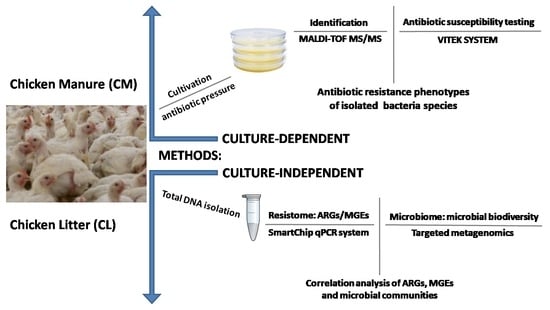
A Comprehensive Study of the Microbiome, Resistome, and Physical and Chemical Characteristics of Chicken Waste from Intensive Farms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Manure Samples
2.2. Bacterial Strain Isolation and Characterization
2.3. DNA Extraction and Metagenomic Sequencing
2.4. qPCR Analysis of the ARG Profile
3. Results and Discussion
3.1. Physicochemical Characterization of Chicken Wastes
3.2. Bacterial Community Composition
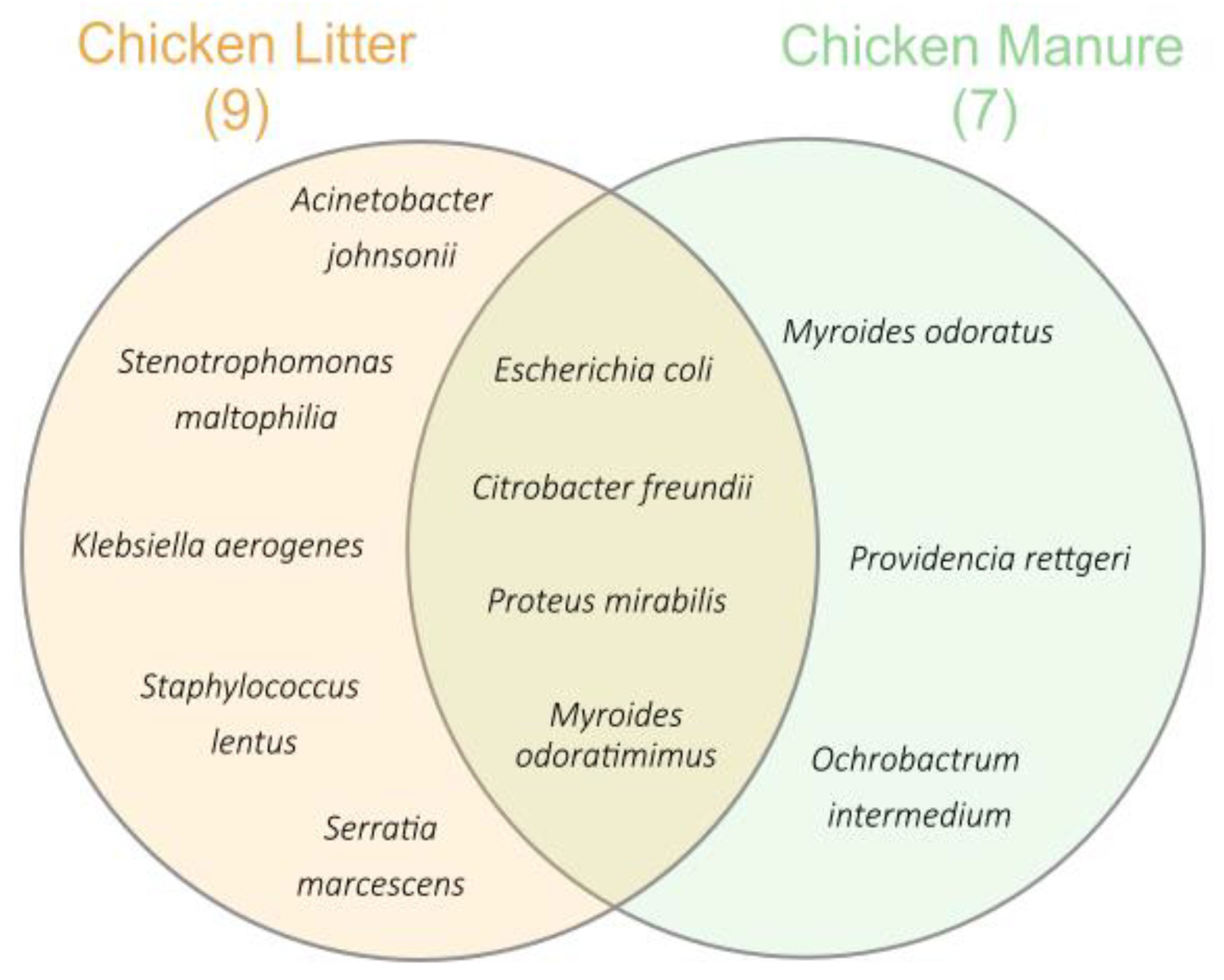
3.2.1. Identification of Bacteria Species and Antibiotic Susceptibility Testing
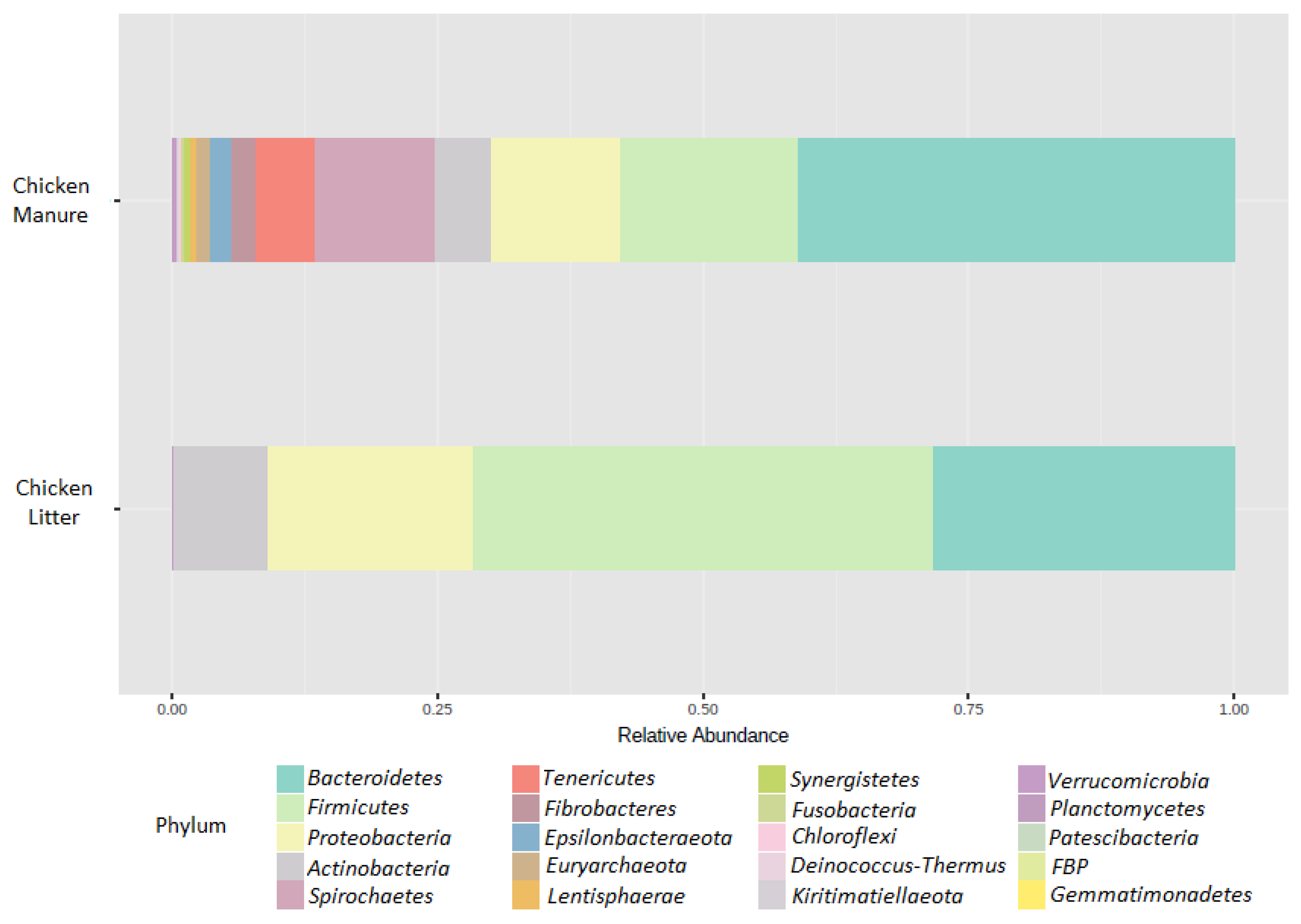
3.2.2. Microbial Community Composition
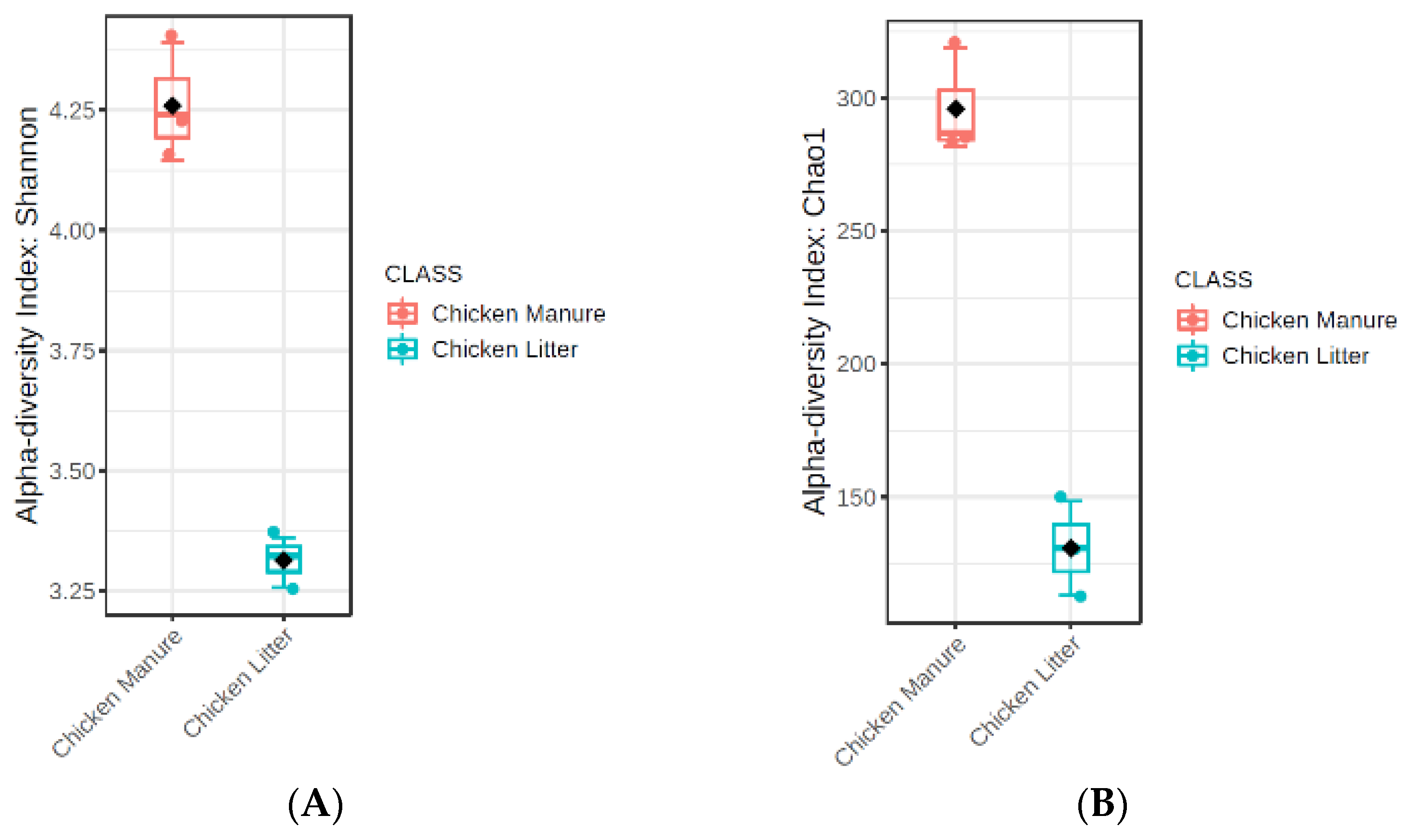
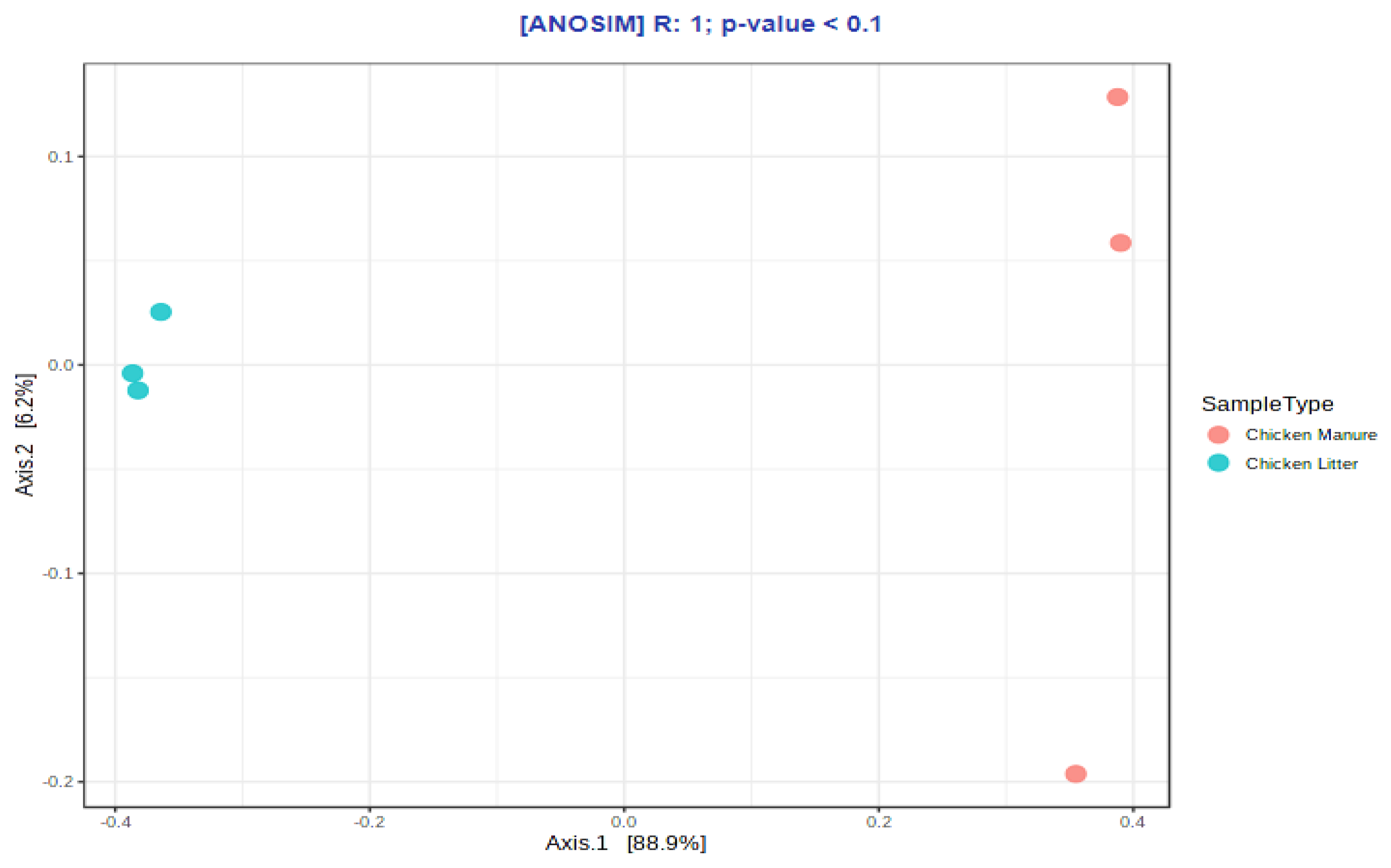
3.2.3. Microbial Diversity
3.2.4. High-Throughput Quantitative PCR Analysis
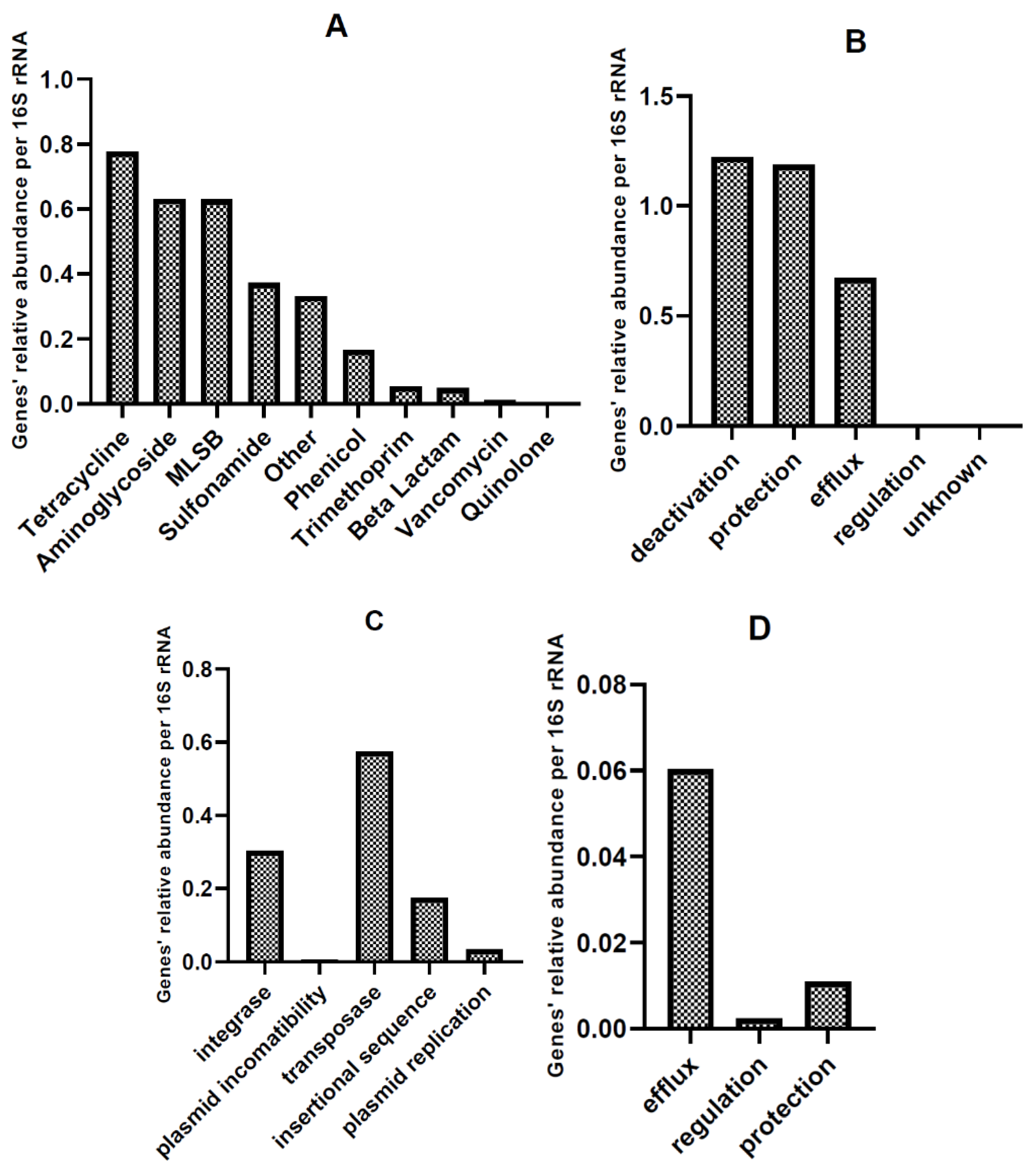
Relative Abundance of AMR and MGE Gene Classes in HT-qPCR Arrays
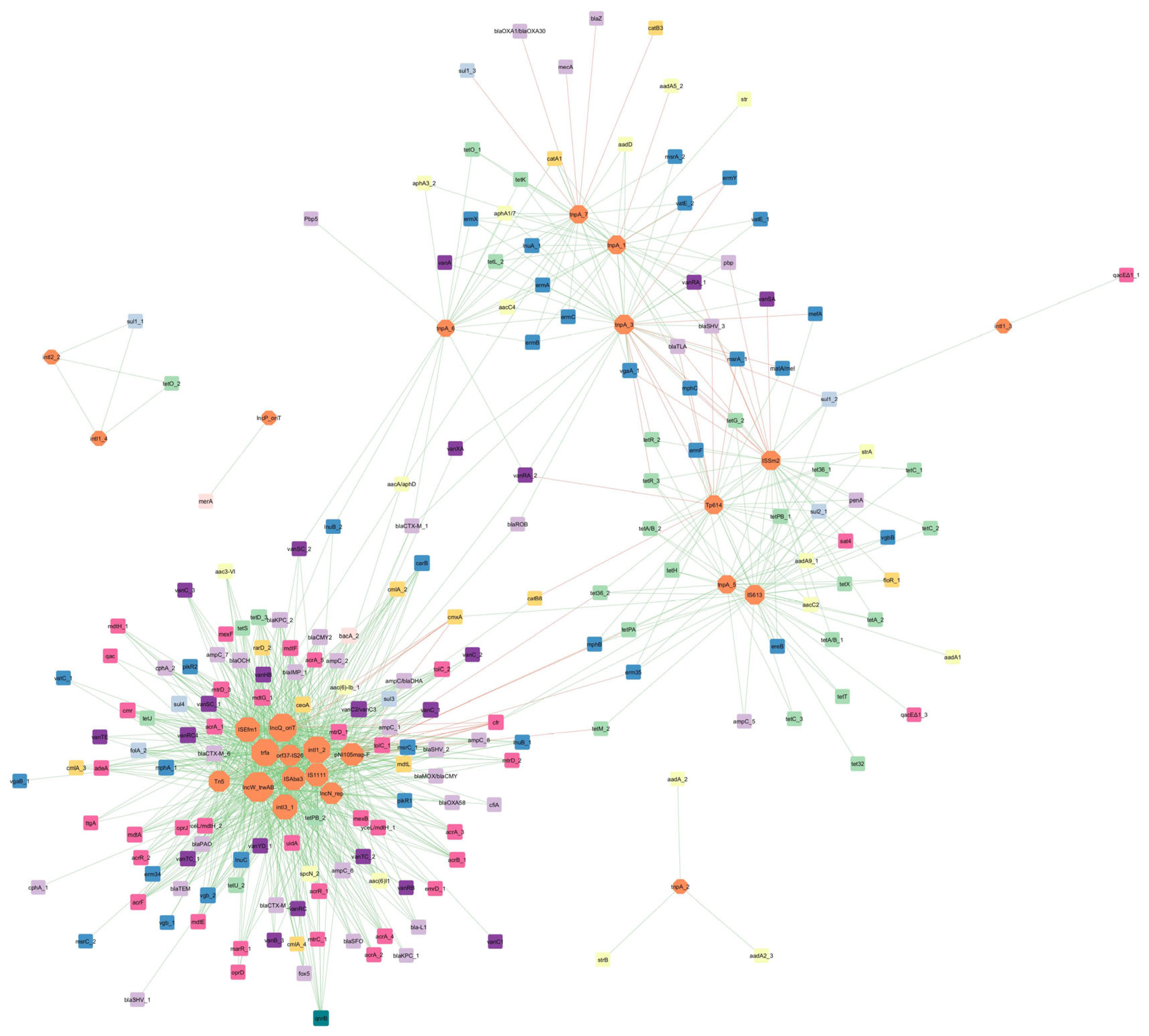
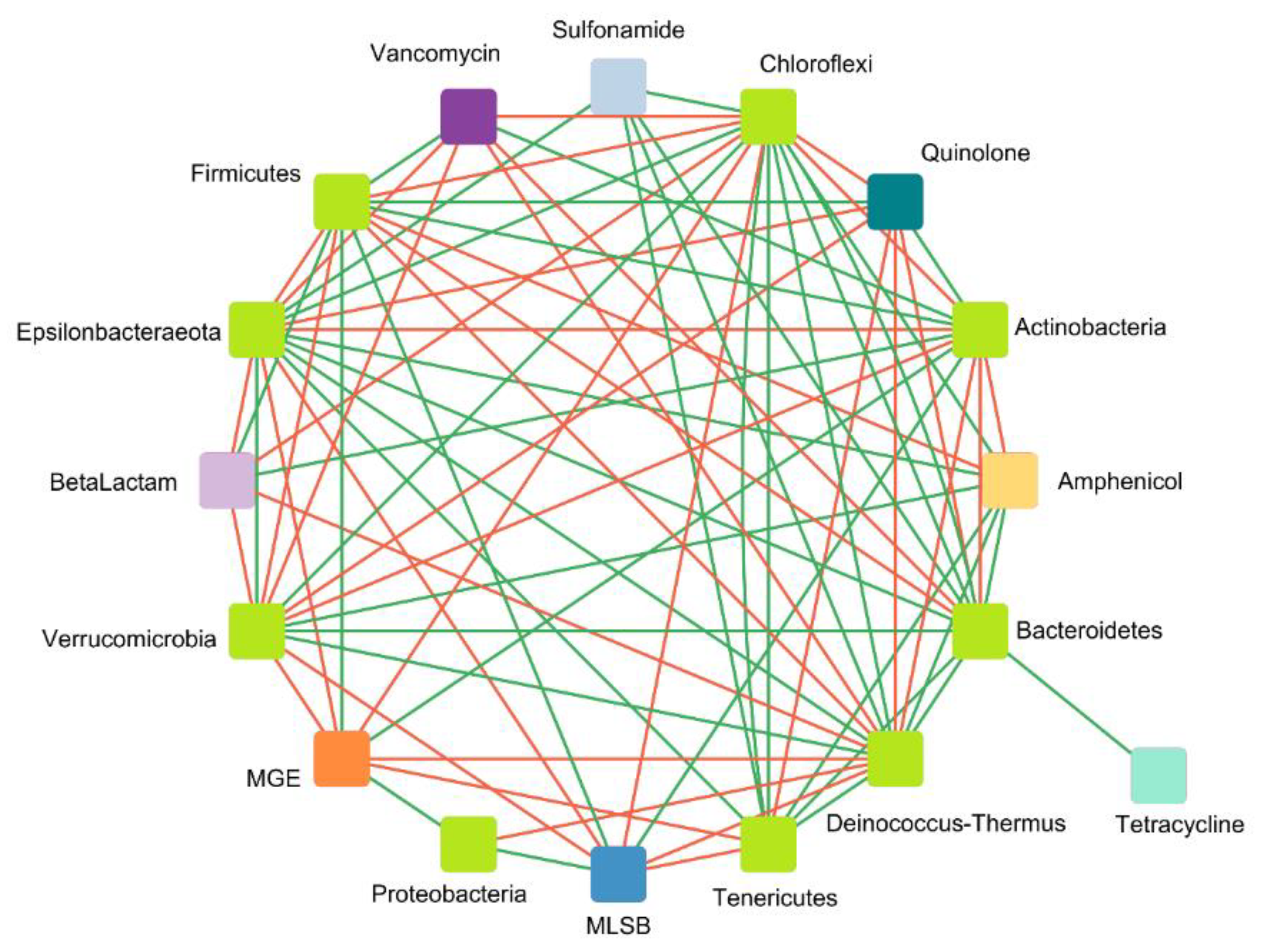
Correlation Analysis of ARGs, MGEs, and Microbial Communities
- Correlation between ARGs and MGEs
- 2.
- Correlation between detected genes and microbial taxa at the phylum level
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO FAOSTAT. Available online: https://www.fao.org/faostat/en/#home (accessed on 23 March 2022).
- Muhammad, J.; Khan, S.; Su, J.Q.; Hesham, A.E.L.; Ditta, A.; Nawab, J.; Ali, A. Antibiotics in Poultry Manure and Their Associated Health Issues: A Systematic Review. J. Soils Sediments Prot. Risk Assess. Remediat. 2020, 20, 486–497. [Google Scholar] [CrossRef]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The Application of Antibiotics in Broiler Production and the Resulting Antibiotic Resistance in Escherichia Coli: A Global Overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, B.J.A.; Lahr, J.; Nibbeling, C.; Jansen, L.J.M.; Bongers, I.E.A.; Wipfler, E.L.; van de Schans, M.G.M. The Persistence of a Broad Range of Antibiotics during Calve, Pig and Broiler Manure Storage. Chemosphere 2018, 204, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Geissen, S.-U.; Gal, C. Carbamazepine and Diclofenac: Removal in Wastewater Treatment Plants and Occurrence in Water Bodies. Chemosphere 2008, 73, 1151–1161. [Google Scholar] [CrossRef]
- Poole, T.; Sheffield, C. Use and Misuse of Antimicrobial Drugs in Poultry and Livestock: Mechanisms of Antimicrobial Resistance. Pak. Vet. J. 2013, 33, 266–271. [Google Scholar]
- Archawakulathep, A.; Kim, C.T.T.; Meunsene, D.; Handijatno, D.; Hassim, H.A.; Rovira, H.R.G.; Myint, K.S.; Baldrias, L.R.; Sothy, M.; Aung, M.; et al. Perspectives on Antimicrobial Resistance in Livestock and Livestock Products in ASEAN Countries. Thai J. Vet. Med. 2014, 44, 5–13. [Google Scholar]
- Rothrock, M.J.; Hiett, K.L.; Guard, J.Y.; Jackson, C.R. Antibiotic Resistance Patterns of Major Zoonotic Pathogens from All-Natural, Antibiotic-Free, Pasture-Raised Broiler Flocks in the Southeastern United States. J. Environ. Qual. 2016, 45, 593–603. [Google Scholar] [CrossRef]
- Franklin, A.M.; Aga, D.S.; Cytryn, E.; Durso, L.M.; McLain, J.E.; Pruden, A.; Roberts, M.C.; Rothrock, M.J.; Snow, D.D.; Watson, J.E.; et al. Antibiotics in Agroecosystems: Introduction to the Special Section. J. Environ. Qual. 2016, 45, 377–393. [Google Scholar] [CrossRef]
- Zalewska, M.; Błażejewska, A.; Czapko, A.; Popowska, M. Antibiotics and Antibiotic Resistance Genes in Animal Manure—Consequences of Its Application in Agriculture. Front. Microbiol. 2021, 12, 610656. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Hu, H.-W.; Gou, M.; Wang, J.-T.; Chen, D.; He, J.-Z. Temporal Succession of Soil Antibiotic Resistance Genes Following Application of Swine, Cattle and Poultry Manures Spiked with or without Antibiotics. Environ. Pollut. 2017, 231, 1621–1632. [Google Scholar] [CrossRef]
- Dróżdż, D.; Wystalska, K.; Malińska, K.; Grosser, A.; Grobelak, A.; Kacprzak, M. Management of Poultry Manure in Poland—Current State and Future Perspectives. J. Environ. Manag. 2020, 264, 110327. [Google Scholar] [CrossRef]
- Wang, L.; Lilburn, M.; Yu, Z. Intestinal Microbiota of Broiler Chickens As Affected by Litter Management Regimens. Front. Microbiol. 2016, 7, 593. [Google Scholar] [CrossRef]
- Lovanh, N.; Cook, K.L.; Rothrock, M.J.; Miles, D.M.; Sistani, K. Spatial Shifts in Microbial Population Structure within Poultry Litter Associated with Physicochemical Properties. Poult. Sci. 2007, 86, 1840–1849. [Google Scholar] [CrossRef]
- Collins, E.; Barker, J.C.; Carr, L.E.; Brodie, H.L.; Martin, J.H. Poultry Waste Management Handbook; Natural Resource, Agriculture, and Engineering Service (NRAES): College Park, MD, USA, 1999. [Google Scholar]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Laconi, A.; Mughini-Gras, L.; Tolosi, R.; Grilli, G.; Trocino, A.; Carraro, L.; Di Cesare, F.; Cagnardi, P.; Piccirillo, A. Microbial Community Composition and Antimicrobial Resistance in Agricultural Soils Fertilized with Livestock Manure from Conventional Farming in Northern Italy. Sci. Total Environ. 2021, 760, 143404. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Da Silva, G.J. Manure as a Potential Hotspot for Antibiotic Resistance Dissemination by Horizontal Gene Transfer Events. Vet. Sci. 2020, 7, 110. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Li, J.; Lv, N.; Liu, F.; Wu, J.; Lin, I.Y.C.; Wu, N.; Weimer, B.C.; Gao, G.F.; et al. The Bacterial Mobile Resistome Transfer Network Connecting the Animal and Human Microbiomes. Appl. Environ. Microbiol. 2016, 82, 6672–6681. [Google Scholar] [CrossRef]
- Chen, Z.; Jiang, X. Microbiological Safety of Chicken Litter or Chicken Litter-Based Organic Fertilizers: A Review. Agriculture 2014, 4, 1–29. [Google Scholar] [CrossRef]
- Lebov, J.; Grieger, K.; Womack, D.; Zaccaro, D.; Whitehead, N.; Kowalcyk, B.; MacDonald, P.D.M. A Framework for One Health Research. One Health 2017, 3, 44–50. [Google Scholar] [CrossRef]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic Resistance Is the Quintessential One Health Issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Biemer, J.J. Antimicrobial Susceptibility Testing by the Kirby-Bauer Disc Diffusion Method. Ann. Clin. Lab. Sci. 1973, 3, 135–140. [Google Scholar]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for Identification of Environmental Bacteria: A Review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef]
- Hogan, C.A.; Watz, N.; Budvytiene, I.; Banaei, N. Rapid Antimicrobial Susceptibility Testing by VITEK®2 Directly from Blood Cultures in Patients with Gram-Negative Rod Bacteremia. Diagn. Microbiol. Infect. Dis. 2019, 94, 116–121. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for Comprehensive Statistical, Functional, and Meta-Analysis of Microbiome Data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Wang, F.-H.; Qiao, M.; Su, J.-Q.; Chen, Z.; Zhou, X.; Zhu, Y.-G. High Throughput Profiling of Antibiotic Resistance Genes in Urban Park Soils with Reclaimed Water Irrigation. Environ. Sci. Technol. 2014, 48, 9079–9085. [Google Scholar] [CrossRef]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.-G. Long-Term Field Application of Sewage Sludge Increases the Abundance of Antibiotic Resistance Genes in Soil. Environ. Int. 2016, 92–93, 1–10. [Google Scholar] [CrossRef]
- Dumas, M.D.; Polson, S.W.; Ritter, D.; Ravel, J.; Gelb, J., Jr.; Morgan, R.; Wommack, K.E. Impacts of Poultry House Environment on Poultry Litter Bacterial Community Composition. PLoS ONE 2011, 6, e24785. [Google Scholar] [CrossRef]
- Dikinya, O.; Mufwanzala, N. Chicken Manure-Enhanced Soil Fertility and Productivity: Effects of Application Rates. J. Soil Sci. Environ. Manag. 2010. [Google Scholar] [CrossRef]
- Regulation (EC) No. 1069/2009 of the European Parliament and of the Council of 21 October 2009 Laying down Health Rules as Regards Animal by-Products and Derived Products Not Intended for Human Consumption and Repealing Regulation (EC) No. 1774/2002 (Animal by-Products Regulation). 2009, Volume 300. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex:32009R1069 (accessed on 15 August 2022).
- Poole, K. At the Nexus of Antibiotics and Metals: The Impact of Cu and Zn on Antibiotic Activity and Resistance. Trends Microbiol. 2017, 25, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Su, J.-Q.; An, X.-L.; Huang, F.-Y.; Rensing, C.; Brandt, K.K.; Zhu, Y.-G. Feed Additives Shift Gut Microbiota and Enrich Antibiotic Resistance in Swine Gut. Sci. Total Environ. 2018, 621, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, A.Z.; Cen, T.; Li, X.; He, M.; Li, D.; Chen, J. Sub-Inhibitory Concentrations of Heavy Metals Facilitate the Horizontal Transfer of Plasmid-Mediated Antibiotic Resistance Genes in Water Environment. Environ. Pollut. 2018, 237, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.D. Impact of Antibiotic Use in the Swine Industry. Curr. Opin. Microbiol. 2014, 19, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Li, X.; Li, L.; Li, S.; Liao, X.; Sun, J.; Liu, Y. Co-Spread of Metal and Antibiotic Resistance within ST3-IncHI2 Plasmids from E. Coli Isolates of Food-Producing Animals. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-Occurrence of Resistance Genes to Antibiotics, Biocides and Metals Reveals Novel Insights into Their Co-Selection Potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef] [PubMed]
- Rogeri, D.; Ernani, P.; Mantovani, A.; Lourenço, K. Composition of Poultry Litter in Southern Brazil. Rev. Bras. Ciência Solo 2016, 40. [Google Scholar] [CrossRef]
- Sarbjit Singh, G.S.; Shamsuddin, M.R.; Aqsha; Lim, S. Characterization of Chicken Manure from Manjung Region. IOP Conf. Ser. Mater. Sci. Eng. 2018, 458, 012084. [Google Scholar] [CrossRef]
- Li, G.; Li, H.; Leffelaar, P.A.; Shen, J.; Zhang, F. Characterization of Phosphorus in Animal Manures Collected from Three (Dairy, Swine, and Broiler) Farms in China. PLoS ONE 2014, 9, e102698. [Google Scholar] [CrossRef]
- Reyer, H.; Oster, M.; Ponsuksili, S.; Trakooljul, N.; Omotoso, A.O.; Iqbal, M.A.; Muráni, E.; Sommerfeld, V.; Rodehutscord, M.; Wimmers, K. Transcriptional Responses in Jejunum of Two Layer Chicken Strains Following Variations in Dietary Calcium and Phosphorus Levels. BMC Genom. 2021, 22, 485. [Google Scholar] [CrossRef]
- Proszkowiec-Weglarz, M.; Angel, R. Calcium and Phosphorus Metabolism in Broilers: Effect of Homeostatic Mechanism on Calcium and Phosphorus Digestibility1 1Presented as a Part of the Informal Nutrition Symposium “Metabolic Responses to Nutrition and Modifiers” at the Poultry Science Association’s Annual Meeting in Athens, Georgia, July 9, 2012. J. Appl. Poult. Res. 2013, 22, 609–627. [Google Scholar] [CrossRef]
- Bedford, M.; Rousseau, X.; Bedford, M.; Rousseau, X. Recent Findings Regarding Calcium and Phytase in Poultry Nutrition. Anim. Prod. Sci. 2017, 57, 2311–2316. [Google Scholar] [CrossRef]
- Shafey, T.M. Calcium Tolerance of Growing Chickens: Effect of Ratio of Dietary Calcium to Available Phosphorus. World’s Poult. Sci. J. 1993, 49, 5–18. [Google Scholar] [CrossRef]
- Mazal, C.; Sieger, B. Staphylococcus Lentus: The Troublemaker. Int. J. Infect. Dis. 2010, 14, e397. [Google Scholar] [CrossRef]
- Meyer, A.; Dang, H.; Roland, W. Myroides spp. Cellulitis and Bacteremia: A Case Report. IDCases 2019, 18, e00638. [Google Scholar] [CrossRef]
- Ktari, S.; Mnif, B.; Koubaa, M.; Mahjoubi, F.; Ben Jemaa, M.; Mhiri, M.N.; Hammami, A. Nosocomial Outbreak of Myroides Odoratimimus Urinary Tract Infection in a Tunisian Hospital. J. Hosp. Infect. 2012, 80, 77–81. [Google Scholar] [CrossRef]
- Ewers, C.; Antão, E.-M.; Diehl, I.; Philipp, H.-C.; Wieler, L.H. Intestine and Environment of the Chicken as Reservoirs for Extraintestinal Pathogenic Escherichia Coli Strains with Zoonotic Potential. Appl. Environ. Microbiol. 2009, 75, 184–192. [Google Scholar] [CrossRef]
- Marano, R.B.M.; Fernandes, T.; Manaia, C.M.; Nunes, O.; Morrison, D.; Berendonk, T.U.; Kreuzinger, N.; Telson, T.; Corno, G.; Fatta-Kassinos, D.; et al. A Global Multinational Survey of Cefotaxime-Resistant Coliforms in Urban Wastewater Treatment Plants. Environ. Int. 2020, 144, 106035. [Google Scholar] [CrossRef]
- Moffo, F.; Mouiche, M.M.M.; Djomgang, H.K.; Tombe, P.; Wade, A.; Kochivi, F.L.; Dongmo, J.B.; Mbah, C.K.; Mapiefou, N.P.; Ngogang, M.P.; et al. Poultry Litter Contamination by Escherichia Coli Resistant to Critically Important Antimicrobials for Human and Animal Use and Risk for Public Health in Cameroon. Antibiotics 2021, 10, 402. [Google Scholar] [CrossRef]
- Agostinho, J.M.A.; Cardozo, M.V.; Borzi, M.M.; Marin, J.M. Antibiotic Resistance and Virulence Factors among Escherichia Coli Isolates from Avian Organic Fertilizer. Cienc. Rural 2020, 50, e20180849. [Google Scholar] [CrossRef]
- Moraru, R.; Pourcher, A.-M.; Jadas-Hecart, A.; Kempf, I.; Ziebal, C.; Kervarrec, M.; Comunal, P.-Y.; Mares, M.; Dabert, P. Changes in Concentrations of Fluoroquinolones and of Ciprofloxacin-Resistant Enterobacteriaceae in Chicken Feces and Manure Stored in a Heap. J. Environ. Qual. 2012, 41, 754–763. [Google Scholar] [CrossRef]
- Atidégla, S.C.; Huat, J.; Agbossou, E.K.; Saint-Macary, H.; Glèlè Kakai, R. Vegetable Contamination by the Fecal Bacteria of Poultry Manure: Case Study of Gardening Sites in Southern Benin. Int. J. Food Sci. 2016, 2016, e4767453. [Google Scholar] [CrossRef]
- Graham, J.P.; Evans, S.L.; Price, L.B.; Silbergeld, E.K. Fate of Antimicrobial-Resistant Enterococci and Staphylococci and Resistance Determinants in Stored Poultry Litter. Environ. Res. 2009, 109, 682–689. [Google Scholar] [CrossRef]
- Thomas, K.V.; Bijlsma, L.; Castiglioni, S.; Covaci, A.; Emke, E.; Grabic, R.; Hernández, F.; Karolak, S.; Kasprzyk-Hordern, B.; Lindberg, R.H.; et al. Comparing Illicit Drug Use in 19 European Cities through Sewage Analysis. Sci. Total Environ. 2012, 432, 432–439. [Google Scholar] [CrossRef]
- Gurmessa, B.; Ashworth, A.J.; Yang, Y.; Savin, M.; Moore, P.A.; Ricke, S.C.; Corti, G.; Pedretti, E.F.; Cocco, S. Variations in Bacterial Community Structure and Antimicrobial Resistance Gene Abundance in Cattle Manure and Poultry Litter. Environ. Res. 2021, 197, 111011. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil PH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Borda-Molina, D.; Vital, M.; Sommerfeld, V.; Rodehutscord, M.; Camarinha-Silva, A. Insights into Broilers’ Gut Microbiota Fed with Phosphorus, Calcium, and Phytase Supplemented Diets. Front. Microbiol. 2016, 7, 2033. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, E14. [Google Scholar] [CrossRef]
- Peng, S.; Wang, Y.; Chen, R.; Lin, X. Chicken Manure and Mushroom Residues Affect Soil Bacterial Community Structure but Not the Bacterial Resistome When Applied at the Same Rate of Nitrogen for 3 Years. Front. Microbiol. 2021, 12, 618693. [Google Scholar] [CrossRef]
- Ziganshin, A.M.; Liebetrau, J.; Pröter, J.; Kleinsteuber, S. Microbial Community Structure and Dynamics during Anaerobic Digestion of Various Agricultural Waste Materials. Appl. Microbiol. Biotechnol. 2013, 97, 5161–5174. [Google Scholar] [CrossRef]
- Deng, W.; Zhang, A.; Chen, S.; He, X.; Jin, L.; Yu, X.; Yang, S.; Li, B.; Fan, L.; Ji, L.; et al. Heavy Metals, Antibiotics and Nutrients Affect the Bacterial Community and Resistance Genes in Chicken Manure Composting and Fertilized Soil. J. Environ. Manag. 2020, 257, 109980. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut Biogeography of the Bacterial Microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Shafey, T.M.; McDonald, M.W.; Dingle, J.G. Effects of Dietary Calcium and Available Phosphorus Concentration on Digesta PH and on the Availability of Calcium, Iron, Magnesium and Zinc from the Intestinal Contents of Meat Chickens. Br. Poult. Sci. 1991, 32, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Ptak, A.; Bedford, M.R.; Świątkiewicz, S.; Żyła, K.; Józefiak, D. Phytase Modulates Ileal Microbiota and Enhances Growth Performance of the Broiler Chickens. PLoS ONE 2015, 10, e0119770. [Google Scholar] [CrossRef] [PubMed]
- Witzig, M.; Silva, A.C.d.; Green-Engert, R.; Hoelzle, K.; Zeller, E.; Seifert, J.; Hoelzle, L.E.; Rodehutscord, M. Spatial Variation of the Gut Microbiota in Broiler Chickens as Affected by Dietary Available Phosphorus and Assessed by T-RFLP Analysis and 454 Pyrosequencing. PLoS ONE 2015, 10, e0143442. [Google Scholar] [CrossRef]
- Peng, S.; Feng, Y.; Wang, Y.; Guo, X.; Chu, H.; Lin, X. Prevalence of Antibiotic Resistance Genes in Soils after Continually Applied with Different Manure for 30 Years. J. Hazard. Mater. 2017, 340, 16–25. [Google Scholar] [CrossRef]
- Qian, X.; Gu, J.; Sun, W.; Wang, X.-J.; Su, J.-Q.; Stedfeld, R. Diversity, Abundance, and Persistence of Antibiotic Resistance Genes in Various Types of Animal Manure Following Industrial Composting. J. Hazard. Mater. 2018, 344, 716–722. [Google Scholar] [CrossRef]
- Xu, Y.; Li, H.; Shi, R.; Lv, J.; Li, B.; Yang, F.; Zheng, X.; Xu, J. Antibiotic Resistance Genes in Different Animal Manures and Their Derived Organic Fertilizer. Environ. Sci. Eur. 2020, 32, 102. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Liu, F.; Cao, J.; Lv, N.; Zhu, B.; Zhang, G.; Gao, G.F. Integrated Metagenomic and Metatranscriptomic Profiling Reveals Differentially Expressed Resistomes in Human, Chicken, and Pig Gut Microbiomes. Environ. Int. 2020, 138, 105649. [Google Scholar] [CrossRef]
- Zhao, L.; Dong, Y.H.; Wang, H. Residues of Veterinary Antibiotics in Manures from Feedlot Livestock in Eight Provinces of China. Sci. Total Environ. 2010, 408, 1069–1075. [Google Scholar] [CrossRef]
- Chen, J.; Fluharty, F.L.; St-Pierre, N.; Morrison, M.; Yu, Z. Technical Note: Occurrence in Fecal Microbiota of Genes Conferring Resistance to Both Macrolide-Lincosamide-Streptogramin B and Tetracyclines Concomitant with Feeding of Beef Cattle with Tylosin. J. Anim. Sci. 2008, 86, 2385–2391. [Google Scholar] [CrossRef]
- Lee, S.; Mir, R.A.; Park, S.H.; Kim, D.; Kim, H.-Y.; Boughton, R.K.; Morris, J.G.; Jeong, K.C. Prevalence of Extended-Spectrum β-Lactamases in the Local Farm Environment and Livestock: Challenges to Mitigate Antimicrobial Resistance. Crit. Rev. Microbiol. 2020, 46, 1–14. [Google Scholar] [CrossRef]
- Binh, C.T.T.; Heuer, H.; Kaupenjohann, M.; Smalla, K. Piggery Manure Used for Soil Fertilization Is a Reservoir for Transferable Antibiotic Resistance Plasmids. FEMS Microbiol. Ecol. 2008, 66, 25–37. [Google Scholar] [CrossRef]
- Moodley, A.; Guardabassi, L. Transmission of IncN Plasmids Carrying BlaCTX-M-1 between Commensal Escherichia Coli in Pigs and Farm Workers. Antimicrob. Agents Chemother. 2009, 53, 1709–1711. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, Y.; Sun, Y.; Ma, L.; Zeng, Q.; Jiang, X.; Li, A.; Zeng, Z.; Zhang, T. Antibiotic-Mediated Changes in the Fecal Microbiome of Broiler Chickens Define the Incidence of Antibiotic Resistance Genes. Microbiome 2018, 6, 34. [Google Scholar] [CrossRef]
- Guo, T.; Lou, C.; Zhai, W.; Tang, X.; Hashmi, M.Z.; Murtaza, R.; Li, Y.; Liu, X.; Xu, J. Increased Occurrence of Heavy Metals, Antibiotics and Resistance Genes in Surface Soil after Long-Term Application of Manure. Sci. Total Environ. 2018, 635, 995–1003. [Google Scholar] [CrossRef]
- Mahmoud, M.A.M.; Abdel-Mohsein, H.S. Hysterical Tetracycline in Intensive Poultry Farms Accountable for Substantial Gene Resistance, Health and Ecological Risk in Egypt- Manure and Fish. Environ. Pollut. 2019, 255, 113039. [Google Scholar] [CrossRef]
- Yoshizawa, N.; Usui, M.; Fukuda, A.; Asai, T.; Higuchi, H.; Okamoto, E.; Seki, K.; Takada, H.; Tamura, Y. Manure Compost Is a Potential Source of Tetracycline-Resistant Escherichia Coli and Tetracycline Resistance Genes in Japanese Farms. Antibiotics 2020, 9, 76. [Google Scholar] [CrossRef]
- Agersø, Y.; Pedersen, A.G.; Aarestrup, F.M. Identification of Tn5397-like and Tn916-like Transposons and Diversity of the Tetracycline Resistance Gene Tet(M) in Enterococci from Humans, Pigs and Poultry. J. Antimicrob. Chemother. 2006, 57, 832–839. [Google Scholar] [CrossRef]
- Leclercq, S.O.; Wang, C.; Zhu, Y.; Wu, H.; Du, X.; Liu, Z.; Feng, J. Diversity of the Tetracycline Mobilome within a Chinese Pig Manure Sample. Appl. Environ. Microbiol. 2016, 82, 6454–6462. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of Sulfonamide Resistance Genes (Sul1, Sul2, and Sul3) in Portuguese Salmonella Enterica Strains and Relation with Integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef]
- Domínguez, M.; Miranda, C.D.; Fuentes, O.; de la Fuente, M.; Godoy, F.A.; Bello-Toledo, H.; González-Rocha, G. Occurrence of Transferable Integrons and Sul and Dfr Genes Among Sulfonamide-and/or Trimethoprim-Resistant Bacteria Isolated From Chilean Salmonid Farms. Front. Microbiol. 2019, 10, 748. [Google Scholar] [CrossRef]
- Møller, A.K.; Barkay, T.; Hansen, M.A.; Norman, A.; Hansen, L.H.; Sørensen, S.J.; Boyd, E.S.; Kroer, N. Mercuric Reductase Genes (MerA) and Mercury Resistance Plasmids in High Arctic Snow, Freshwater and Sea-Ice Brine. FEMS Microbiol. Ecol. 2014, 87, 52–63. [Google Scholar] [CrossRef]
- Popowska, M.; Krawczyk-Balska, A. Broad-Host-Range IncP-1 Plasmids and Their Resistance Potential. Front. Microbiol. 2013, 4, 44. [Google Scholar] [CrossRef]
- Zhou, Y.; Awasthi, S.K.; Liu, T.; Verma, S.; Zhang, Z.; Pandey, A.; Varjani, S.; Li, R.; Taherzadeh, M.J.; Awasthi, M.K. Patterns of Heavy Metal Resistant Bacterial Community Succession Influenced by Biochar Amendment during Poultry Manure Composting. J. Hazard. Mater. 2021, 420, 126562. [Google Scholar] [CrossRef]
- Qiu, X.; Zhou, G.; Wang, H.; Wu, X. The Behavior of Antibiotic-Resistance Genes and Their Relationships with the Bacterial Community and Heavy Metals during Sewage Sludge Composting. Ecotoxicol. Environ. Saf. 2021, 216, 112190. [Google Scholar] [CrossRef]
- Liao, H.; Zhao, Q.; Cui, P.; Chen, Z.; Yu, Z.; Geisen, S.; Friman, V.-P.; Zhou, S. Efficient Reduction of Antibiotic Residues and Associated Resistance Genes in Tylosin Antibiotic Fermentation Waste Using Hyperthermophilic Composting. Environ. Int. 2019, 133, 105203. [Google Scholar] [CrossRef]


| Bacteria Species | Screening Selective Agar + Antibiotic (Concentration) Using the EUCAST Breakpoints | Selecting for |
|---|---|---|
| Klebsiella pneumoniae | Simmons Citrate Agar + inositol Imipenem (16 mg/L) | Carbapenem resistance |
| Klebsiella pneumoniae | Simmons Citrate Agar + inositol Cefotaxime (4 mg/L) | Extended-spectrum beta-lactamase |
| Escherichia coli | Eosin Methylene Blue Agar Imipenem (16 mg/L) | Carbapenem resistance |
| MacConkey Agar Imipenem (16 mg/L) | ||
| Escherichia coli | Eosin Methylene Blue Agar Cefotaxime (4 mg/L) | Extended-spectrum beta-lactamase |
| MacConkey Agar Cefotaxime (4 mg/L) | ||
| Enterococcus sp. | Brilliance VRE Agar | Vancomycin resistance |
| Acinetobacter baumannii | CHROMagar Acinetobacter Imipenem (16 mg/L) | Carbapenem resistance |
| Pseudomonas aeruginosa | Cetrimide Agar Imipenem (16 mg/L) | Carbapenem resistance |
| Staphylococcus aureus | Mannitol salt Agar Oxacillin (4 mg/L) | Methicillin resistance |
| Staphylococcus aureus | Mannitol Salt Agar Vancomycin (4 mg/L) | Vancomycin resistance |
| Parameter | Soil | Chicken Manure (CM) | Chicken Litter (CL) | |
|---|---|---|---|---|
| pH | 6–6.5 | 8 | 6.5 | |
| heavy metals [mg/kg] | mercury (Hg) | <0.05 | <0.05 | <0.05 |
| chrome (Cr) | 5.56 | <2.00 | <2.00 | |
| zinc (Zn) | 19.7 | 616 | 430 | |
| cadmium (Cd) | <0.4 | 0.687 | 0.478 | |
| copper (Cu) | 3.41 | 86.7 | 66.3 | |
| nickel (Ni) | <3.00 | 6.27 | 9.2 | |
| lead (Pb) | 7.74 | <5.00 | <5.00 | |
| macronutrients [mg/kg] | calcium (Ca) | 110 | 1140.0 | 6670 |
| magnesium (Mg) | 57.7 | 945 | 613 | |
| total phosphorus (P) | 205 | 2430 | 1510 | |
| ammoniacal nitrogen (NH4-N) [mg/kg] | <0.00233 | 3200 | 2400 | |
| total nitrogen (N) [%wt] | 0.0926 | 1.22 | 2.78 | |
| dry organic matter (OM) [%] | 2.2 | 18.4 | 60.2 | |
| Medium | Antibiotic | Temperature [°C] | Chicken Waste | [cfu/mL] |
|---|---|---|---|---|
| Mannitol Salt Agar | Vancomycin | 25 | CM | 3.1 × 106 |
| 25 | CL | 2.1 × 105 | ||
| 37 | CM | 1.8 × 105 | ||
| 37 | CL | 6.5 × 104 | ||
| Oxacillin | 25 | CM | ---- | |
| 25 | CL | 4.6 × 105 | ||
| 37 | CM | 5.1 × 106 | ||
| 37 | CL | 4.1 × 107 | ||
| Eosin Methylene Blue Agar | Cefotaxime | 25 | CM | 7.2 × 108 |
| 25 | CL | 5.1 × 105 | ||
| 37 | CM | 3.9 × 105 | ||
| 37 | CL | 1.9 × 104 | ||
| Imipenem | 25 | CM | 1.2 × 107 | |
| 25 | CL | 3.3 × 105 | ||
| 37 | CM | ---- | ||
| 37 | CL | 1.0 × 103 | ||
| MacConkey Agar | Cefotaxime | 25 | CM | 3.2 × 105 |
| 25 | CL | 4.1 × 104 | ||
| 37 | CM | 4.7 × 106 | ||
| 37 | CL | 3.8 × 105 | ||
| Imipenem | 25 | CM | 2.2 × 103 | |
| 25 | CL | 1.5 × 102 | ||
| 37 | CM | 4.1 × 104 | ||
| 37 | CL | 1.5 × 103 | ||
| Simmons Citrate Agar | Cefotaxime | 25 | CM | 3.3 × 106 |
| 25 | CL | 2.4 × 105 | ||
| 37 | CM | 1.4 × 102 | ||
| 37 | CL | --- | ||
| Imipenem | 25 | CM | 4.2 × 106 | |
| 25 | CL | ---- | ||
| 37 | CM | 6.2 × 106 | ||
| 37 | CL | ---- | ||
| Cetrimide Agar | Imipenem | 25 | CM | 1.2 × 104 |
| 25 | CL | 2.1 × 106 | ||
| 37 | CM | 1.1 × 105 | ||
| 37 | CL | 1.0 × 104 | ||
| CHROMagar™ Acinetobacter | Imipenem | 25 | CM | ---- |
| 25 | CL | ---- | ||
| 37 | CM | ---- | ||
| 37 | CL | ---- | ||
| Brilliance VRE Agar | Vancomycin | 25 | CM | ----- |
| 25 | CL | 2.4 × 103 | ||
| 37 | CM | 1.3 × 102 | ||
| 37 | CL | ---- |
| Species | No. of Strains/All Isolates | Source | Phenotype | MDR (Yes/No) | |
|---|---|---|---|---|---|
| Resistance | Intermediate Resistance | ||||
| Acinetobacter johnsonii | 1/1 | CL | TZP | CIP(I), PRL(I) | No |
| Citrobacter freundii | 1/1 | CL, CM | AMC, CXM, CAZ, CN, TOB, CTX, CIP, CXM-AK | TZP(I) | Yes |
| Escherichia coli | 1/23 | CL | AMC, CXM, CAZ, CN, TOB, CTX, CIP, CXM-AK | IMP(I) | Yes |
| 4/23 | CL, CM | AMC, TZP, CXM, CAZ, CN, TOB, CTX, CIP, CXM-AK | Yes | ||
| 8/23 | CL | AMC, CXM, CAZ, CN, TOB, CTX, CIP, CXM-AK | TZP(I) | Yes | |
| 1/23 | CL | AMC, CXM, CAZ, CN, TOB, CTX, FEP, CXM-AK | TZP(I) | Yes | |
| 8/23 | CL, CM | AMC, CXM, CAZ, CN, TOB, CTX, CIP, CXM-AK | Yes | ||
| 1/23 | CL | AMC, CXM, CAZ, MEM, CN, TOB, CTX, CIP, CXM-AK | TZP(I) | Yes | |
| Klebsiella aerogenes (Enterobacter aerogenes) | 1/3 | CL | AMC(I) | No | |
| 2/3 | CL | AMC | CXM(I) | No | |
| Myroides odoratimimus | 2/13 | CL, CM | TZP, CN, AK, TOB, CIP, SXT, ATM, LEV | Yes | |
| 1/13 | CL, CM | CN, AK, TOB, CIP, SXT, PRL, ATM, LEV | TZP(I), IMP(I) | Yes | |
| 2/13 | CL, CM | CN, AK, TOB, CIP, SXT, PRL, ATM, LEV | TZP(I) | Yes | |
| 1/13 | CL, CM | CN, AK, TOB, SXT, PRL, ATM | TZP(I) | Yes | |
| 2/13 | CL, CM | CN, AK, TOB, CIP, SXT, PRL, ATM, LEV | Yes | ||
| 1/13 | CL, CM | TZP, CN, AK, TOB, SXT, PRL, ATM | IMP(I), CIP(I) | Yes | |
| 1/13 | CL, CM | CN, TOB, CIP, SXT, PRL, ATM, LEV | TZP(I), AK(I) | Yes | |
| 2/13 | CL, CM | CN, AK, TOB, SXT, PRL, ATM | TZP(I), CIP(I) | Yes | |
| Myroides odoratus | 2/2 | CM | TZP, IMP, MEM, CN, AK, TOB, CIP, SXT, PRL, ATM, LEV | Yes | |
| Providencia rettgeri | 1/3 | CM | AMC, CIP, TGC, CT | CXM(I), IMP(I) | Yes |
| 1/3 | CM | AMC, TGC, CT, SXT | CXM(I), IMP(I), CTX(I) | Yes | |
| 1/3 | CM | AMC, TZP, CIP, TGC | IMP(I), FEP(I) | Yes | |
| Serratia marcescens | 1/5 | CL | AMC, CXM, CIP, CXM-AK, CT | TGC(I) | Yes |
| 3/5 | CL | AMC, CXM, CIP, CXM-AK, CT | Yes | ||
| 1/5 | CL | AMC, CXM, CXM-AK | Yes | ||
| Staphylococcus lentus | 2/19 | CL | CIP, LEV, E, TE, DA | STX(I) | Yes |
| 2/19 | CL | CN, CIP, STX, LEV, E, TE, DA | Yes | ||
| 3/19 | CL | CIP, LEV, OB, TE, DA | Yes | ||
| 2/19 | CL | CN, CIP, STX, LEV, LZD, E, TE, DA | Yes | ||
| 2/19 | CL | TE | CIP(I), LEV(I), DA(I) | Yes | |
| 2/19 | CL | TE | CIP(I), LEV(I) | Yes | |
| 1/19 | CL | CN, CIP, SXT, LEV, OB, E, TE, DA | Yes | ||
| 1/19 | CL | CIP, TGC, LEV, LZD, E, TE, DA | Yes | ||
| 1/19 | CL | CN, CIP, SXT, LEV, E, TE, DA | Yes | ||
| 1/19 | CL | CIP, TGC, LEV, LZD, TE, DA | Yes | ||
| 1/19 | CL | CIP, LEV, ER, TET, DA | Yes | ||
| 1/19 | CL | CIP, SXT, LEV, ER, TET, DA | Yes | ||
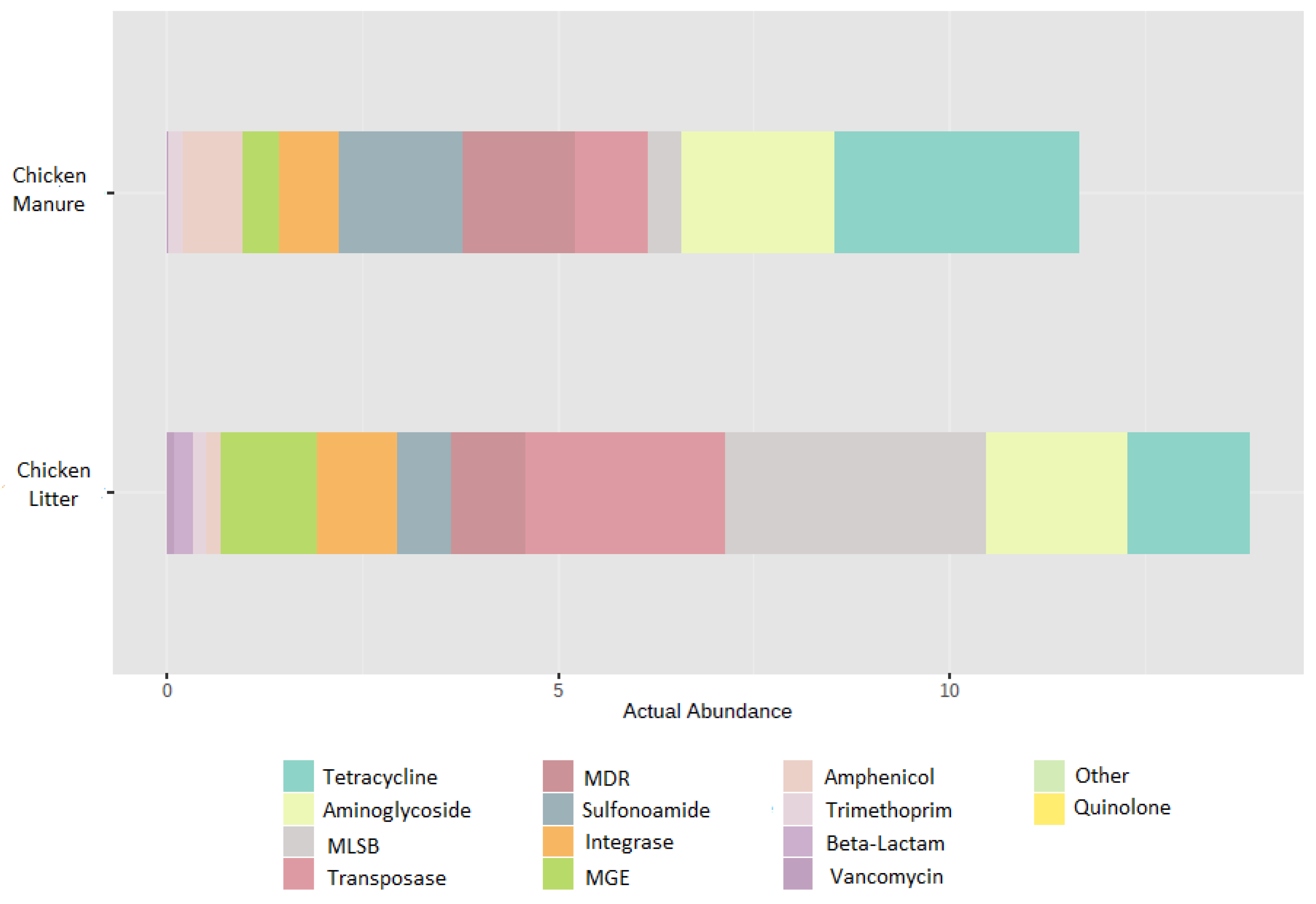
| Gene Group | CL (Mean) | CM (Mean) | Chicken Waste (Mean) |
|---|---|---|---|
| MGE | 1.2556307984 | 0.3211213793 | 0.7883760889 |
| Tetracycline | 0.5144201057 | 1.0377003019 | 0.7760602038 |
| Aminoglycoside | 0.6050128022 | 0.6581070783 | 0.6315599403 |
| MLSB | 1.1137056246 | 0.1455982502 | 0.6296519374 |
| Sulfonamide | 0.2256385199 | 0.5193078503 | 0.3724731851 |
| Other | 0.2076692490 | 0.4556390180 | 0.3316541335 |
| Integrons | 0.3429847709 | 0.2639967037 | 0.3034907373 |
| Phenicol | 0.0691178113 | 0.2602892051 | 0.1647035082 |
| MDR | 0.1157465734 | 0.0317774276 | 0.0737620005 |
| Trimethoprim | 0.0476679473 | 0.0578028990 | 0.0527354232 |
| Beta Lactam | 0.0909582761 | 0.0055880528 | 0.0482731645 |
| Vancomycin | 0.0196980994 | 0.0006651844 | 0.0101816419 |
| Quinolone | 0.0006499213 | 0.0000000000 | 0.0003249607 |
| Gene | CM (Mean) |
|---|---|
| tetPB_2 | 0.2751246220 |
| tnpA_2 | 0.2459280497 |
| cmxA | 0.2216566087 |
| qacE∆1_1 | 0.2021822017 |
| tetM_3 | 0.1993868087 |
| tetX | 0.1452556967 |
| qacE∆1_2 | 0.1434562960 |
| tetM_1 | 0.1427971303 |
| sul2_2 | 0.1301656963 |
| sul1_1 | 0.1214871360 |
| Gene | CL (Mean) |
|---|---|
| lnuA_1 | 0.6792154677 |
| tnpA_6 | 0.4736300717 |
| ISEfm1-Entero | 0.3201372400 |
| ermB | 0.2429636293 |
| tetM_3 | 0.1942361840 |
| intI1_1 | 0.1446150687 |
| tnpA_2 | 0.1364071333 |
| tnpA_1 | 0.1259366313 |
| tetM_1 | 0.1186746973 |
| sul1_1 | 0.1135728363 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Błażejewska, A.; Zalewska, M.; Grudniak, A.; Popowska, M. A Comprehensive Study of the Microbiome, Resistome, and Physical and Chemical Characteristics of Chicken Waste from Intensive Farms. Biomolecules 2022, 12, 1132. https://doi.org/10.3390/biom12081132
Błażejewska A, Zalewska M, Grudniak A, Popowska M. A Comprehensive Study of the Microbiome, Resistome, and Physical and Chemical Characteristics of Chicken Waste from Intensive Farms. Biomolecules. 2022; 12(8):1132. https://doi.org/10.3390/biom12081132
Chicago/Turabian StyleBłażejewska, Aleksandra, Magdalena Zalewska, Anna Grudniak, and Magdalena Popowska. 2022. "A Comprehensive Study of the Microbiome, Resistome, and Physical and Chemical Characteristics of Chicken Waste from Intensive Farms" Biomolecules 12, no. 8: 1132. https://doi.org/10.3390/biom12081132
APA StyleBłażejewska, A., Zalewska, M., Grudniak, A., & Popowska, M. (2022). A Comprehensive Study of the Microbiome, Resistome, and Physical and Chemical Characteristics of Chicken Waste from Intensive Farms. Biomolecules, 12(8), 1132. https://doi.org/10.3390/biom12081132