MiR-29a Family as a Key Regulator of Skeletal Muscle Dysplasia in a Porcine Model of Intrauterine Growth Retardation
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Treatment
2.2. Small RNA Sequencing
2.3. Prediction and Functional Annotation of Target Genes
2.4. Real-Time Quantitative PCR
2.5. Cell Culture and Transfection
2.6. Luciferase Reporter Assay
2.7. Statistical Analysis
3. Results
3.1. Skeletal Muscle Characteristics in Pigs with Intrauterine Growth Retardation
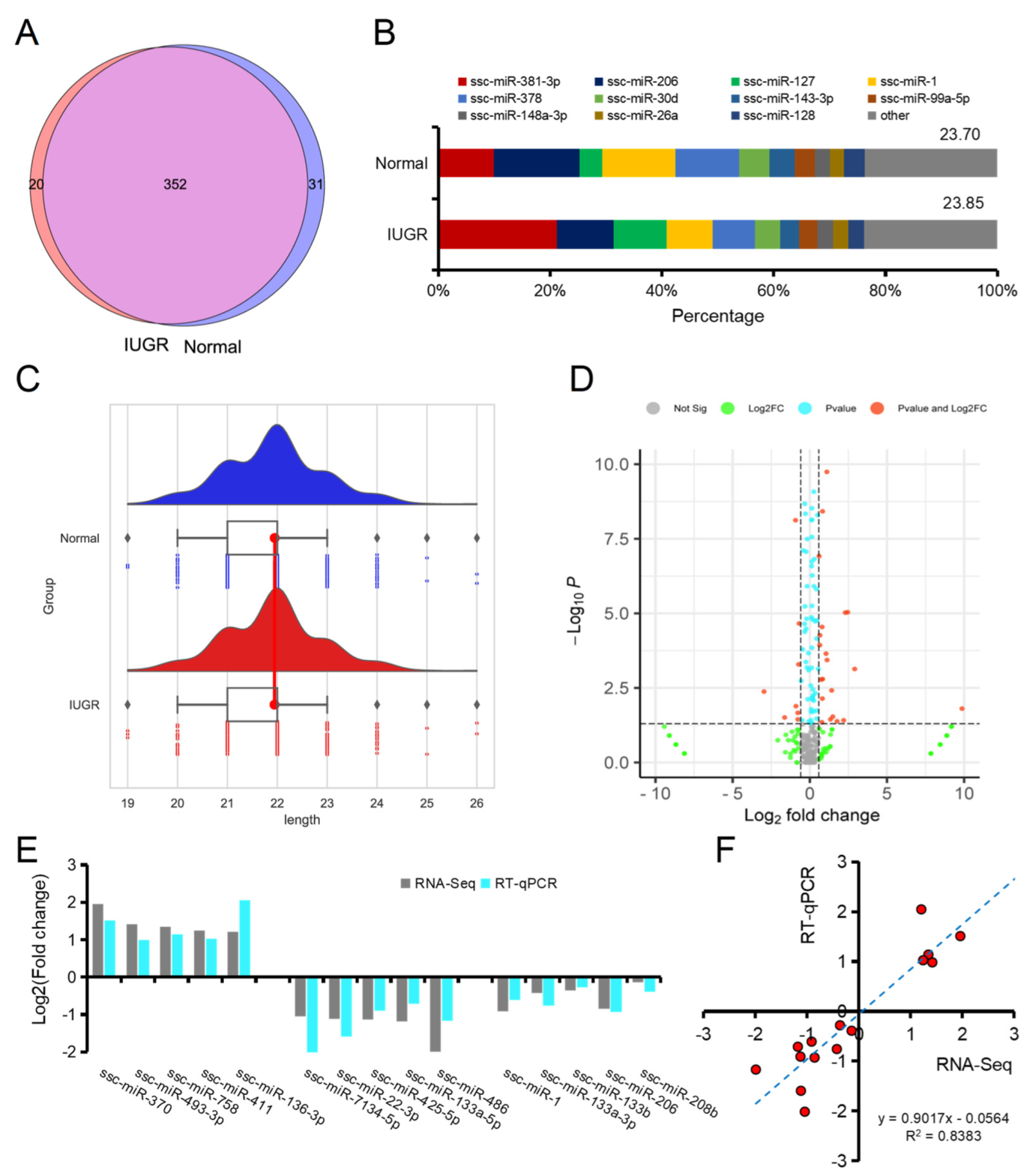
3.2. Characterization of miRNAs in Normal and IUGR Pig Skeletal Muscle
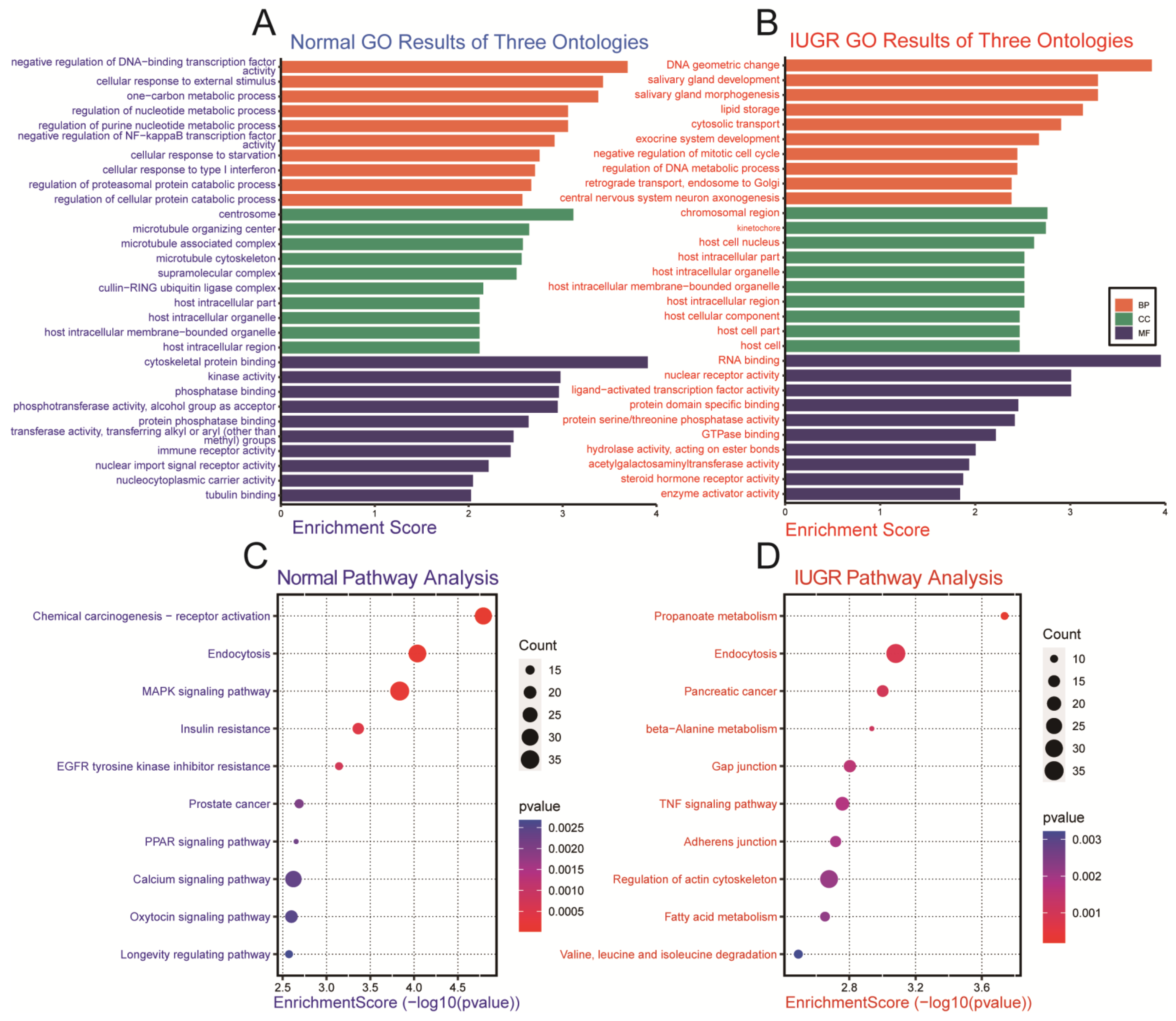
3.3. Functional Enrichment Analysis of Differentially Expressed miRNAs
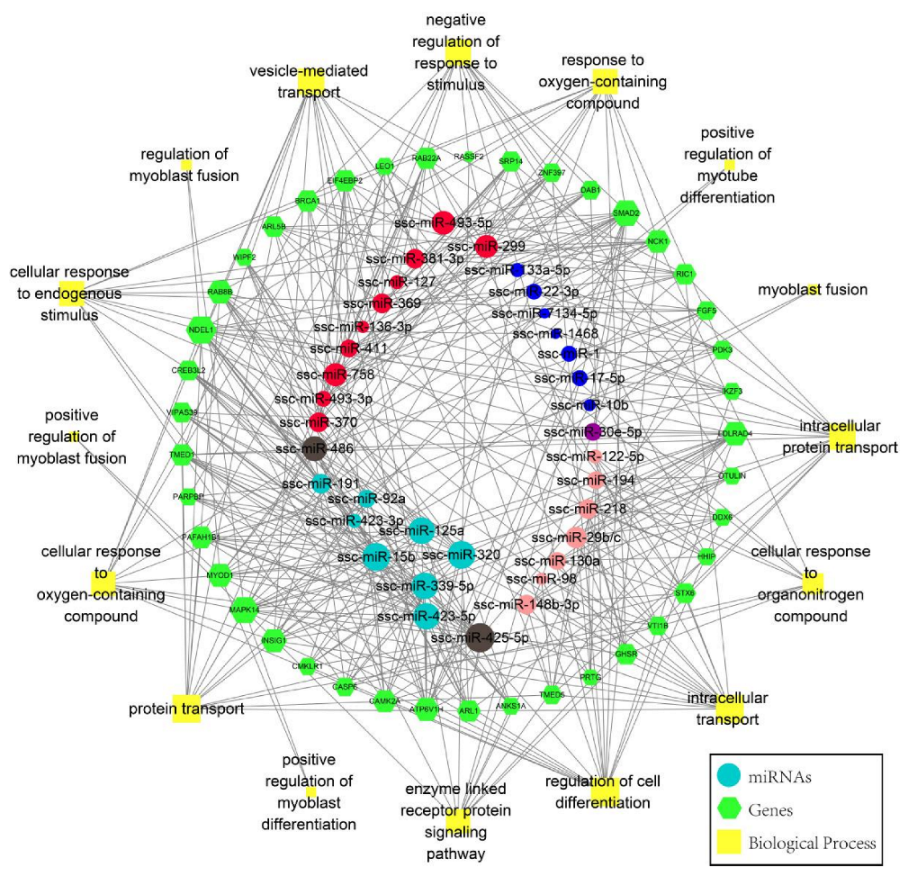
3.4. Cord Blood and Skeletal Muscle miRNAs Regulatory Network in Normal and IUGR Pigs
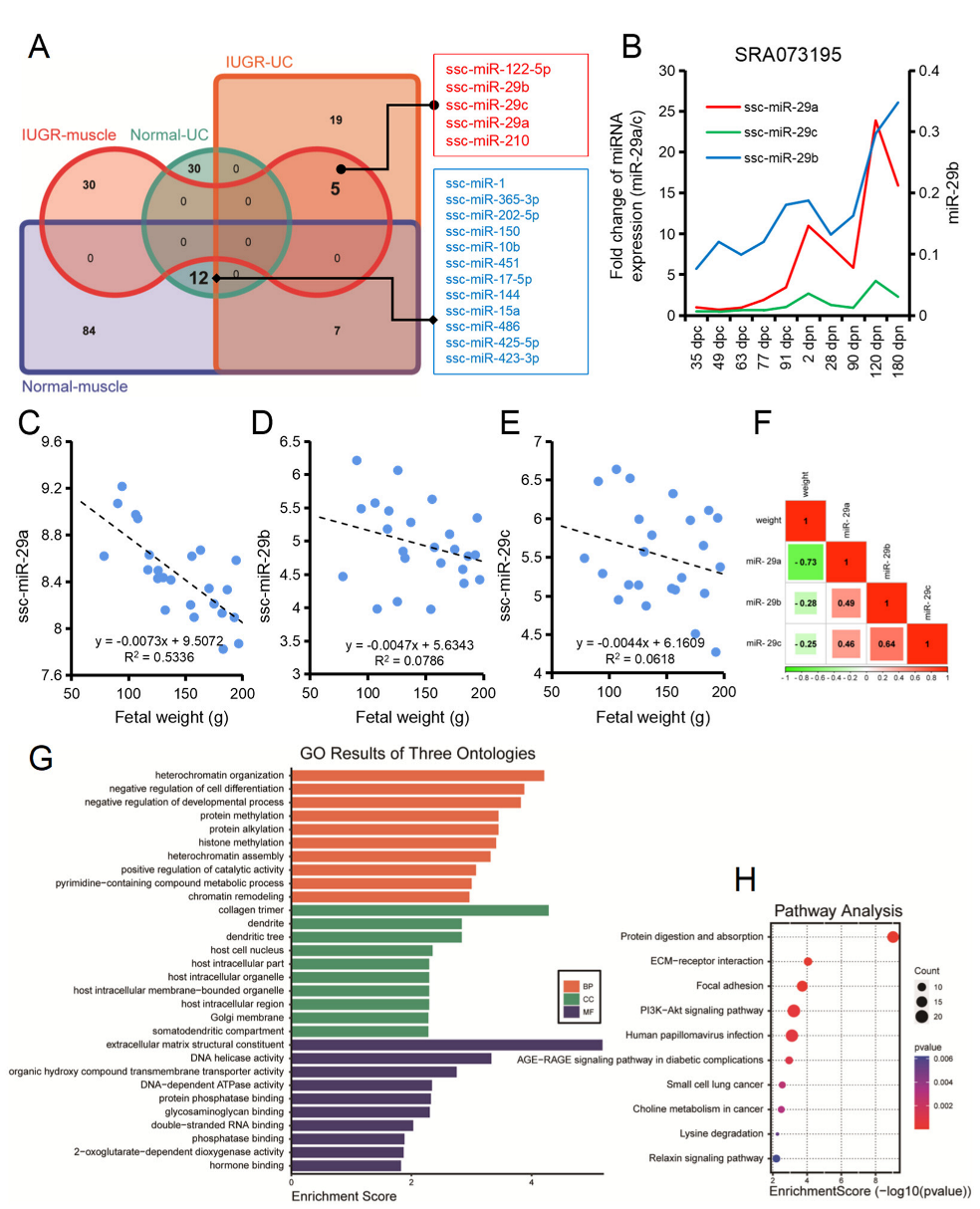
3.5. IGF1 and CCND1 as Common Target Genes of the miR-29 Family
3.6. MiR-29 Family Is Involved in the Regulation of Umbilical Cord Blood miRNAs on Skeletal Muscle Development
4. Discussion
4.1. Characterization of miRNAs in Skeletal Muscle of Normal and IUGR Neonatal Piglets
4.2. Characterization of miRNAs in Skeletal Muscle of Normal and IUGR Neonatal Piglets
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Zhang, R.; Zhou, L.; He, J.; Huang, Q.; Siyal, F.; Zhang, L.; Zhong, X.; Wang, T. Intrauterine growth retardation promotes fetal intestinal autophagy in rats via the mechanistic target of rapamycin pathway. J. Reprod. Dev. 2017, 63, 547–554. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Onis, M.; Blössner, M.; Villar, J. Level and patterns of intrauterine growth retardation in developing countries. Eur. J. Clin. Nutr. 1998, 52 (Suppl. S1), S5–S15. [Google Scholar] [PubMed]
- Hughes, I. Management of fetal endocrine disorders. Growth Horm. IGF Res. Off. J. Growth Horm. Res. Soc. Int. IGF Res. Soc. 2003, 13 (Suppl. A), S55–S61. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.; Wallace, J.; Spencer, T. Board-invited review: Intrauterine growth retardation: Implications for the animal sciences. J. Anim. Sci. 2006, 84, 2316–2337. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Gómez, M.; Garcia-Contreras, C.; Pesantez-Pacheco, J.; Torres-Rovira, L.; Heras-Molina, A.; Astiz, S.; Óvilo, C.; Isabel, B.; Gonzalez-Bulnes, A. Differential effects of litter size and within-litter birthweight on postnatal traits of fatty pigs. Animals 2020, 10, 870. [Google Scholar] [CrossRef] [PubMed]
- Năstase, L.; Cretoiu, D.; Stoicescu, S. Skeletal muscle damage in intrauterine growth restriction. Adv. Exp. Med. Biol. 2018, 1088, 93–106. [Google Scholar] [PubMed]
- Brown, L.; Hay, W. Impact of placental insufficiency on fetal skeletal muscle growth. Mol. Cell. Endocrinol. 2016, 435, 69–77. [Google Scholar] [CrossRef]
- Pedersen, B.; Febbraio, M. Muscles, exercise and obesity: Skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 2012, 8, 457–465. [Google Scholar] [CrossRef]
- Wang, T.; Liu, C.; Feng, C.; Wang, X.; Lin, G.; Zhu, Y.; Yin, J.; Li, D.; Wang, J. Iugr alters muscle fiber development and proteome in fetal pigs. Front. Biosci. 2013, 18, 598–607. [Google Scholar] [CrossRef]
- Yates, D.; Macko, A.; Nearing, M.; Chen, X.; Rhoads, R.; Limesand, S. Developmental programming in response to intrauterine growth restriction impairs myoblast function and skeletal muscle metabolism. J. Pregnancy 2012, 2012, 631038. [Google Scholar] [CrossRef]
- Wilson, S.J.; Mcewan, J.C.; Sheard, P.W.; Harris, A.J. Early stages of myogenesis in a large mammal: Formation of successive generations of myotubes in sheep tibialis cranialis muscle. J. Muscle Res. Cell Motil. 1992, 13, 534–550. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Chen, Y.; Liu, X.; Ye, S.; Yu, K.; Huang, Z.; Yu, J.; Zhou, X.; Chen, H.; Mo, D. Integrative analysis of porcine micrornaome during skeletal muscle development. PLoS ONE 2013, 8, e72418. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.; Calin, G. Micrornas in body fluids—the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef]
- Hu, Y.; Rao, S.; Wang, Z.; Cao, J.; Tan, Y.; Luo, J.; Li, H.; Zhang, W.; Chen, C.; Xie, H. Exosomes from human umbilical cord blood accelerate cutaneous wound healing through mir-21-3p-mediated promotion of angiogenesis and fibroblast function. Theranostics 2018, 8, 169–184. [Google Scholar] [CrossRef]
- Weitzel, R.; Lesniewski, M.; Haviernik, P.; Kadereit, S.; Leahy, P.; Greco, N.; Laughlin, M. Microrna 184 regulates expression of nfat1 in umbilical cord blood cd4+ t cells. Blood 2009, 113, 6648–6657. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Nakaoka, T.; Yamashita, N. Profiling of immune-related microrna expression in human cord blood and adult peripheral blood cells upon proinflammatory stimulation. Eur. J. Haematol. 2012, 88, 31–38. [Google Scholar] [CrossRef]
- Floris, I.; Kraft, J.; Altosaar, I. Roles of microrna across prenatal and postnatal periods. Int. J. Mol. Sci. 2016, 17, 1994. [Google Scholar] [CrossRef]
- Niu, Y.; He, J.; Ahmad, H.; Shen, M.; Zhao, Y.; Gan, Z.; Zhang, L.; Zhong, X.; Wang, C.; Wang, T. Dietary curcumin supplementation increases antioxidant capacity, upregulates nrf2 and hmox1 levels in the liver of piglet model with intrauterine growth retardation. Nutrients 2019, 11, 2978. [Google Scholar] [CrossRef]
- Ali, A.; Murani, E.; Hadlich, F.; Liu, X.; Wimmers, K.; Ponsuksili, S. Prenatal skeletal muscle transcriptome analysis reveals novel microrna-mrna networks associated with intrauterine growth restriction in pigs. Cells 2021, 10, 1007. [Google Scholar] [CrossRef]
- Luo, J.; Fan, Y.; Shen, L.; Niu, L.; Zhao, Y.; Jiang, D.; Zhu, L.; Jiang, A.; Tang, Q.; Ma, J.; et al. The pro-angiogenesis of exosomes derived from umbilical cord blood of intrauterine growth restriction pigs was repressed associated with mirnas. Int. J. Biol. Sci. 2018, 14, 1426–1436. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.; Nam, J.-W.; Bartel, D. Predicting effective microrna target sites in mammalian mrnas. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microrna/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.; Lempicki, R. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Gan, M.; Zheng, T.; Shen, L.-Y.; Tan, Y.; Fan, Y.; Shuai, S.; Bai, L.; Li, X.; Wang, J.; Zhang, S.; et al. Genistein reverses isoproterenol-induced cardiac hypertrophy by regulating mir-451/timp2. Biomed. Pharmacother. 2019, 112, 108618. [Google Scholar] [CrossRef]
- Lin, G.; Wang, X.; Wu, G.; Feng, C.; Zhou, H.; Li, D.; Wang, J. Improving amino acid nutrition to prevent intrauterine growth restriction in mammals. Amino Acids 2014, 46, 1605–1623. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Gan, M.; Zhang, S.; Ma, J.; Tang, G.; Jiang, Y.; Li, M.; Wang, J.; Li, X.; Che, L.; et al. Transcriptome analyses reveal adult metabolic syndrome with intrauterine growth restriction in pig models. Front. Genet. 2018, 9, 291. [Google Scholar] [CrossRef]
- Sakaue, S.; Hirata, J.; Maeda, Y.; Kawakami, E.; Nii, T.; Kishikawa, T.; Ishigaki, K.; Terao, C.; Suzuki, K.; Akiyama, M.; et al. Integration of genetics and mirna-target gene network identified disease biology implicated in tissue specificity. Nucleic Acids Res. 2018, 46, 11898–11909. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related micrornas are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef]
- Li, R.; Sun, Q.; Jia, Y.; Cong, R.; Ni, Y.; Yang, X.; Jiang, Z.; Zhao, R. Coordinated mirna/mrna expression profiles for understanding breed-specific metabolic characters of liver between erhualian and large white pigs. PLoS ONE 2012, 7, e38716. [Google Scholar] [CrossRef]
- Luo, W.; Nie, Q.; Zhang, X. Micrornas involved in skeletal muscle differentiation. J. Genet. Genom. = Yi Chuan Xue Bao 2013, 40, 107–116. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, J.; Liu, W.; Wang, H.; Zhao, L.; Liu, S.; Li, P.; Zhang, S.; Sun, C.; Wu, Y.; et al. Microrna-378 promotes autophagy and inhibits apoptosis in skeletal muscle. Proc. Natl. Acad. Sci. USA 2018, 115, E10849–E10858. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, D.; Kumar, A.; Poojary, M.; Scaria, V.; Datta, M. Rna sequencing reveals potential interacting networks between the altered transcriptome and ncrnome in the skeletal muscle of diabetic mice. Biosci. Rep. 2021, 41, BSR20210495. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.; D’Souza, R.; Schierding, W.; Zeng, N.; Ramzan, F.; O’Sullivan, J.; Poppitt, S.; Cameron-Smith, D. Identification of human skeletal muscle mirna related to strength by high-throughput sequencing. Physiol. Genom. 2018, 50, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Soriano-Arroquia, A.; McCormick, R.; Molloy, A.; McArdle, A.; Goljanek-Whysall, K. Age-related changes in mir-143-3p:Igfbp5 interactions affect muscle regeneration. Aging Cell 2016, 15, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Ping, J.; Ali, S.; Zhen, G.; Juan, L.; Kang, J.; Ziyi, P.; Huixian, L.; Zhihui, Z. Role of micrornas in myogenesis and their effects on meat quality in pig—A review. Asian-Australas J. Anim. Sci. 2020, 33, 1873–1884. [Google Scholar] [CrossRef]
- Margolis, L.; Berryman, C.; Murphy, N.; Carrigan, C.; Young, A.; Carbone, J.; Pasiakos, S. Pi3k-akt-foxo1 pathway targeted by skeletal muscle microrna to suppress proteolytic gene expression in response to carbohydrate intake during aerobic exercise. Physiol. Rep. 2018, 6, e13931. [Google Scholar] [CrossRef]
- Mármol-Sánchez, E.; Ramayo-Caldas, Y.; Quintanilla, R.; Cardoso, T.; González-Prendes, R.; Tibau, J.; Amills, M. Co-expression network analysis predicts a key role of micrornas in the adaptation of the porcine skeletal muscle to nutrient supply. J. Anim. Sci. Biotechnol. 2020, 11, 10. [Google Scholar] [CrossRef]
- Martínez-Rivera, V.; Cárdenas-Monroy, C.; Millan-Catalan, O.; González-Corona, J.; Huerta-Pacheco, N.; Martínez-Gutiérrez, A.; Villavicencio-Queijeiro, A.; Pedraza-Lara, C.; Hidalgo-Miranda, A.; Bravo-Gómez, M.; et al. Dysregulation of mir-381-3p and mir-23b-3p in skeletal muscle could be a possible estimator of early post-mortem interval in rats. PeerJ 2021, 9, e11102. [Google Scholar] [CrossRef]
- Vienberg, S.; Geiger, J.; Madsen, S.; Dalgaard, L. Micrornas in metabolism. Acta Physiol. 2017, 219, 346–361. [Google Scholar] [CrossRef]
- Harafuji, N.; Schneiderat, P.; Walter, M.; Chen, Y. Mir-411 is up-regulated in fshd myoblasts and suppresses myogenic factors. Orphanet J. Rare Dis. 2013, 8, 55. [Google Scholar] [CrossRef]
- Hitachi, K.; Nakatani, M.; Tsuchida, K. Myostatin signaling regulates akt activity via the regulation of mir-486 expression. Int. J. Biochem. Cell Biol. 2014, 47, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, B.; Cao, Y.; Li, W.; Dai, W.; Zhang, L.; Zhang, X.; Ning, C.; Li, H.; Yao, Y.; et al. Mir22hglong non-coding rna -derived mir-22-3p promotes skeletal muscle differentiation and regeneration by inhibiting hdac4. Mol. Ther. Nucleic Acids 2021, 24, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ouyang, H.; Wang, Z.; Chen, B.; Nie, Q. A novel circular rna generated by fgfr2 gene promotes myoblast proliferation and differentiation by sponging mir-133a-5p and mir-29b-1-5p. Cells 2018, 7, 199. [Google Scholar] [CrossRef]
- Pendleton, A.; Wesolowski, S.; Regnault, T.; Lynch, R.; Limesand, S. Dimming the powerhouse: Mitochondrial dysfunction in the liver and skeletal muscle of intrauterine growth restricted fetuses. Front. Endocrinol. 2021, 12, 612888. [Google Scholar] [CrossRef]
- Du, A.; Huang, S.; Zhao, X.; Zhang, Y.; Zhu, L.; Ding, J.; Xu, C. Endoplasmic reticulum stress contributes to acetylcholine receptor degradation by promoting endocytosis in skeletal muscle cells. J. Neuroimmunol. 2016, 290, 109–114. [Google Scholar] [CrossRef]
- Mark, H.; Cooksey, R.C.; Deborah, J.; Glendon, P.; Neidigh, J.L.; Bryan, W.; Gulve, E.A.; Mcclain, D.A. Activation of the hexosamine signaling pathway in adipose tissue results in decreased serum adiponectin and skeletal muscle insulin resistance. Endocrinology 2004, 145, 2118–2128. [Google Scholar]
- You, J.; Chen, J. Autophagy-dependent regulation of skeletal muscle regeneration and strength by a rhogef. Autophagy 2021, 17, 1044–1045. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Han, S.; Shen, X.; He, H.; Chen, Y.; Zhao, J.; Wei, Y.; Wang, Y.; Zhu, Q.; Li, D.; et al. Islr regulates skeletal muscle atrophy via igf1-pi3k/akt-foxo signaling pathway. Cell Tissue Res. 2020, 381, 479–492. [Google Scholar] [CrossRef]
- Keren, A.; Tamir, Y.; Bengal, E. The p38 mapk signaling pathway: A major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 2006, 252, 224–230. [Google Scholar] [CrossRef]
- Calkins, K.; Thamotharan, S.; Dai, Y.; Shin, B.; Kalhan, S.; Devaskar, S. Early dietary restriction in rats alters skeletal muscle tuberous sclerosis complex, ribosomal s6 and mitogen-activated protein kinase. Nutr. Res. 2018, 54, 93–104. [Google Scholar] [CrossRef]
- Juracek, J.; Piler, P.; Janku, P.; Radova, L.; Slaby, O. Identification of microrna signatures in umbilical cord blood associated with maternal characteristics. PeerJ 2019, 7, e6981. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ning, R.; Zacharek, A.; Cui, C.; Cui, X.; Yan, T.; Venkat, P.; Zhang, Y.; Chopp, M. Mir-126 contributes to human umbilical cord blood cell-induced neurorestorative effects after stroke in type-2 diabetic mice. Stem Cells 2016, 34, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Mok, G.; Lozano-Velasco, E.; Münsterberg, A. Micrornas in skeletal muscle development. Semin. Cell Dev. Biol. 2017, 72, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Diniz, G.; Wang, D. Regulation of skeletal muscle by micrornas. Compr. Physiol. 2016, 6, 1279–1294. [Google Scholar] [PubMed]
- Kriegel, A.J.; Liu, Y.; Fang, Y.; Ding, X.; Liang, M. The mir-29 family: Genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol. Genom. 2012, 44, 237–244. [Google Scholar] [CrossRef]
- Wei, W.; He, H.; Zhang, W.; Zhang, H.; Bai, J.; Liu, H.; Cao, J.; Chang, K.; Li, X.; Zhao, S. Mir-29 targets akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013, 4, e668. [Google Scholar] [CrossRef]
- Xu, M.; Tian, G.; Hao, C.; Shi, M.; Zha, D.; Liang, K. Microrna-29 targets fgf2 and inhibits the proliferation, migration and invasion of nasopharyngeal carcinoma cells via pi3k/akt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5215–5222. [Google Scholar]
- Li, J.; Chan, M.; Yu, Y.; Bei, Y.; Chen, P.; Zhou, Q.; Cheng, L.; Chen, L.; Ziegler, O.; Rowe, G.; et al. Mir-29b contributes to multiple types of muscle atrophy. Nat. Commun. 2017, 8, 15201. [Google Scholar] [CrossRef]
- Gan, M.; Shen, L.; Wang, S.; Guo, Z.; Zheng, T.; Tan, Y.; Fan, Y.; Liu, L.; Chen, L.; Jiang, A.; et al. Genistein inhibits high fat diet-induced obesity through mir-222 by targeting btg2 and adipor1. Food Funct. 2020, 11, 2418–2426. [Google Scholar] [CrossRef]
- Panda, A.; Abdelmohsen, K.; Martindale, J.; Di Germanio, C.; Yang, X.; Grammatikakis, I.; Noh, J.; Zhang, Y.; Lehrmann, E.; Dudekula, D.; et al. Novel rna-binding activity of myf5 enhances ccnd1/cyclin d1 mrna translation during myogenesis. Nucleic Acids Res. 2016, 44, 2393–2408. [Google Scholar] [CrossRef]
- Thorn, S.; Sekar, S.; Lavezzi, J.; O’Meara, M.; Brown, L.; Hay, W.; Rozance, P. A physiological increase in insulin suppresses gluconeogenic gene activation in fetal sheep with sustained hypoglycemia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 303, R861–R869. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caliebe, J.; Broekman, S.; Boogaard, M.; Bosch, C.; Ruivenkamp, C.; Oostdijk, W.; Kant, S.; Binder, G.; Ranke, M.; Wit, J.; et al. Igf1, igf1r and shox mutation analysis in short children born small for gestational age and short children with normal birth size (idiopathic short stature). Horm. Res. Paediatr. 2012, 77, 250–260. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Ma, J.; Pan, H.; Gan, M.; Shen, L. MiR-29a Family as a Key Regulator of Skeletal Muscle Dysplasia in a Porcine Model of Intrauterine Growth Retardation. Biomolecules 2022, 12, 1193. https://doi.org/10.3390/biom12091193
Zhu Y, Ma J, Pan H, Gan M, Shen L. MiR-29a Family as a Key Regulator of Skeletal Muscle Dysplasia in a Porcine Model of Intrauterine Growth Retardation. Biomolecules. 2022; 12(9):1193. https://doi.org/10.3390/biom12091193
Chicago/Turabian StyleZhu, Yan, Jianfeng Ma, Hongmei Pan, Mailin Gan, and Linyuan Shen. 2022. "MiR-29a Family as a Key Regulator of Skeletal Muscle Dysplasia in a Porcine Model of Intrauterine Growth Retardation" Biomolecules 12, no. 9: 1193. https://doi.org/10.3390/biom12091193
APA StyleZhu, Y., Ma, J., Pan, H., Gan, M., & Shen, L. (2022). MiR-29a Family as a Key Regulator of Skeletal Muscle Dysplasia in a Porcine Model of Intrauterine Growth Retardation. Biomolecules, 12(9), 1193. https://doi.org/10.3390/biom12091193








