Reduced Expression of KRT17 Predicts Poor Prognosis in HER2high Breast Cancer
Abstract
:1. Introduction
2. Methods and Materials
2.1. Data Collection
2.2. Patients Sample Collection
2.3. Cell Lines and Cell Culture
2.4. Total RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
2.5. Breast Cancer Tissue Microarray (TMA) and IHC Detection
2.6. Analysis of Correlations with Survival and Clinical Stage
2.7. Identification of Differentially Expressed Genes (DEGs) and Functional Enrichment Analysis
2.8. Immune Cell Infiltration Analysis
2.9. Protein–Protein Interaction (PPI) Network Analysis and Functional Module Analysis
2.10. Statistical Analysis
3. Results
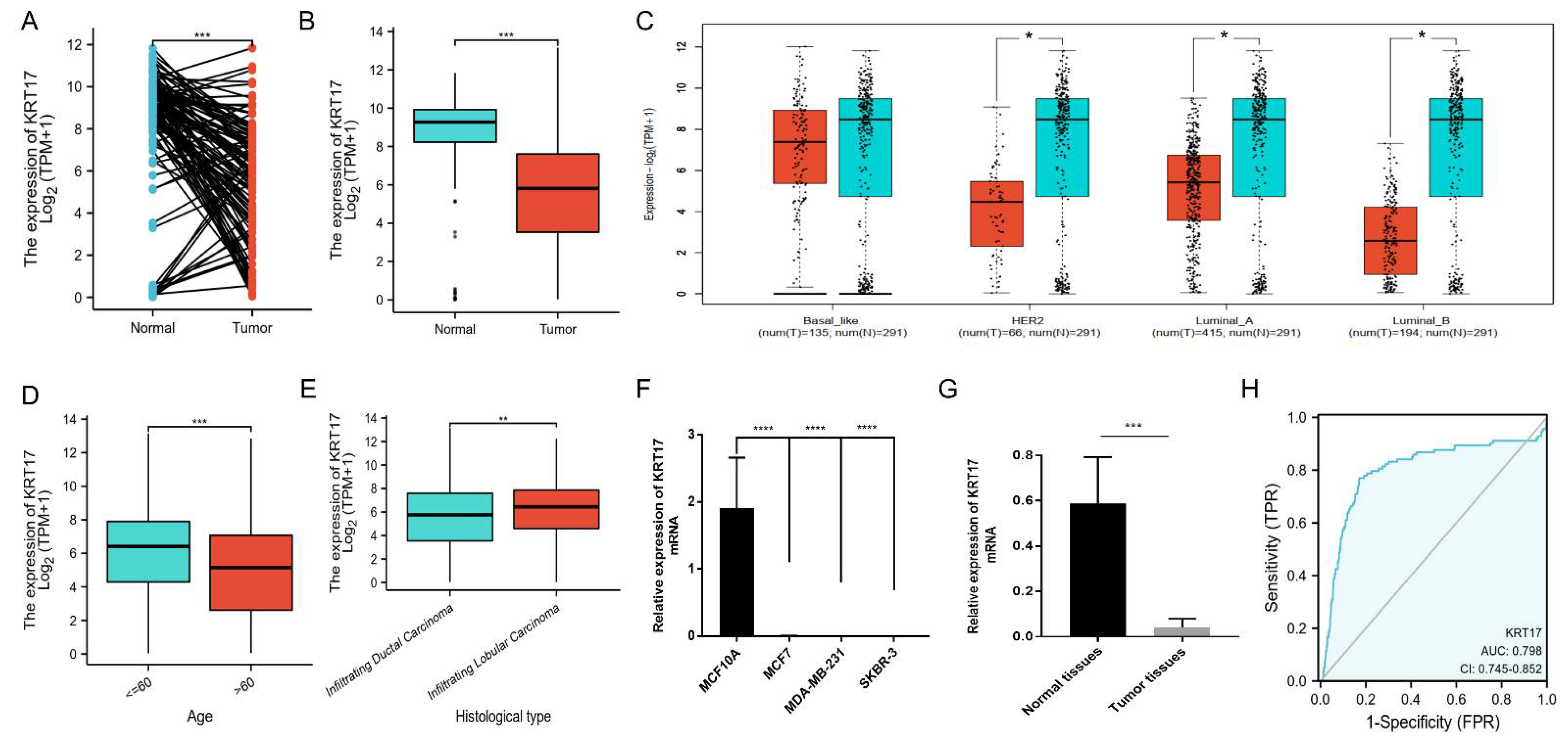
3.1. Downregulated Expression of KRT17 in BC
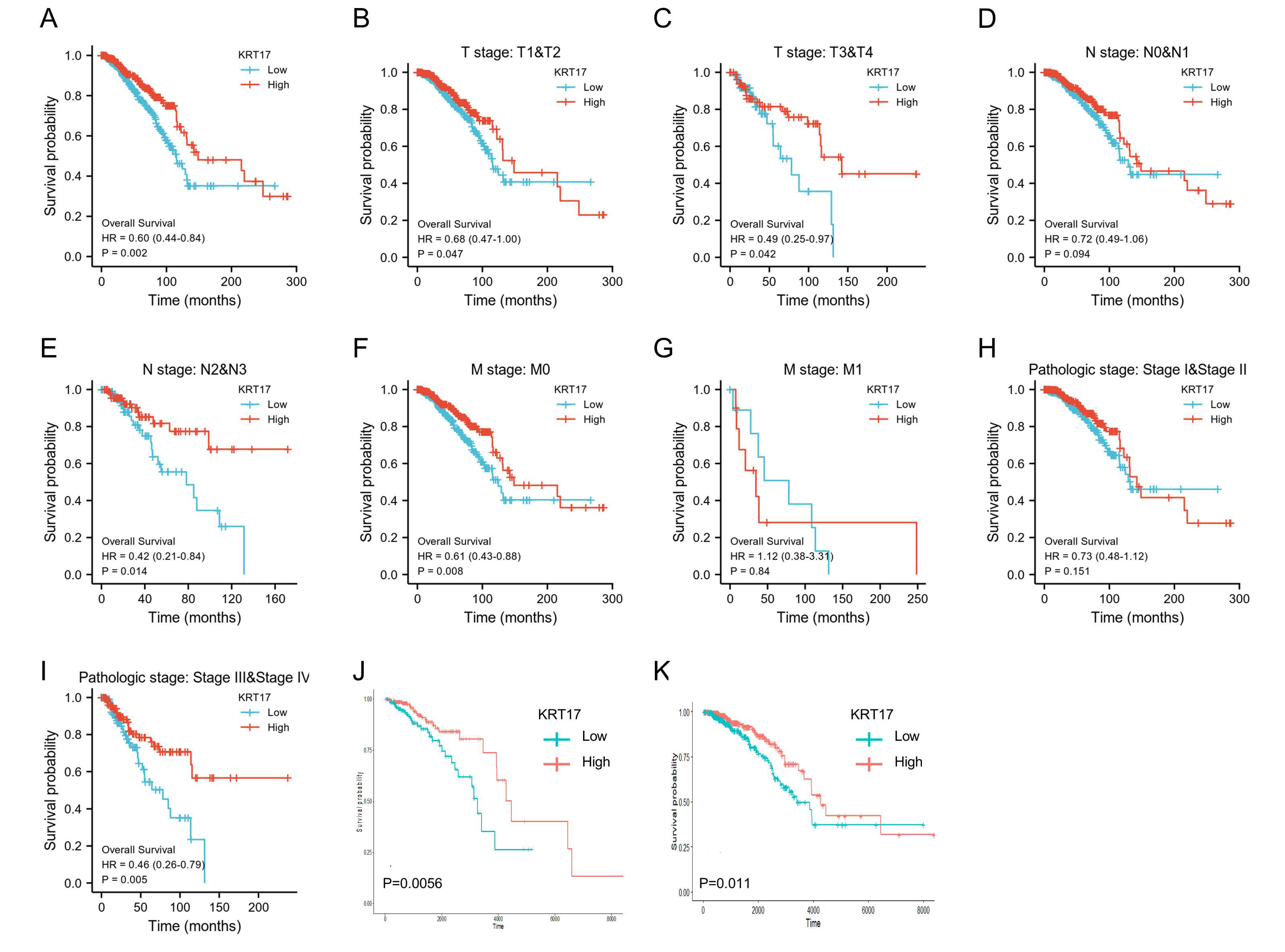
3.2. Association between KRT17 Expression and BC Patient Prognosis
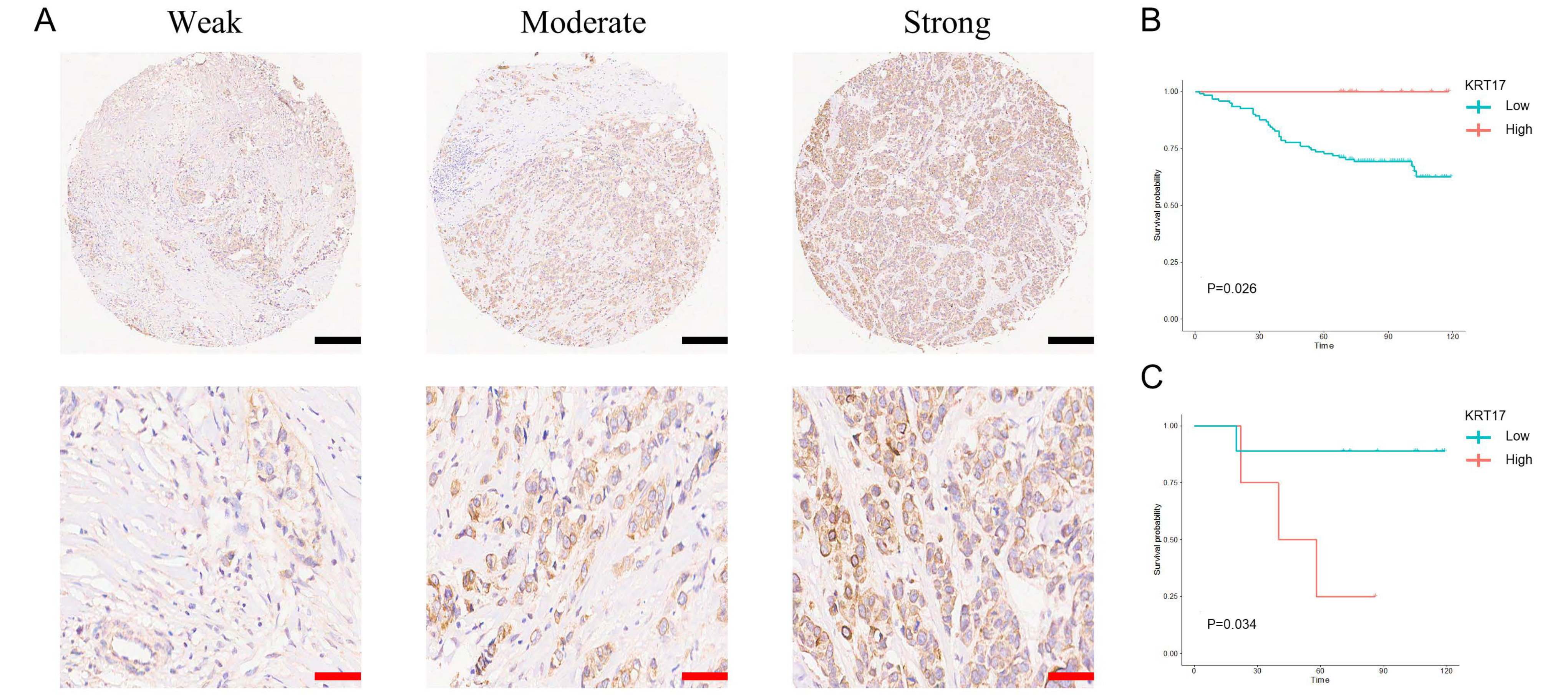
3.3. Expression of KRT17 in BC Tissues and Its Effect on BC Patient Prognosis
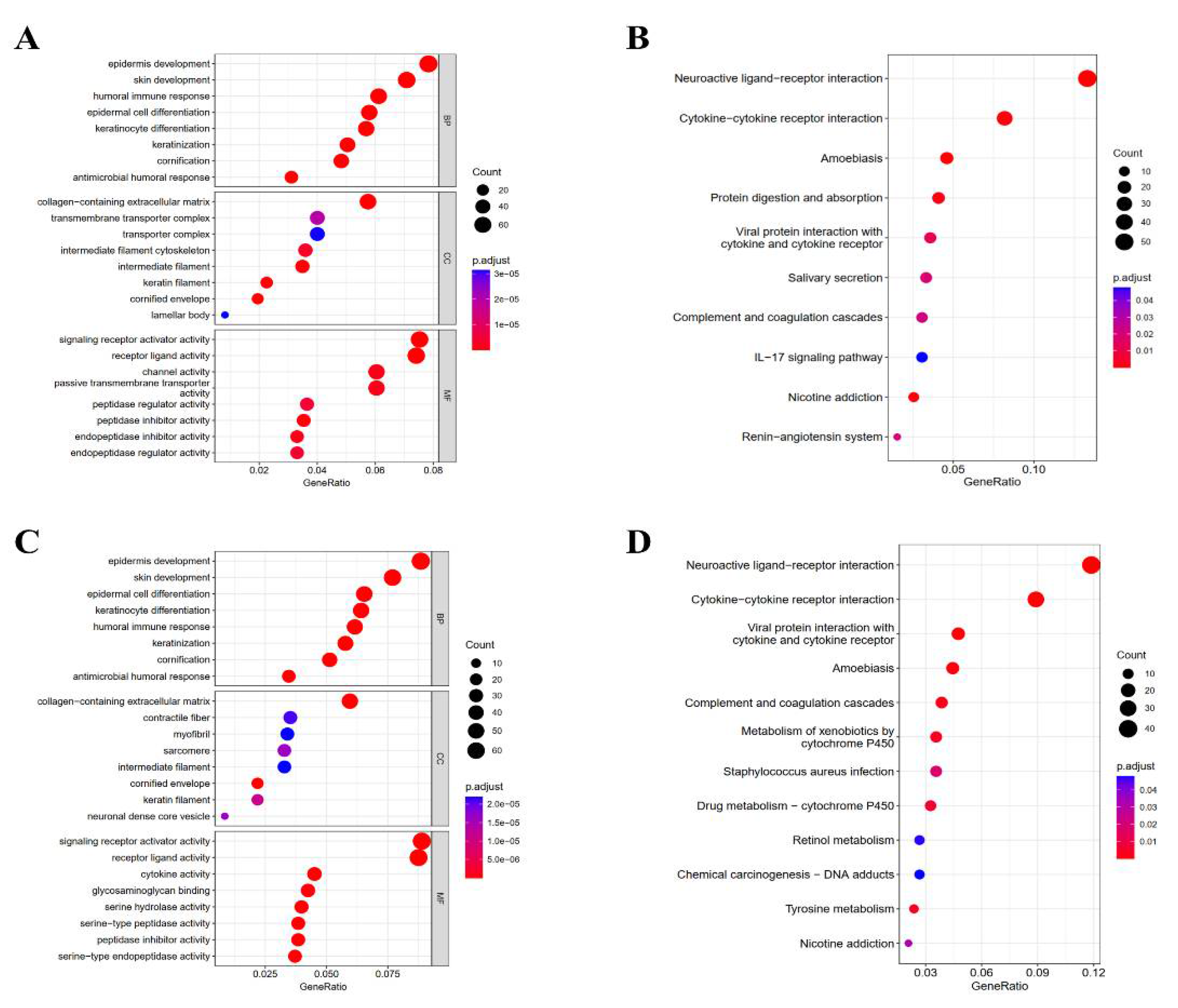
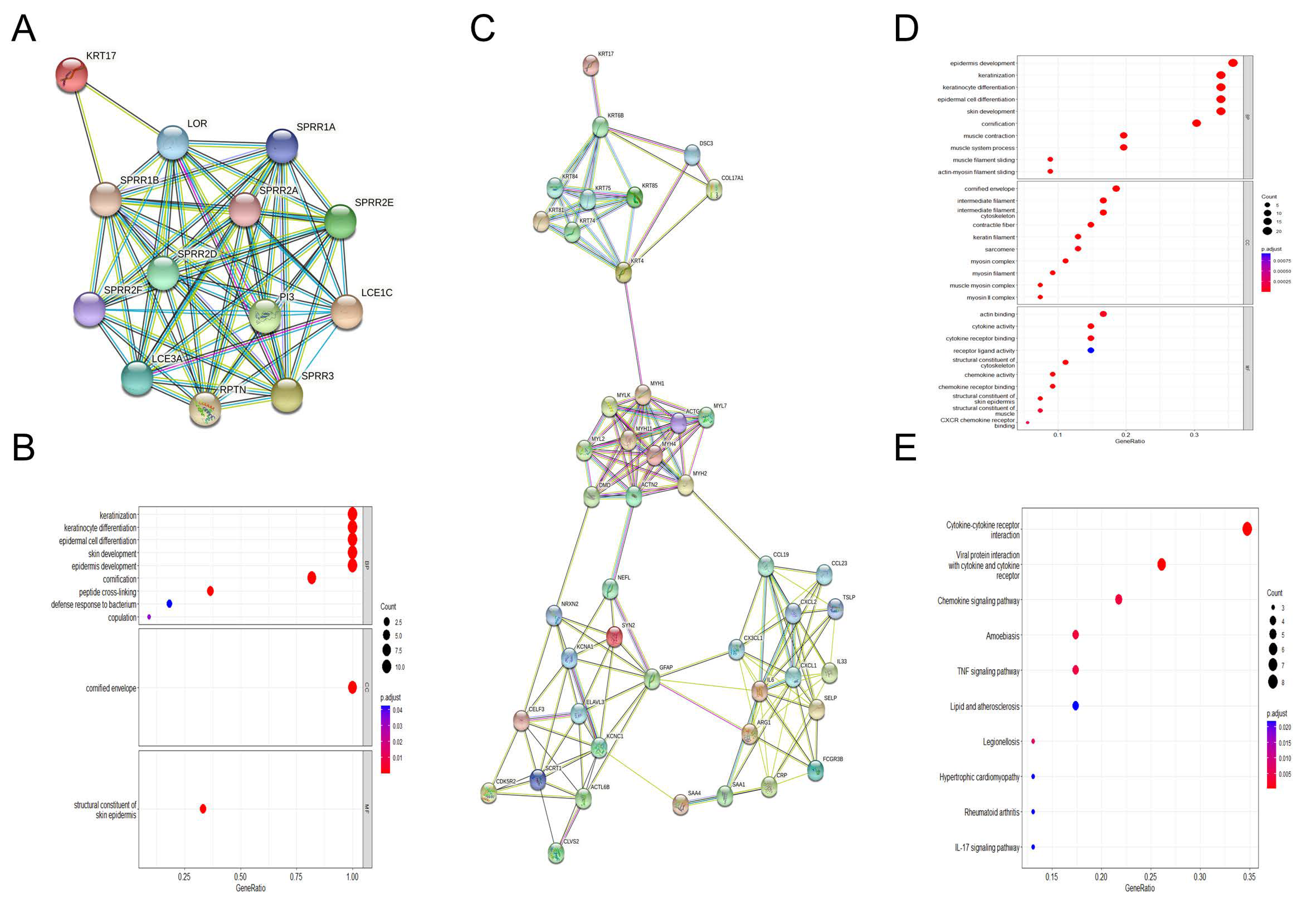
3.4. Enrichment Analysis of KRT17-Related Genes
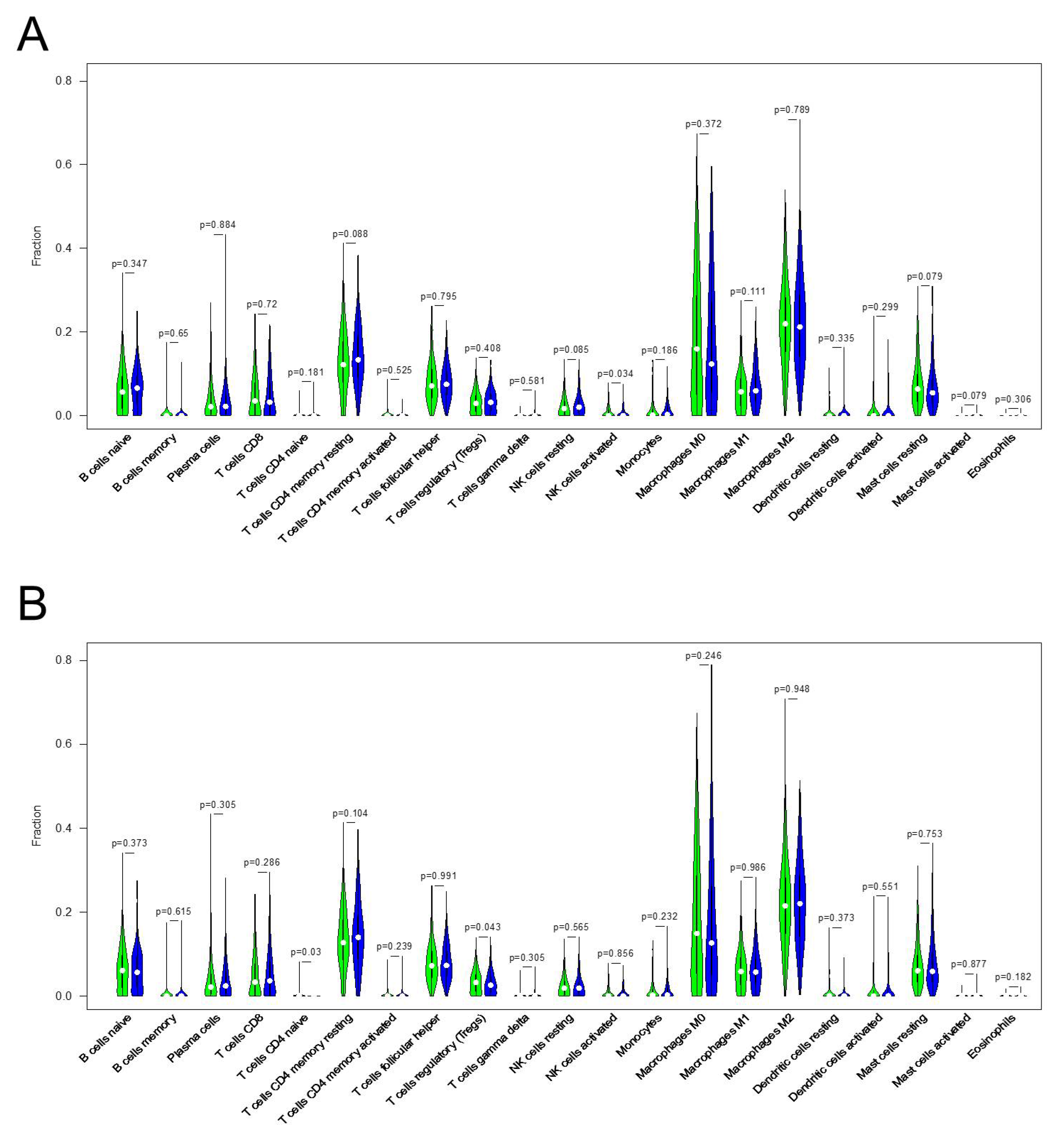
3.5. Correlation between KRT17 Expression and the Levels of Infiltrating Immune Cells in HER2high and ERhigh BC
3.6. PPI Network and Functional Module Analyses in HER2high BC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sperduto, P.W.; Mesko, S.; Li, J.; Cagney, D.; Aizer, A.; Lin, N.U.; Nesbit, E.; Kruser, T.J.; Chan, J.; Braunstein, S.; et al. Estrogen/progesterone receptor and HER2 discordance between primary tumor and brain metastases in breast cancer and its effect on treatment and survival. Neuro Oncol. 2020, 22, 1359–1367. [Google Scholar] [CrossRef] [PubMed]
- Tolaney, S.M.; Najita, J.; Sperinde, J.; Huang, W.; Chen, W.Y.; Savoie, J.; Fornier, M.; Winer, E.P.; Bunnell, C.; Krop, I.E. A phase II study of ixabepilone and trastuzumab for metastatic HER2-positive breast cancer. Ann. Oncol. 2013, 24, 1841–1847. [Google Scholar] [CrossRef] [PubMed]
- Tavares, S.; Vieira, A.F.; Taubenberger, A.V.; Araujo, M.; Martins, N.P.; Bras-Pereira, C.; Polonia, A.; Herbig, M.; Barreto, C.; Otto, O.; et al. Actin stress fiber organization promotes cell stiffening and proliferation of pre-invasive breast cancer cells. Nat. Commun. 2017, 8, 15237. [Google Scholar] [CrossRef]
- Tischler, J.; Crew, K.D.; Chung, W.K. Cases in Precision Medicine: The Role of Tumor and Germline Genetic Testing in Breast Cancer Management. Ann. Intern. Med. 2019, 171, 925–930. [Google Scholar] [CrossRef]
- Soudy, R.; Etayash, H.; Bahadorani, K.; Lavasanifar, A.; Kaur, K. Breast Cancer Targeting Peptide Binds Keratin 1: A New Molecular Marker for Targeted Drug Delivery to Breast Cancer. Mol. Pharm. 2017, 14, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.W.; Pritchard, J.R.; Leighow, S.M.; Aminov, B.I.; Beppu, L.; Kim, D.S.; Hodgson, J.G.; Rivera, V.M.; Loeb, L.A.; Radich, J.P. Single-Molecule Sequencing Reveals Patterns of Preexisting Drug Resistance That Suggest Treatment Strategies in Philadelphia-Positive Leukemias. Clin. Cancer Res. 2018, 24, 5321–5334. [Google Scholar] [CrossRef]
- Ehrlich, F.; Lachner, J.; Hermann, M.; Tschachler, E.; Eckhart, L. Convergent Evolution of Cysteine-Rich Keratins in Hard Skin Appendages of Terrestrial Vertebrates. Mol. Biol. Evol. 2020, 37, 982–993. [Google Scholar] [CrossRef]
- Cruz, R.; Pereira-Castro, I.; Almeida, M.T.; Moreira, A.; Cabanes, D.; Sousa, S. Epithelial Keratins Modulate cMet Expression and Signaling and Promote InlB-Mediated Listeria monocytogenes Infection of HeLa Cells. Front. Cell. Infect. Microbiol. 2018, 8, 146. [Google Scholar] [CrossRef]
- DePianto, D.J.; Heiden, J.A.V.; Morshead, K.B.; Sun, K.H.; Modrusan, Z.; Teng, G.; Wolters, P.J.; Arron, J.R. Molecular mapping of interstitial lung disease reveals a phenotypically distinct senescent basal epithelial cell population. JCI Insight 2021, 6, e143626. [Google Scholar] [CrossRef]
- Hu, H.; Xu, D.H.; Huang, X.X.; Zhu, C.C.; Xu, J.; Zhang, Z.Z.; Zhao, G. Keratin17 Promotes Tumor Growth and is Associated with Poor Prognosis in Gastric Cancer. J. Cancer 2018, 9, 346–357. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Ni, X.F.; Tang, H.; Zhang, J.; Zheng, C.; Lin, J.; Wang, C.; Sun, L.; Chen, B. KRT17 Functions as a Tumor Promoter and Regulates Proliferation, Migration and Invasion in Pancreatic Cancer via mTOR/S6k1 Pathway. Cancer Manag. Res. 2020, 12, 2087–2095. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xu, H.; Ji, H.; Zhai, B.; Zhu, J.; Gao, M.; Zhu, H.; Wang, X. Low Expression of Keratin17 is Related to Poor Prognosis in Bladder Cancer. OncoTargets Ther. 2021, 14, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Merkin, R.D.; Vanner, E.A.; Romeiser, J.L.; Shroyer, A.L.W.; Escobar-Hoyos, L.F.; Li, J.; Powers, R.S.; Burke, S.; Shroyer, K.R. Keratin 17 is overexpressed and predicts poor survival in estrogen receptor-negative/human epidermal growth factor receptor-2-negative breast cancer. Hum. Pathol. 2017, 62, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Mockler, D.; Escobar-Hoyos, L.F.; Akalin, A.; Romeiser, J.; Shroyer, A.L.; Shroyer, K.R. Keratin 17 Is a Prognostic Biomarker in Endocervical Glandular Neoplasia. Am. J. Clin. Pathol. 2017, 148, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Regenbogen, E.; Mo, M.; Romeiser, J.; Shroyer, A.L.W.; Escobar-Hoyos, L.F.; Burke, S.; Shroyer, K.R. Elevated expression of keratin 17 in oropharyngeal squamous cell carcinoma is associated with decreased survival. Head Neck 2018, 40, 1788–1798. [Google Scholar] [CrossRef]
- Wang, Y.F.; Lang, H.Y.; Yuan, J.; Wang, J.; Wang, R.; Zhang, X.H.; Zhang, J.; Zhao, T.; Li, Y.R.; Liu, J.Y.; et al. Overexpression of keratin 17 is associated with poor prognosis in epithelial ovarian cancer. Tumour Biol. 2013, 34, 1685–1689. [Google Scholar] [CrossRef]
- Roa-Peña, L.; Leiton, C.V.; Babu, S.; Pan, C.H.; Vanner, E.A.; Akalin, A.; Bandovic, J.; Moffitt, R.A.; Shroyer, K.R.; Escobar-Hoyos, L.F. Keratin 17 identifies the most lethal molecular subtype of pancreatic cancer. Sci. Rep. 2019, 9, 11239. [Google Scholar] [CrossRef]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e411. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Karantza, V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene 2011, 30, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Maruthappu, T.; Chikh, A.; Fell, B.; Delaney, P.J.; Brooke, M.A.; Levet, C.; Moncada-Pazos, A.; Ishida-Yamamoto, A.; Blaydon, D.; Waseem, A.; et al. Rhomboid family member 2 regulates cytoskeletal stress-associated Keratin 16. Nat. Commun. 2017, 8, 14174. [Google Scholar] [CrossRef]
- Chivu-Economescu, M.; Dragu, D.L.; Necula, L.G.; Matei, L.; Enciu, A.M.; Bleotu, C.; Diaconu, C.C. Knockdown of KRT17 by siRNA induces antitumoral effects on gastric cancer cells. Gastric Cancer 2017, 20, 948–959. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Cooper, L.A.; Demicco, E.G.; Saltz, J.H.; Powell, R.T.; Rao, A.; Lazar, A.J. PanCancer insights from The Cancer Genome Atlas: The pathologist’s perspective. J. Pathol. 2018, 244, 512–524. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Napoli, M.; Ackerman, H.D.; Raulji, P.M.; Montey, N.; Flores, E.R.; Brohl, A.S. Regulation of MYO18B mRNA by a network of C19MC miRNA-520G, IFN-gamma, CEBPB, p53 and bFGF in hepatocellular carcinoma. Sci. Rep. 2020, 10, 12371. [Google Scholar] [CrossRef]
- Shi, X.; Jin, L.; Dang, E.; Chang, T.; Feng, Z.; Liu, Y.; Wang, G. IL-17A upregulates keratin 17 expression in keratinocytes through STAT1- and STAT3-dependent mechanisms. J. Investig. Dermatol. 2011, 131, 2401–2408. [Google Scholar] [CrossRef]
- Jiang, C.K.; Flanagan, S.; Ohtsuki, M.; Shuai, K.; Freedberg, I.M.; Blumenberg, M. Disease-activated transcription factor: Allergic reactions in human skin cause nuclear translocation of STAT-91 and induce synthesis of keratin K17. Mol. Cell. Biol. 1994, 14, 4759–4769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horlock, C.; Shakib, F.; Mahdavi, J.; Jones, N.S.; Sewell, H.F.; Ghaemmaghami, A.M. Analysis of proteomic profiles and functional properties of human peripheral blood myeloid dendritic cells, monocyte-derived dendritic cells and the dendritic cell-like KG-1 cells reveals distinct characteristics. Genome Biol. 2007, 8, R30. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zou, M.; Liu, Y.; Que, K.; Wang, Y.; Liu, C.; Gong, J.; You, Y. Keratin 17 Suppresses Cell Proliferation and Epithelial-Mesenchymal Transition in Pancreatic Cancer. Front. Med. 2020, 7, 572494. [Google Scholar] [CrossRef] [PubMed]
- Nunez, N.G.; Tosello Boari, J.; Ramos, R.N.; Richer, W.; Cagnard, N.; Anderfuhren, C.D.; Niborski, L.L.; Bigot, J.; Meseure, D.; De La Rochere, P.; et al. Tumor invasion in draining lymph nodes is associated with Treg accumulation in breast cancer patients. Nat. Commun. 2020, 11, 3272. [Google Scholar] [CrossRef] [PubMed]
- Block, M.S.; Dietz, A.B.; Gustafson, M.P.; Kalli, K.R.; Erskine, C.L.; Youssef, B.; Vijay, G.V.; Allred, J.B.; Pavelko, K.D.; Strausbauch, M.A.; et al. Th17-inducing autologous dendritic cell vaccination promotes antigen-specific cellular and humoral immunity in ovarian cancer patients. Nat. Commun. 2020, 11, 5173. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, S.; Liu, W.; Yong, L.; Liu, D.; Lin, X.; Huang, Y.; Wang, H.; Cai, F. Reduced Expression of KRT17 Predicts Poor Prognosis in HER2high Breast Cancer. Biomolecules 2022, 12, 1183. https://doi.org/10.3390/biom12091183
Tang S, Liu W, Yong L, Liu D, Lin X, Huang Y, Wang H, Cai F. Reduced Expression of KRT17 Predicts Poor Prognosis in HER2high Breast Cancer. Biomolecules. 2022; 12(9):1183. https://doi.org/10.3390/biom12091183
Chicago/Turabian StyleTang, Shasha, Wenjing Liu, Liyun Yong, Dongyang Liu, Xiaoyan Lin, Yuan Huang, Hui Wang, and Fengfeng Cai. 2022. "Reduced Expression of KRT17 Predicts Poor Prognosis in HER2high Breast Cancer" Biomolecules 12, no. 9: 1183. https://doi.org/10.3390/biom12091183
APA StyleTang, S., Liu, W., Yong, L., Liu, D., Lin, X., Huang, Y., Wang, H., & Cai, F. (2022). Reduced Expression of KRT17 Predicts Poor Prognosis in HER2high Breast Cancer. Biomolecules, 12(9), 1183. https://doi.org/10.3390/biom12091183




