Comparative Transcriptome Analysis Reveals Hormone Signal Transduction and Sucrose Metabolism Related Genes Involved in the Regulation of Anther Dehiscence in Photo-Thermo-Sensitive Genic Male Sterile Wheat
Abstract
:1. Introduction
2. Results
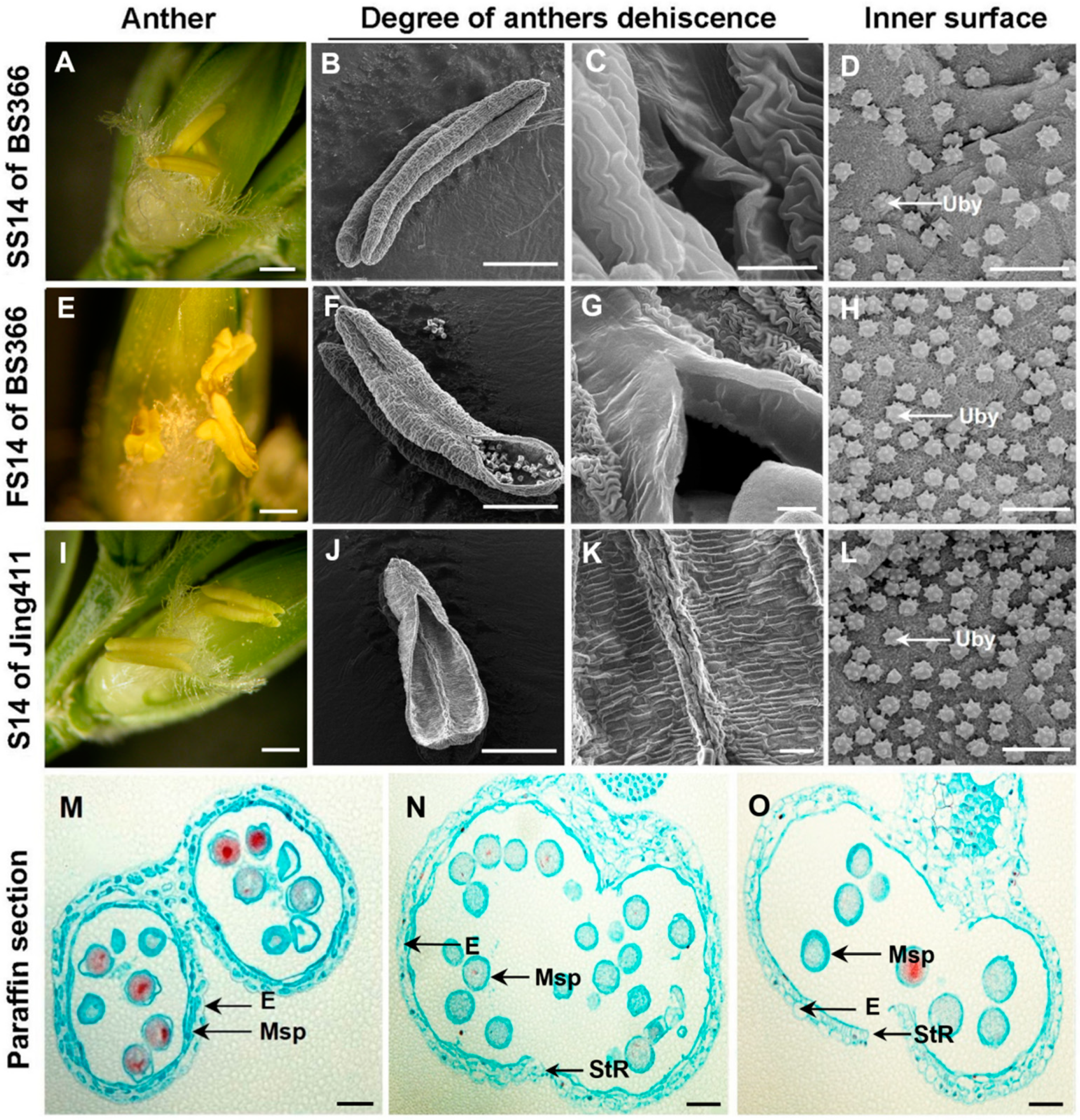
2.1. Phenotypic Characterization of Anther Dehiscence
2.2. Overview of RNA-Seq Data Analysis
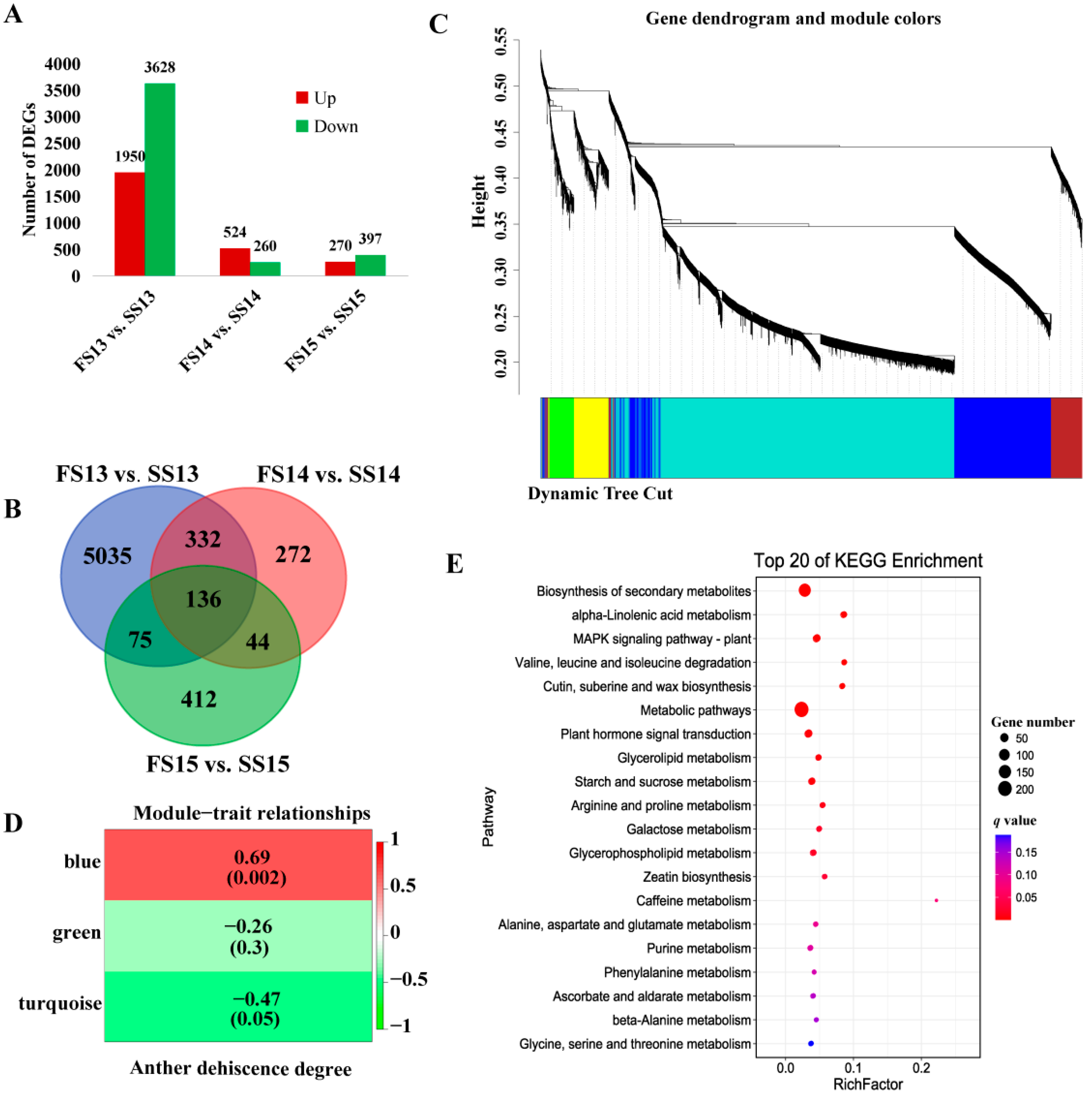
2.3. Identification of Differentially Expressed Genes and Weighted Gene Co-Expression Network Analysis for DEGs
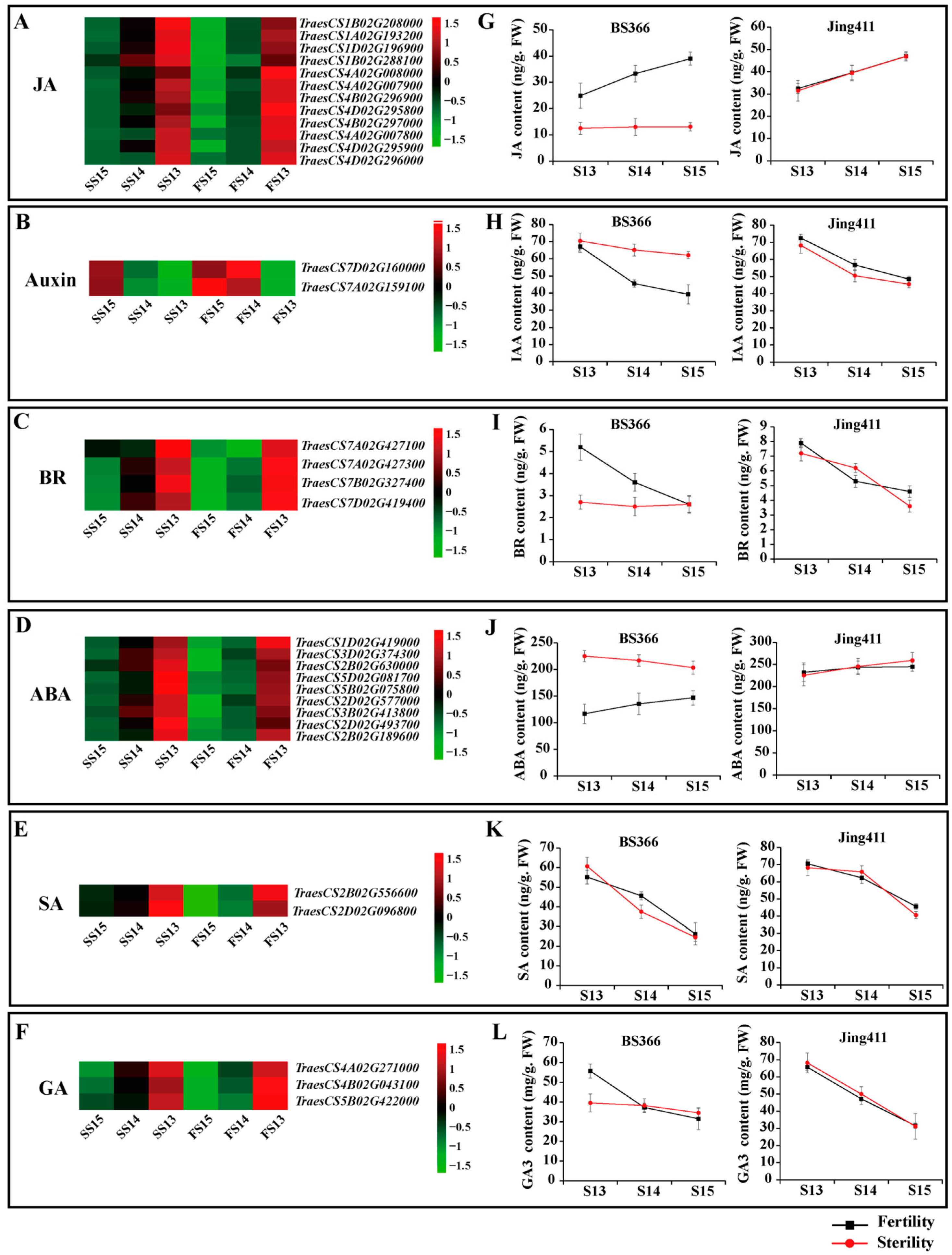
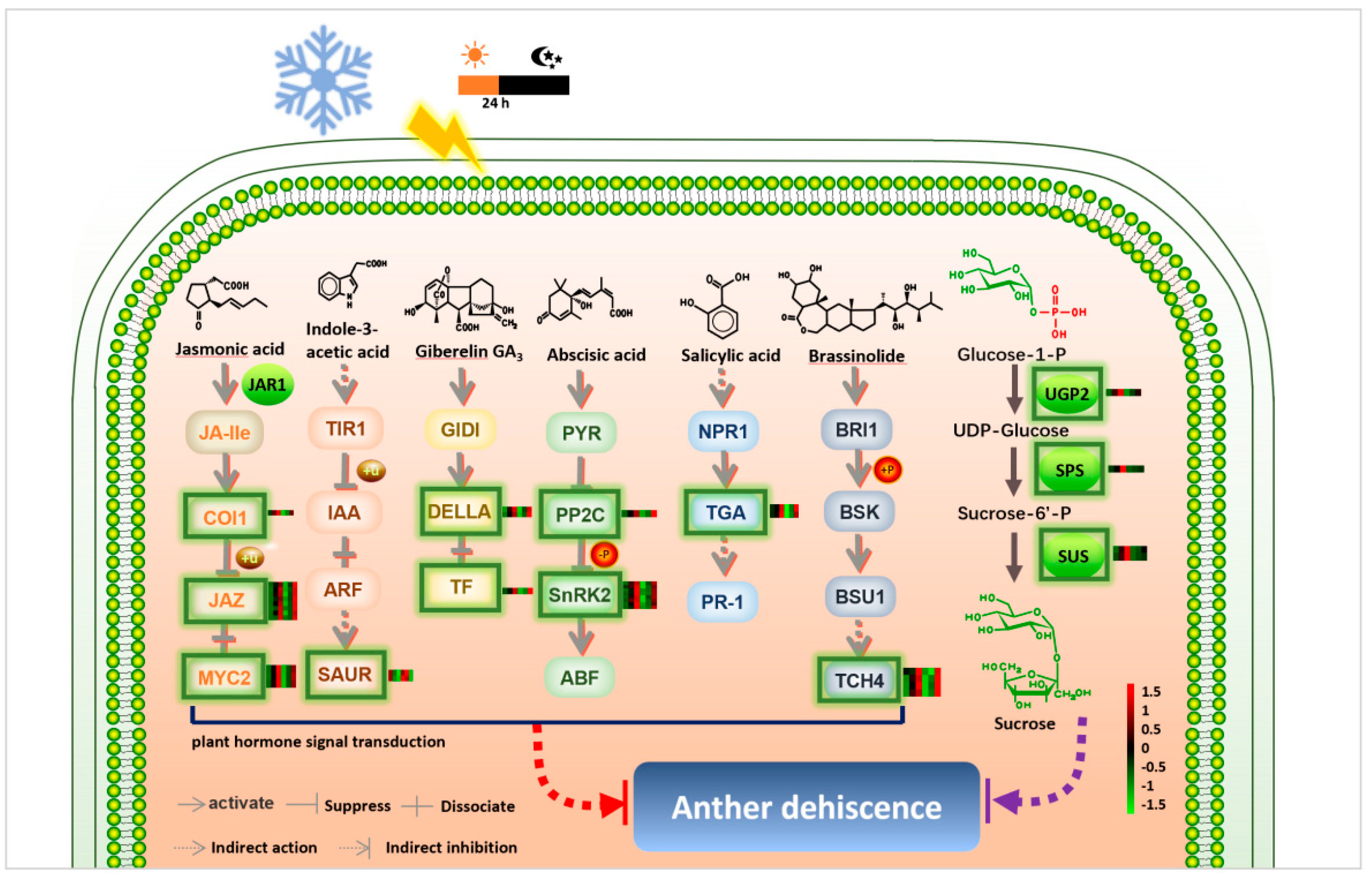
2.4. Expression Patterns of DEGs of the Plant Hormone Signal Transduction Pathway and Endogenous Hormone Measurements
2.5. Expression Patterns of DEGs of Starch and Sucrose Metabolism Pathway and the Sucrose Content of Anthers Measurements
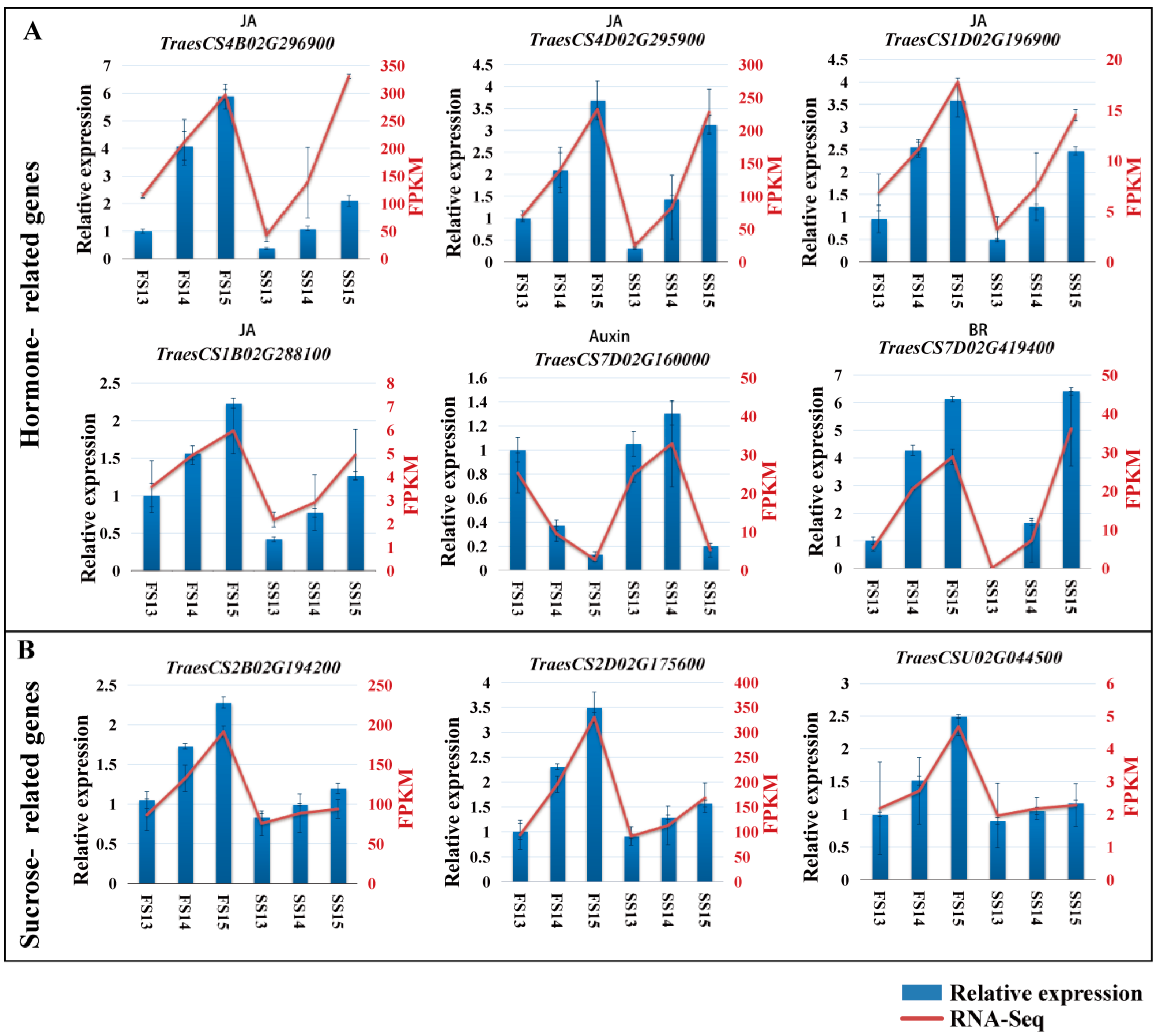
2.6. Validation of DEGs by qRT-PCR
3. Discussion
Putative Anther Dehiscence Related Male Sterile Network in Wheat PTGMS Line
4. Materials and Methods
4.1. Plant Material
4.2. Phenotypic Characterization at the Dehiscence Stage
4.3. Measurement of Plant Endogenous Hormones
4.4. Measure the Concentration of Sucrose
4.5. RNA Extraction and Sequencing
4.6. Bioinformatic Analysis of Transcriptome Data
4.7. qRT-PCR Analysis
4.8. A putative Model of Anther Dehiscence in Wheat PTGMS Line
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| BR | Brassinolide |
| DEGs | Differentially expressed genes |
| FPKM | Fragments per kilobase of exon model per million reads mapped |
| GA | Gibberellin acid |
| IAA | Indoleacetic acid |
| JA | Jasmonic acid |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PTGMS | Photo-thermo-sensitive genic male sterility |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| SEM | Scanning electron microscope |
| SA | Salicylic acid |
| WGCNA | Weighted gene co-expression network analysis |
References
- Wise, R.P.; Pring, D.R. Nuclear-mediated mitochondrial gene regulation and male fertility in higher plants: Light at the end of the tunnel? Proc. Natl. Acad. Sci. USA 2002, 99, 10240–10242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, Z.A.; Song, J.; Taylor, B.; Yang, C. The final split: The regulation of anther dehiscence. J. Exp. Bot. 2011, 62, 1633–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonner, L.J.; Dickinson, H.G. Anther dehiscence in Lycopersicon esculentum. New Phytol. 1990, 115, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16, S46–S60. [Google Scholar] [CrossRef]
- Bürger, M.; Chory, J. Stressed Out About Hormones: How plants orchestrate immunity. Cell Host Microbe 2019, 26, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.-M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [Green Version]
- Kim, J. Four shades of detachment: Regulation of floral organ abscission. Plant Signal. Behav. 2014, 9, e976154. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef] [Green Version]
- Bai, J.; Wang, Y.; Wang, P.; Yuan, S.; Gao, J.; Duan, W.; Wang, N.; Zhang, F.; Zhang, W.; Qin, M.; et al. Genome-wide identification and analysis of the COI gene family in wheat (Triticum aestivum L.). BMC Genom. 2018, 19, 754. [Google Scholar] [CrossRef]
- He, Y.; Liu, C.; Zhu, L.; Fu, M.; Sun, Y.; Zeng, H. Jasmonic acid plays a pivotal role in pollen development and fertility regulation in different types of P(T)GMS rice lines. Int. J. Mol. Sci. 2021, 22, 7926. [Google Scholar] [CrossRef]
- Park, J.-H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Tian, J.; Liu, Y.; Chen, X.; Li, S.; Persson, S.; Lu, D.; Chen, M.; Luo, Z.; Zhang, D.; et al. Ectopic expression of OsJAZ6, which interacts with OsJAZ1, alters JA signaling and spikelet development in rice. Plant J. 2021, 108, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Wang, Y.; Guo, L.; Guo, X.; Guo, H.; Yuan, S.; Duan, W.; Liu, Z.; Zhao, C.; Zhang, F.; et al. Genomic identification and characterization of MYC family genes in wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genom. 2017, 18, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cecchetti, V.; Altamura, M.M.; Falasca, G.; Costantino, P.; Cardarelli, M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 2008, 20, 1760–1774. [Google Scholar] [CrossRef] [Green Version]
- Cecchetti, V.; Celebrin, D.; Napoli, N.; Ghelli, R.; Brunetti, P.; Costantino, P.; Cardarelli, M. An auxin maximum in the middle layer controls stamen development and pollen maturation in Arabidopsis. New Phytol. 2017, 213, 1194–1207. [Google Scholar] [CrossRef] [Green Version]
- Salinas-Grenet, H.; Herrera-Vásquez, A.; Parra, S.; Cortez, A.; Gutiérrez, L.; Pollmann, S.; León, G.; Blanco-Herrera, F. Modulation of auxin levels in pollen grains affects stamen development and anther dehiscence in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2480. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Chen, Y.; Liu, L.; See, Y.H.B.; Mao, C.; Gan, Y.; Yu, H. OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nat. Plants 2018, 4, 495–504. [Google Scholar] [CrossRef]
- Yao, X.; Chen, J.; Zhou, J.; Yu, H.; Ge, C.; Zhang, M.; Gao, X.; Dai, X.; Yang, Z.-N.; Zhao, Y. An essential role for miRNA167 in maternal control of embryonic and seed development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef] [Green Version]
- Jewell, J.B.; Browse, J. Epidermal jasmonate perception is sufficient for all aspects of jasmonate-mediated male fertility in Arabidopsis. Plant J. 2016, 85, 634–647. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Hao, M.; Mei, D.; Zaman, Q.U.; Sang, S.; Wang, H.; Wang, W.; Fu, L.; Cheng, H.; Hu, Q. Transcriptome and hormone comparison of three cytoplasmic male sterile systems in Brassica napus. Int. J. Mol. Sci. 2018, 19, 4022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dill, A.; Sun, T.-p. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 2001, 159, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liu, Y.; Ali, M.; Ye, L.; Pan, C.; Li, M.; Zhao, X.; Yu, F.; Zhao, X.; Lu, G. Phytochrome interacting factor 3 regulates pollen mitotic division through auxin signalling and sugar metabolism pathways in tomato. New Phytol. 2022, 234, 560–577. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Z.; Wu, S.; Wan, X. The essential roles of sugar metabolism for pollen development and male fertility in plants. Crop J. 2021, 9, 1223–1236. [Google Scholar] [CrossRef]
- Goetz, M.; Guivarćh, A.; Hirsche, J.; Bauerfeind, M.A.; González, M.-C.; Hyun, T.K.; Eom, S.H.; Chriqui, D.; Engelke, T.; Großkinsky, D.K.; et al. Metabolic control of tobacco pollination by sugars and invertases. Plant Physiol. 2017, 173, 984–997. [Google Scholar] [CrossRef] [Green Version]
- Keijzer, C.J. The processes of anther dehiscence and pollen dispersal. New Phytol. 1987, 105, 487–498. [Google Scholar] [CrossRef]
- Hirose, T.; Hashida, Y.; Aoki, N.; Okamura, M.; Yonekura, M.; Ohto, C.; Terao, T.; Ohsugi, R. Analysis of gene-disruption mutants of a sucrose phosphate synthase gene in rice, OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant Sci. 2014, 225, 102–106. [Google Scholar] [CrossRef]
- Cho, J.-I.; Ryoo, N.; Ko, S.; Lee, S.-K.; Lee, J.; Jung, K.-H.; Lee, Y.-H.; Bhoo, S.H.; Winderickx, J.; An, G.; et al. Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 2006, 224, 598–611. [Google Scholar] [CrossRef]
- Brummell, D.A.; Hall, B.D.; Bennett, A.B. Antisense suppression of tomato endo-1,4-β-glucanase Cel2 mRNA accumulation increases the force required to break fruit abscission zones but does not affect fruit softening. Plant Mol. Biol. 1999, 40, 615–622. [Google Scholar] [CrossRef]
- Moon, S.; Kim, S.R.; Zhao, G.; Yi, J.; Yoo, Y.; Jin, P.; Lee, S.W.; Jung, K.H.; Zhang, D.; An, G. Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol. 2013, 161, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, P.; Lv, J.; Cheng, Y.; Cui, J.; Zhao, H.; Hu, S. Global dynamic transcriptome programming of rapeseed (Brassica napus L.) anther at different development stages. PLoS ONE 2016, 11, e0154039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Bai, J.; Yuan, S.; Guo, L.; Liu, Z.; Guo, H.; Zhang, T.; Duan, W.; Li, Y.; Zhao, C.; et al. Genome Wide identification and characterization of wheat GH9 genes reveals their roles in pollen development and anther dehiscence. Int. J. Mol. Sci. 2022, 23, 6324. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zhang, L.; Xu, C.; Yuan, S.; Zhang, F.; Zheng, Y.; Zhao, C. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 2012, 159, 721–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Liu, Z.; Zhang, L.; Zhao, C.; Yuan, S.; Zhang, F. Organization of actin cytoskeleton during meiosis I in a wheat thermo-sensitive genic male sterile line. Protoplasma 2013, 250, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zhang, L.; Yang, D.I.; Zhao, C.; Zheng, Y. Cold stress contributes to aberrant cytokinesis during male meiosis I in a wheat thermosensitive genic male sterile line. Plant Cell Environ. 2011, 34, 389–405. [Google Scholar] [CrossRef]
- Shih, C.-F.; Hsu, W.-H.; Peng, Y.-J.; Yang, C.-H. The NAC-like gene ANTHER INDEHISCENCE FACTOR acts as a repressor that controls anther dehiscence by regulating genes in the jasmonate biosynthesis pathway in Arabidopsis. J. Exp. Bot. 2014, 65, 621–639. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Chen, Y.; Charnikhova, T.; Mulder, P.P.J.; Heijmans, J.; Hoogenboom, A.; Agalou, A.; Michel, C.; Morel, J.-B.; Dreni, L.; et al. OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol. Biol. 2014, 86, 19–33. [Google Scholar] [CrossRef]
- Hori, Y.; Kurotani, K.-i.; Toda, Y.; Hattori, T.; Takeda, S. Overexpression of the JAZ factors with mutated jas domains causes pleiotropic defects in rice spikelet development. Plant Signal. Behav. 2014, 9, e970414. [Google Scholar] [CrossRef] [Green Version]
- Marciniak, K.; Przedniczek, K. Anther dehiscence is regulated by gibberellic acid in yellow lupine (Lupinus luteus L.). BMC Plant Biol. 2021, 21, 314. [Google Scholar] [CrossRef]
- Zhu, T.; Li, Z.; An, X.; Long, Y.; Xue, X.; Xie, K.; Ma, B.; Zhang, D.; Guan, Y.; Niu, C.; et al. Normal structure and function of endothecium chloroplasts maintained by ZmMs33-mediated lipid biosynthesis in tapetal cells are critical for anther development in maize. Mol. Plant 2020, 13, 1624–1643. [Google Scholar] [CrossRef]
- Xu, F.-Q.; Li, X.-R.; Ruan, Y.-L. RNAi-mediated suppression of hexokinase gene OsHXK10 in rice leads to non-dehiscent anther and reduction of pollen germination. Plant Sci. 2008, 175, 674–684. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Wasternack, C.; Xie, D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 2014, 21, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, L.; Zhuo, F.; Zhang, X.; Ren, M.; Li, F. Progress on jasmonic acid signaling in plant stress resistant. J. Agric. Sci. Technol. 2015, 17, 17–24. [Google Scholar]
- Zhang, Y.; Lan, H.; Shao, Q.; Wang, R.; Chen, H.; Tang, H.; Zhang, H.; Huang, J. An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa). J. Exp. Bot. 2016, 67, 315–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-I.; Ishimizu, T.; Suwabe, K.; Sudo, K.; Masuko, H.; Hakozaki, H.; Nou, I.-S.; Suzuki, G.; Watanabe, M. UDP-Glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 981–996. [Google Scholar] [CrossRef] [PubMed]
- Browne, R.G.; Iacuone, S.; Li, S.F.; Dolferus, R.; Parish, R.W. Anther morphological development and stage determination in Triticum aestivum. Front. Plant Sci. 2018, 9, 228. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Geng, X.; Liu, Z.; Ye, J.; Xu, M.; Zhang, L.; Song, X. A sterility induction trait in the genic male sterility wheat line 4110S induced by high temperature and its cytological response. Crop Sci. 2018, 58, 1866–1876. [Google Scholar] [CrossRef]
- Wang, N.; Fisher, D.B. Sucrose Release into the Endosperm Cavity of Wheat Grains Apparently Occurs by Facilitated Diffusion across the Nucellar Cell Membranes. Plant Physiol. 1995, 109, 579–585. [Google Scholar] [CrossRef]
- Wang, Y.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Zhang, F.; Gao, S.; Liu, L.; Pang, B.; Zhang, L.; et al. Comparative transcriptome analysis identifies genes involved in the regulation of the pollen cytoskeleton in a genic male sterile wheat line. Plant Growth Regul. 2018, 86, 133–147. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Li, M.-C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or normalized counts? a comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhang, Y.-H.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.-D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef] [Green Version]
- Harshitha, R.; Arunraj, D.R. Real-time quantitative PCR: A tool for absolute and relative quantification. Biochem. Mol. Biol. Educ. 2021, 49, 800–812. [Google Scholar] [CrossRef] [PubMed]

| Sample | Clean Reads (M) | Clean Bases (G) | Q20 (%) | GC (%) | Total Mapped | Unique Mapped |
|---|---|---|---|---|---|---|
| FS13-1 | 70.1261M | 10.5189G | 96.57 | 52.07 | 67,399,956 (96.11%) | 62,945,694 (89.76%) |
| FS13-2 | 69.2820M | 10.3923G | 96.21 | 52.66 | 66,795,338 (96.41%) | 62,356,024 (90.00%) |
| FS13-3 | 68.0320M | 10.2048G | 96.59 | 52.42 | 65,494,818 (96.27%) | 61,426,150 (90.29%) |
| FS14-1 | 67.8116M | 10.1717G | 96.12 | 53.43 | 65,422,558 (96.48%) | 60,944,936 (89.87%) |
| FS14-2 | 67.4691M | 10.1204G | 96.37 | 52.56 | 65,022,032 (96.37%) | 60,901,854 (90.27%) |
| FS14-3 | 69.3257M | 10.3989G | 96 | 52.99 | 66,711,060 (96.23%) | 62,484,910 (90.13%) |
| FS15-1 | 67.7965M | 10.1695G | 95.72 | 53.46 | 65,353,484 (96.40%) | 61,362,940 (90.51%) |
| FS15-2 | 69.3829M | 10.4074G | 96.1 | 53.35 | 66,889,454 (96.41%) | 62,665,108 (90.32%) |
| FS15-3 | 71.1352M | 10.6703G | 96.37 | 53.42 | 68,578,804 (96.41%) | 64,320,258 (90.42%) |
| SS13-1 | 69.1204M | 10.3681G | 96.46 | 51.5 | 66,420,770 (96.09%) | 62,481,716 (90.40%) |
| SS13-2 | 69.3102M | 10.3965G | 96.92 | 50.86 | 66,429,536 (95.84%) | 62,370,380 (89.99%) |
| SS13-3 | 67.8598M | 10.1790G | 96.96 | 51.49 | 65,341,338 (96.29%) | 61,350,318 (90.41%) |
| SS14-1 | 71.0547M | 10.6582G | 96.46 | 51.73 | 68,495,044 (96.40%) | 64,330,860 (90.54%) |
| SS14-2 | 68.4178M | 10.2627G | 95.8 | 52.68 | 65,398,718 (95.59%) | 61,288,724 (89.58%) |
| SS14-3 | 63.8187M | 9.5728G | 95.87 | 52.49 | 61,324,110 (96.09%) | 57,427,232 (89.99%) |
| SS15-1 | 64.4363M | 9.6654G | 95.69 | 52.87 | 61,858,806 (96.00%) | 57,772,684 (89.66%) |
| SS15-2 | 69.6364M | 10.4455G | 95.65 | 53.26 | 66,919,980 (96.10%) | 62,606,924 (89.91%) |
| SS15-3 | 69.5348M | 10.4302G | 95.86 | 52.76 | 66,832,700 (96.11%) | 62,456,070 (89.82%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Yuan, S.; Liu, Z.; Luo, L.; Guo, H.; Li, Y.; Bai, J.; Zhao, C.; Zhang, L. Comparative Transcriptome Analysis Reveals Hormone Signal Transduction and Sucrose Metabolism Related Genes Involved in the Regulation of Anther Dehiscence in Photo-Thermo-Sensitive Genic Male Sterile Wheat. Biomolecules 2022, 12, 1149. https://doi.org/10.3390/biom12081149
Zhang T, Yuan S, Liu Z, Luo L, Guo H, Li Y, Bai J, Zhao C, Zhang L. Comparative Transcriptome Analysis Reveals Hormone Signal Transduction and Sucrose Metabolism Related Genes Involved in the Regulation of Anther Dehiscence in Photo-Thermo-Sensitive Genic Male Sterile Wheat. Biomolecules. 2022; 12(8):1149. https://doi.org/10.3390/biom12081149
Chicago/Turabian StyleZhang, Tianbao, Shaohua Yuan, Zihan Liu, Liqing Luo, Haoyu Guo, Yanmei Li, Jianfang Bai, Changping Zhao, and Liping Zhang. 2022. "Comparative Transcriptome Analysis Reveals Hormone Signal Transduction and Sucrose Metabolism Related Genes Involved in the Regulation of Anther Dehiscence in Photo-Thermo-Sensitive Genic Male Sterile Wheat" Biomolecules 12, no. 8: 1149. https://doi.org/10.3390/biom12081149
APA StyleZhang, T., Yuan, S., Liu, Z., Luo, L., Guo, H., Li, Y., Bai, J., Zhao, C., & Zhang, L. (2022). Comparative Transcriptome Analysis Reveals Hormone Signal Transduction and Sucrose Metabolism Related Genes Involved in the Regulation of Anther Dehiscence in Photo-Thermo-Sensitive Genic Male Sterile Wheat. Biomolecules, 12(8), 1149. https://doi.org/10.3390/biom12081149






