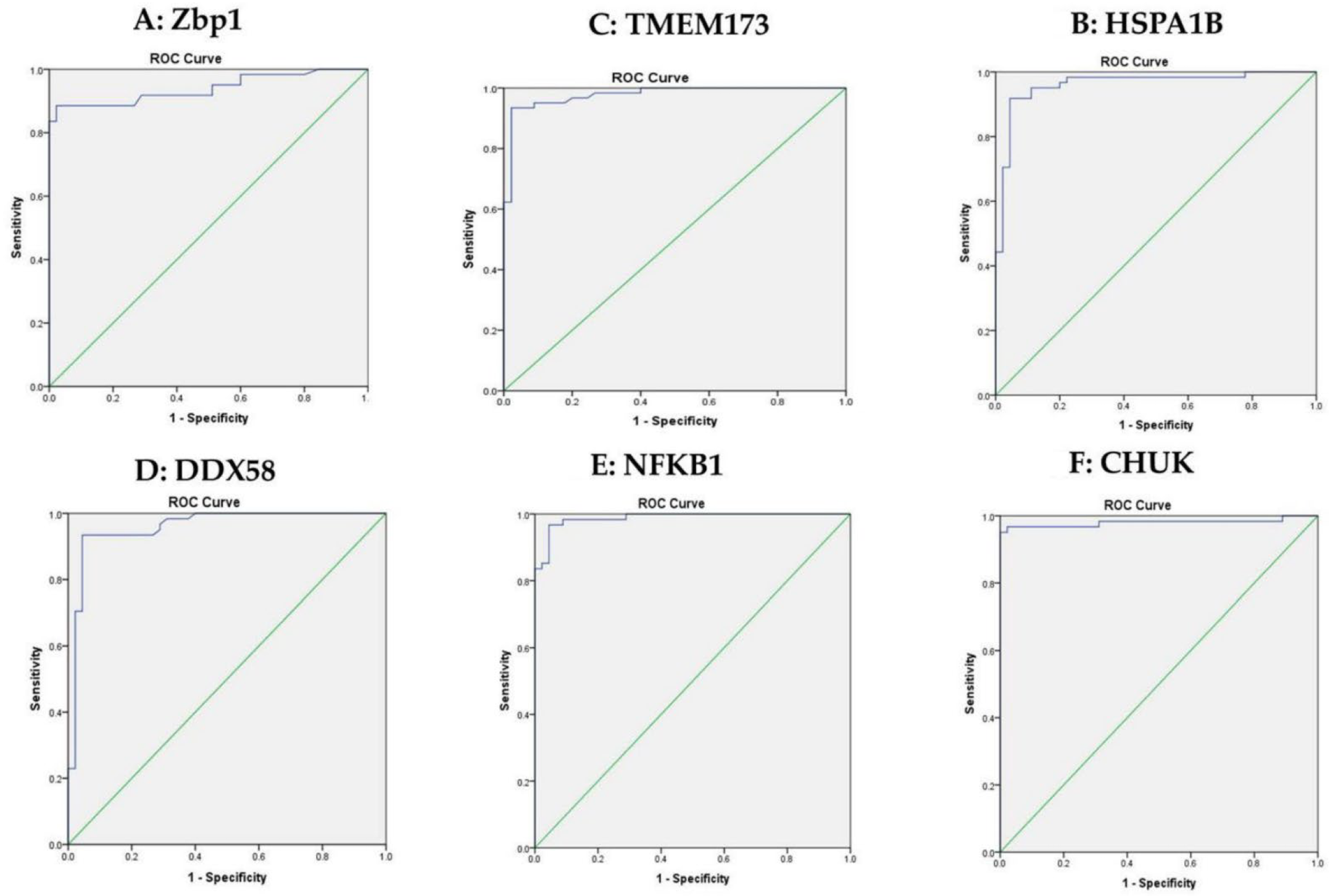

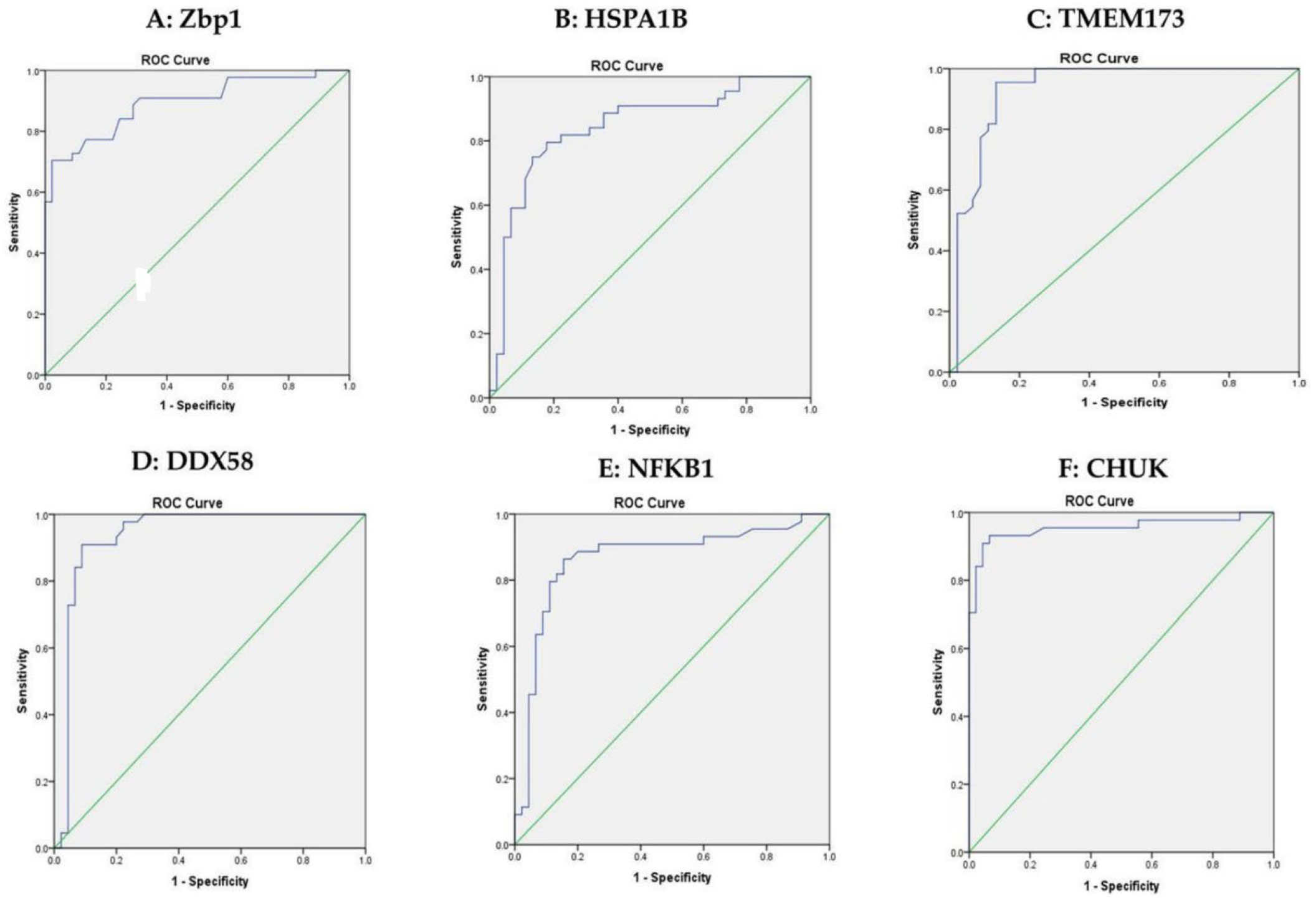
| mRNAs | Healthy Controls | Impaired Glucose Level | T2DM | χ2(c) | p | |||
|---|---|---|---|---|---|---|---|---|
| Median | Mean Rank | Median | Mean Rank | Median | Mean Rank | |||
| RQ ZBP1 | 1 | 31.37 | 5.3 | 73.37 | 56 | 109.5 | 83.9 | 0.000 ** |
| RQ HSPA1B | 1.3 | 32.3 | 4.6 | 68.0 | 17.1 | 112.8 | 90.9 | 0.000 ** |
| RQ TMEM173 | 0.1 | 27.3 | 4.0 | 69.6 | 78.0 | 115.3 | 107.5 | 0.000 ** |
| RQ DDX58 | 0.1 | 28.4 | 8.4 | 71.0 | 80.0 | 113.5 | 99.9 | 0.000 ** |
| RQ NFKB1 | 0.5 | 29.7 | 4.3 | 65.4 | 99.0 | 116.6 | 106.9 | 0.000 ** |
| RQ CHUK | 0.4 | 26.2 | 12.5 | 76.7 | 45.0 | 110.9 | 98.6 | 0.000 ** |
| Bad Glycemic Control HbA1c ≥ 7 | Good Glycemic Control HbA1c < 7 | U(d) | p | |||
|---|---|---|---|---|---|---|
| Median | Mean Rank | Median | Mean Rank | |||
| Zbp1 | 88 | 33 | 29.5 | 17.4 | 351 | 0.0241 ** |
| HSPA1B | 18.2 | 32.59 | 15.03 | 28.18 | 367 | 0.352 |
| TMEM173 | 69.01 | 31.45 | 83.5 | 30.20 | 411.5 | 0.793 |
| DDX58 | 89 | 32.08 | 71.5 | 29.09 | 387 | 0.528 |
| NFKB1 | 122.8 | 33.36 | 66.5 | 19.82 | 437 | 0.05 * |
| CHUK | 44 | 30.79 | 110.9 | 31.36 | 421 | 0.904 |
| Insulin Resistance HOMA-IR ≥ 2.5 | Insulin Sensitive HOMA-IR < 2.5 | U(d) | p | |||
|---|---|---|---|---|---|---|
| Median | Mean Rank | Median | Mean Rank | |||
| Zbp1 | 56 | 31.5 | 55.1 | 27.7 | 185.5 | 0.57 |
| HSPA1B | 88.2 | 32.5 | 12.4 | 17.3 | 134.5 | 0.05 * |
| TMEM173 | 78 | 30.5 | 105.5 | 34.3 | 185.5 | 0.57 |
| DDX58 | 66.6 | 30.2 | 94 | 36.1 | 171 | 0.38 |
| NFKB1 | 89 | 30.5 | 174.8 | 34.2 | 186.5 | 0.58 |
| CHUK | 45 | 30.2 | 138 | 36.1 | 171.5 | 0.39 |
| Group | RQ (Zbp1) | RQ (HSPA1B) | RQ (TMEM173) | RQ (DDX58) | RQ (NFKB1) | RQ (CHUK) | ||
|---|---|---|---|---|---|---|---|---|
| All groups | RQ (Zbp1) | Correlation Coefficient | 1 | 0.578 ** | 0.606 ** | 0.603 ** | 0.642 ** | 0.621 ** |
| Sig. | -------- | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| RQ (HSPA1B) | Correlation Coefficient | 0.578 ** | 1 | 0.626 ** | 0.654 ** | 0.616 ** | 0.601 ** | |
| Sig. | 0.000 | -------- | 0.000 | 0.000 | 0.000 | 0.000 | ||
| RQ (TMEM173) | Correlation Coefficient | 0.606 ** | 0.626 ** | 1 | 0.740 ** | 0.730 ** | 0.739 ** | |
| Sig. | 0.000 | 0.000 | -------- | 0.000 | 0.000 | 0.000 | ||
| RQ (DDX58) | Correlation Coefficient | 0.603 ** | 0.654 ** | 0.740 ** | 1 | 0.691 ** | 0.652 ** | |
| Sig. | 0.000 | 0.000 | 0.000 | -------- | 0.000 | 0.000 | ||
| RQ (NFKB1) | Correlation Coefficient | 0.642 ** | 0.616 ** | 0.730 ** | 0.691 ** | 1 | 0.751 ** | |
| Sig. | 0.000 | 0.000 | 0.000 | 0.000 | -------- | 0.000 | ||
| RQ (CHUK) | Correlation Coefficient | 0.621 ** | 0.601 ** | 0.739 ** | 0.652 ** | 0.751 ** | 1 | |
| Sig. | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | -------- | ||
| Pre-DM group and T2DM | RQ (Zbp1) | Correlation Coefficient | 1 | 0.318 ** | 0.374 ** | 0.384 ** | 0.463 ** | 0.360 ** |
| Sig. | -------- | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| RQ (HSPA1B) | Correlation Coefficient | 0.318 ** | 1 | 0.368 ** | 0.475 ** | 0.403 ** | 0.351 ** | |
| Sig. | 0.001 | -------- | 0.000 | 0.000 | 0.000 | 0.000 | ||
| RQ (TMEM173) | Correlation Coefficient | 0.374 ** | 0.368 ** | 1 | 0.433 ** | 0.539 ** | 0.466 ** | |
| Sig. | 0.000 | 0.000 | -------- | 0.000 | 0.000 | 0.000 | ||
| RQ (DDX58) | Correlation Coefficient | 0.384 ** | 0.475 ** | 0.433 ** | 1 | 0.499 ** | 0.297 ** | |
| Sig. | 0.000 | 0.000 | 0.000 | -------- | 0.000 | 0.002 | ||
| RQ (NFKB1) | Correlation Coefficient | 0.463 ** | 0.403 ** | 0.539 ** | 0.499 ** | 1 | 0.542 ** | |
| Sig. | 0.000 | 0.000 | 0.000 | 0.000 | -------- | 0.000 | ||
| RQ (CHUK) | Correlation Coefficient | 0.360 ** | 0.351 ** | 0.466 ** | 0.297 ** | 0.542 ** | 1 | |
| Sig. | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | -------- | ||
| RQ (Zbp1) | RQ (HSPA1B) | RQ (TMEM173) | RQ (DDX58) | RQ (NFKB1) | RQ (CHUK) | FSG | HbA1c | HOMA_IR | HOMA-B | BMI | Total_Cholesterol | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RQ (Zbp1) | ||||||||||||
| RQ (HSPA1B) | 0.578 ** | |||||||||||
| RQ (TMEM173) | 0.606 ** | 0.626 ** | ||||||||||
| RQ (DDX58) | 0.603 ** | 0.654 ** | 0.740 ** | |||||||||
| RQ (NFKB1) | 0.642 ** | 0.616 ** | 0.730 ** | 0.691 ** | ||||||||
| RQ (CHUK) | 0.621 ** | 0.601 ** | 0.739 ** | 0.652 ** | 0.751 ** | |||||||
| FSG | 0.571 ** | 0.604 ** | 0.657 ** | 0.638 ** | 0.681 ** | 0.615 ** | ||||||
| HbA1c | 0.505 ** | 0.588 ** | 0.567 ** | 0.533 ** | 0.652 ** | 0.484 ** | 0.671 ** | |||||
| HOMA_IR | 0.555 ** | 0.572 ** | 0.477 ** | 0.489 ** | 0.550 ** | 0.438 ** | 0.576 ** | 0.572 ** | ||||
| HOMA-B | −0.655 ** | −0.676 ** | −0.701 ** | −0.724 ** | −0.734 ** | −0.666 ** | −0.739 ** | −0.704 ** | −708 ** | |||
| BMI | 0.500 ** | 0.546 ** | 0.578 ** | 0.551 ** | 0.512 ** | 0.543 ** | 0.491 ** | 0.420 ** | 0.428 ** | −0.495 ** | ||
| Total_Cholesterol | 0.569 ** | 0.606 ** | 0.633 ** | 0.590 ** | 0.658 ** | 0.612 ** | 0.607 ** | 0.656 ** | 0.673 ** | −0.733 ** | 0.747 ** | |
| LDLc | 0.575 ** | 0.669 ** | 0.655 ** | 0.640 ** | 0.702 ** | 0.622 * | 0.670 ** | 0.655 ** | 0.678 ** | −0.744 ** | 0.710 ** | 0.879 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, H.S.; Boshra, M.S.; Agwa, S.H.A.; Hakeem, M.S.A.; Meteini, M.S.E.; Matboli, M. Identification of a Multi-Messenger RNA Signature as Type 2 Diabetes Mellitus Candidate Genes Involved in Crosstalk between Inflammation and Insulin Resistance. Biomolecules 2022, 12, 1230. https://doi.org/10.3390/biom12091230
Ali HS, Boshra MS, Agwa SHA, Hakeem MSA, Meteini MSE, Matboli M. Identification of a Multi-Messenger RNA Signature as Type 2 Diabetes Mellitus Candidate Genes Involved in Crosstalk between Inflammation and Insulin Resistance. Biomolecules. 2022; 12(9):1230. https://doi.org/10.3390/biom12091230
Chicago/Turabian StyleAli, Hebatalla Said, Mariam Sameh Boshra, Sara H. A. Agwa, Mohamed S. Abdel Hakeem, Mahmoud Shawky El Meteini, and Marwa Matboli. 2022. "Identification of a Multi-Messenger RNA Signature as Type 2 Diabetes Mellitus Candidate Genes Involved in Crosstalk between Inflammation and Insulin Resistance" Biomolecules 12, no. 9: 1230. https://doi.org/10.3390/biom12091230
APA StyleAli, H. S., Boshra, M. S., Agwa, S. H. A., Hakeem, M. S. A., Meteini, M. S. E., & Matboli, M. (2022). Identification of a Multi-Messenger RNA Signature as Type 2 Diabetes Mellitus Candidate Genes Involved in Crosstalk between Inflammation and Insulin Resistance. Biomolecules, 12(9), 1230. https://doi.org/10.3390/biom12091230








