Biology of the Extracellular Proteasome
Abstract
:1. Introduction
2. Integrated Analysis of Proteomic Data from Body Fluids
3. Plasma Circulating Proteasomes
4. Proteasomes Encapsulated within Extracellular Vesicles
5. Experimental Strategies for Investigating Extracellular Proteasomes
6. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 2014, 1843, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciechanover, A. Intracellular protein degradation: From a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Best Pract. Res. Clin. Haematol. 2017, 30, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Wada, M.; Kosaka, M.; Saito, S.; Sano, T.; Tanaka, K.; Ichihara, A. Serum concentration and localization in tumor cells of proteasomes in patients with hematologic malignancy and their pathophysiologic significance. J. Lab. Clin. Med. 1993, 121, 215–223. [Google Scholar] [PubMed]
- Saitoh, Y.; Sawada, H.; Yokosawa, H. High-molecular-weight protease complexes (proteasomes) of sperm of the ascidian, Halocynthia roretzi: Isolation, characterization, and physiological roles in fertilization. Dev. Biol. 1993, 158, 238–244. [Google Scholar] [CrossRef]
- Sutovsky, P. Sperm proteasome and fertilization. Reproduction 2011, 142, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Dieude, M.; Bell, C.; Turgeon, J.; Beillevaire, D.; Pomerleau, L.; Yang, B.; Hamelin, K.; Qi, S.; Pallet, N.; Beland, C.; et al. The 20S proteasome core, active within apoptotic exosome-like vesicles, induces autoantibody production and accelerates rejection. Sci. Transl. Med. 2015, 7, 318ra200. [Google Scholar] [CrossRef] [Green Version]
- Mueller, O.; Anlasik, T.; Wiedemann, J.; Thomassen, J.; Wohlschlaeger, J.; Hagel, V.; Keyvani, K.; Schwieger, I.; Dahlmann, B.; Sure, U.; et al. Circulating extracellular proteasome in the cerebrospinal fluid: A study on concentration and proteolytic activity. J. Mol. Neurosci. 2012, 46, 509–515. [Google Scholar] [CrossRef]
- Sixt, S.U.; Beiderlinden, M.; Jennissen, H.P.; Peters, J. Extracellular proteasome in the human alveolar space: A new housekeeping enzyme? Am. J. Physiol. Lung Cell Mol. Physiol. 2007, 292, L1280–L1288. [Google Scholar] [CrossRef] [Green Version]
- Tengowski, M.W.; Feng, D.; Sutovsky, M.; Sutovsky, P. Differential expression of genes encoding constitutive and inducible 20S proteasomal core subunits in the testis and epididymis of theophylline- or 1,3-dinitrobenzene-exposed rats. Biol. Reprod. 2007, 76, 149–163. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, V.; Yaniv, K.; Sharon, M. Beyond cells: The extracellular circulating 20S proteasomes. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166041. [Google Scholar] [CrossRef]
- Brinkman, J.E.; Dorius, B.; Sharma, S. Physiology, Body Fluids; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Huang, L.; Shao, D.; Wang, Y.; Cui, X.; Li, Y.; Chen, Q.; Cui, J. Human body-fluid proteome: Quantitative profiling and computational prediction. Brief. Bioinform. 2020, 22, 315–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tobias, A.; Ballard, B.D.; Mohiuddin, S.S. Physiology, Water Balance; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Rousseau, A.; Bertolotti, A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 697–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar Deshmukh, F.; Yaffe, D.; Olshina, M.A.; Ben-Nissan, G.; Sharon, M. The contribution of the 20S proteasome to proteostasis. Biomolecules 2019, 9, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 426, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Ben-Nissan, G.; Sharon, M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 2014, 4, 862–884. [Google Scholar] [CrossRef] [Green Version]
- Cascio, P. PA28g: New insights on an ancient proteasome activator. Biomolecules 2021, 11, 228. [Google Scholar] [CrossRef]
- Pickering, A.M.; Davies, K.J.A. Differential roles of proteasome and immunoproteasome regulators Pa28αβ, Pa28γ and Pa200 in the degradation of oxidized proteins. Arch. Biochem. Biophys. 2012, 523, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Sahu, I.; Mali, S.; Sulkshane, P.; Rozenberg, A.; Xu, C.; Morag, R.; Sahoo, M.P.; Singh, S.; Ding, Z.; Wang, Y.; et al. Signature activities of 20S proteasome include degradation of the ubiquitin-tag with the protein under hypoxia. bioRxiv, 2019; preprint. [Google Scholar] [CrossRef]
- Fabre, B.; Lambour, T.; Delobel, J.; Amalric, F.; Monsarrat, B.; Burlet-Schiltz, O.; Bousquet-Dubouch, M.P. Subcellular distribution and dynamics of active proteasome complexes unraveled by a workflow combining in vivo complex cross-linking and quantitative proteomics. Mol. Cell Proteom. 2013, 12, 687–699. [Google Scholar] [CrossRef] [Green Version]
- Fabre, B.; Lambour, T.; Garrigues, L.; Ducoux-Petit, M.; Amalric, F.; Monsarrat, B.; Burlet-Schiltz, O.; Bousquet-Dubouch, M.P. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J. Proteome Res. 2014, 13, 3027–3037. [Google Scholar] [CrossRef]
- Olshina, M.A.; Arkind, G.; Kumar Deshmukh, F.; Fainer, I.; Taranavsky, M.; Hayat, D.; Ben-Dor, S.; Ben-Nissan, G.; Sharon, M. Regulation of the 20S proteasome by a novel family of inhibitory proteins. Antioxid Redox Signal 2020, 32, 636–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raynes, R.; Pomatto, L.C.; Davies, K.J. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol. Asp. Med. 2016, 50, 41–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol Transl. Sci. 2012, 109, 75–112. [Google Scholar]
- Gaczynska, M.; Rock, K.L.; Spies, T.; Goldberg, A.L. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex-encoded genes for LMP2 and LMP. Proc. Natl. Acad. Sci. USA 1994, 91, 9213–9217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebstein, F.; Kloetzel, P.M.; Kruger, E.; Seifert, U. Emerging roles of immunoproteasomes beyond MHC class I antigen processing. Cell Mol. Life Sci. 2012, 69, 2543–2558. [Google Scholar] [CrossRef]
- Opitz, E.; Koch, A.; Klingel, K.; Schmidt, F.; Prokop, S.; Rahnefeld, A.; Sauter, M.; Heppner, F.L.; Volker, U.; Kandolf, R.; et al. Impairment of immunoproteasome function by beta5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog. 2011, 7, e1002233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.P.; Shen, Z.; Van Kaer, L.; Becker, L.C. Ischemic preconditioning-induced cardioprotection is lost in mice with immunoproteasome subunit low molecular mass polypeptide-2 deficiency. FASEB J. 2008, 22, 4248–4257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrington, D.A.; Husom, A.D.; Thompson, L.V. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005, 19, 644–646. [Google Scholar] [CrossRef]
- Klare, N.; Seeger, M.; Janek, K.; Jungblut, P.R.; Dahlmann, B. Intermediate-type 20S proteasomes in HeLa cells: “asymmetric” subunit composition, diversity and adaptation. J. Mol. Biol. 2007, 373, 1–10. [Google Scholar] [CrossRef]
- Dahlmann, B.; Ruppert, T.; Kuehn, L.; Merforth, S.; Kloetzel, P.M. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J. Mol. Biol. 2000, 303, 643–653. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Didiot, M.C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef] [PubMed]
- Harshman, S.W.; Canella, A.; Ciarlariello, P.D.; Agarwal, K.; Branson, O.E.; Rocci, A.; Cordero, H.; Phelps, M.A.; Hade, E.M.; Dubovsky, J.A.; et al. Proteomic characterization of circulating extracellular vesicles identifies novel serum myeloma associated markers. J. Proteom. 2016, 136, 89–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bec, N.; Bonhoure, A.; Henry, L.; Berry, L.; Larroque, C.; Coux, O.; Stoebner, P.E.; Vidal, M. Proteasome 19S RP and translation preinitiation complexes are secreted within exosomes upon serum starvation. Traffic 2019, 20, 516–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Chen, X.; Pan, Q.; Wang, Y.; Su, S.; Jiang, C.; Li, Y.; Xu, N.; Wu, L.; Lou, X.; et al. A Comprehensive proteomics analysis reveals a secretory path- and status-dependent signature of exosomes released from tumor-associated macrophages. J. Proteome Res. 2015, 14, 4319–4331. [Google Scholar] [CrossRef]
- Menneteau, T.; Fabre, B.; Garrigues, L.; Stella, A.; Zivkovic, D.; Roux-Dalvai, F.; Mouton-Barbosa, E.; Beau, M.; Renoud, M.L.; Amalric, F.; et al. Mass spectrometry-based absolute quantification of 20S proteasome status for controlled ex-vivo expansion of human adipose-derived mesenchymal stromal/stem cells. Mol. Cell Proteom. 2019, 18, 744–759. [Google Scholar] [CrossRef] [Green Version]
- Dekel, E.; Yaffe, D.; Rosenhek-Goldian, I.; Ben-Nissan, G.; Ofir-Birin, Y.; Morandi, M.I.; Ziv, T.; Sisquella, X.; Pimentel, M.A.; Nebl, T.; et al. 20S proteasomes secreted by the malaria parasite promote its growth. Nat. Commun. 2021, 12, 1172. [Google Scholar] [CrossRef]
- Majetschak, M.; Sorell, L.T.; Patricelli, T.; Seitz, D.H.; Knoferl, M.W. Detection and possible role of proteasomes in the bronchoalveolar space of the injured lung. Physiol. Res. 2009, 58, 363–372. [Google Scholar] [CrossRef]
- Albright, J.M.; Romero, J.; Saini, V.; Sixt, S.U.; Bird, M.D.; Kovacs, E.J.; Gamelli, R.L.; Peters, J.; Majetschak, M. Proteasomes in human bronchoalveolar lavage fluid after burn and inhalation injury. J. Burn Care Res. 2009, 30, 948–956. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, H.H.; Beck, H.C.; Kristensen, L.P.; Burton, M.; Csepany, T.; Simo, M.; Dioszeghy, P.; Sejbaek, T.; Grebing, M.; Heegaard, N.H.; et al. The urine proteome profile is different in neuromyelitis optica compared to multiple sclerosis: A clinical proteome study. PLoS ONE 2015, 10, e0139659. [Google Scholar] [CrossRef]
- Zhao, M.; Li, M.; Yang, Y.; Guo, Z.; Sun, Y.; Shao, C.; Li, M.; Sun, W.; Gao, Y. A comprehensive analysis and annotation of human normal urinary proteome. Sci. Rep. 2017, 7, 3024. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Liu, H.; Sun, W.; Ding, B.; Zhao, Y.; Chen, P.; Zhu, L.; Li, Z.; Li, N.; et al. Urine proteome of COVID-19 patients. Urine 2020, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Prikryl, P.; Satrapova, V.; Frydlova, J.; Hruskova, Z.; Zima, T.; Tesar, V.; Vokurka, M. Mass spectrometry-based proteomic exploration of the small urinary extracellular vesicles in ANCA-associated vasculitis in comparison with total urine. J. Proteom. 2021, 233, 104067. [Google Scholar] [CrossRef] [PubMed]
- Huo, S.; Wang, H.; Yan, M.; Xu, P.; Song, T.; Li, C.; Tian, R.; Chen, X.; Bao, K.; Xie, Y.; et al. Urinary proteomic characteristics of hyperuricemia and their possible links with the occurrence of its concomitant diseases. ACS Omega 2021, 6, 9500–9508. [Google Scholar] [CrossRef] [PubMed]
- Zoeger, A.; Blau, M.; Egerer, K.; Feist, E.; Dahlmann, B. Circulating proteasomes are functional and have a subtype pattern distinct from 20S proteasomes in major blood cells. Clin. Chem. 2006, 52, 2079–2086. [Google Scholar] [CrossRef] [PubMed]
- Majetschak, M.; Zedler, S.; Romero, J.; Albright, J.M.; Kraft, R.; Kovacs, E.J.; Faist, E.; Gamelli, R.L. Circulating proteasomes after burn injury. J. Burn. Care Res. Off. Publ. Am. Burn. Assoc. 2010, 31, 243–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, Y.; Lee, S.Y.; Choi, W.H.; Park, J.C.; Lee, D.H.; Kim, Y.K.; Lee, J.H.; Lee, J.Y.; Lee, M.J.; Kim, Y.H. Proteasome activity in the plasma as a novel biomarker in mild cognitive impairment with chronic tinnitus. J. Alzheimers Dis. 2020, 78, 195–205. [Google Scholar] [CrossRef]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma proteome profiling to assess human health and disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Farrah, T.; Deutsch, E.W.; Omenn, G.S.; Campbell, D.S.; Sun, Z.; Bletz, J.A.; Mallick, P.; Katz, J.E.; Malmstrom, J.; Ossola, R.; et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell Proteom. 2011, 10, M110.006353. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Hincapie, M.; Pitteri, S.J.; Hanash, S.; Schalkwijk, J.; Hogan, J.M.; Wang, H.; Hancock, W.S. A proteomics platform combining depletion, multi-lectin affinity chromatography (M-LAC), and isoelectric focusing to study the breast cancer proteome. Anal. Chem. 2011, 83, 4845–4854. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, H.; Kim, S.Y.; Kim, Y.; Lee, J.S.; Dan, K.; Seong, M.W.; Han, D. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci. Rep. 2020, 10, 22418. [Google Scholar] [CrossRef]
- Liu, C.W.; Bramer, L.; Webb-Robertson, B.J.; Waugh, K.; Rewers, M.J.; Zhang, Q. Temporal expression profiling of plasma proteins reveals oxidative stress in early stages of Type 1 Diabetes progression. J. Proteom. 2018, 172, 100–110. [Google Scholar] [CrossRef]
- Harel, M.; Oren-Giladi, P.; Kaidar-Person, O.; Shaked, Y.; Geiger, T. Proteomics of microparticles with SILAC Quantification (PROMIS-Quan): A novel proteomic method for plasma biomarker quantification. Mol. Cell Proteom. 2015, 14, 1127–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sixt, S.U.; Dahlmann, B. Extracellular, circulating proteasomes and ubiquitin—Incidence and relevance. Biochim. Biophys. Acta Mol. Basis Dis. 2008, 1782, 817–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavabre-Bertrand, T.; Henry, L.; Carillo, S.; Guiraud, I.; Ouali, A.; Dutaud, D.; Aubry, L.; Rossi, J.F.; Bureau, J.P. Plasma proteasome level is a potential marker in patients with solid tumors and hemopoietic malignancies. Cancer 2001, 92, 2493–2500. [Google Scholar] [CrossRef]
- Chan, F.K.; Moriwaki, K.; De Rosa, M.J. Detection of necrosis by release of lactate dehydrogenase activity. Methods Mol. Biol 2013, 979, 65–70. [Google Scholar] [PubMed] [Green Version]
- Stoebner, P.E.; Lavabre-Bertrand, T.; Henry, L.; Guiraud, I.; Carillo, S.; Dandurand, M.; Joujoux, J.M.; Bureau, J.P.; Meunier, L. High plasma proteasome levels are detected in patients with metastatic malignant melanoma. Br. J. Dermatol. 2005, 152, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Tsimokha, A.S.; Zaykova, J.J.; Bottrill, A.; Barlev, N.A. Extracellular proteasomes are deficient in 19s subunits as revealed by itraq quantitative proteomics. J. Cell Physiol. 2017, 232, 842–851. [Google Scholar] [CrossRef]
- Dianzani, C.; Vecchio, D.; Clemente, N.; Chiocchetti, A.; Martinelli Boneschi, F.; Galimberti, D.; Dianzani, U.; Comi, C.; Mishto, M.; Liepe, J. Untangling Extracellular Proteasome-Osteopontin Circuit Dynamics in Multiple Sclerosis. Cells 2019, 8, 262. [Google Scholar] [CrossRef] [Green Version]
- Bochmann, I.; Ebstein, F.; Lehmann, A.; Wohlschlaeger, J.; Sixt, S.U.; Kloetzel, P.M.; Dahlmann, B. T lymphocytes export proteasomes by way of microparticles: A possible mechanism for generation of extracellular proteasomes. J. Cell Mol. Med. 2014, 18, 59–68. [Google Scholar] [CrossRef]
- Maruyama, H.; Hirayama, K.; Yamashita, M.; Ohgi, K.; Tsujimoto, R.; Takayasu, M.; Shimohata, H.; Kobayashi, M. Serum 20S proteasome levels are associated with disease activity in MPO-ANCA-associated microscopic polyangiitis. BMC Rheumatol. 2020, 4, 36. [Google Scholar] [CrossRef]
- Ma, W.; Kantarjian, H.; O’Brien, S.; Jilani, I.; Zhang, X.; Estrov, Z.; Ferrajoli, A.; Keating, M.; Giles, F.; Albitar, M. Enzymatic activity of circulating proteasomes correlates with clinical behavior in patients with chronic lymphocytic leukemia. Cancer 2008, 112, 1306–1312. [Google Scholar] [CrossRef]
- Ma, W.; Kantarjian, H.; Bekele, B.; Donahue, A.C.; Zhang, X.; Zhang, Z.J.; O’Brien, S.; Estey, E.; Estrov, Z.; Cortes, J.; et al. Proteasome enzymatic activities in plasma as risk stratification of patients with acute myeloid leukemia and advanced-stage myelodysplastic syndrome. Clin. Cancer Res. 2009, 15, 3820–3826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldziej, A.; Bolkun, L.; Galar, M.; Kalita, J.; Ostrowska, H.; Romaniuk, W.; Kloczko, J. Assessment of proteasome concentration and chymotrypsin-like activity in plasma of patients with newly diagnosed multiple myeloma. Leuk. Res. 2014, 38, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Tylicka, M.; Matuszczak, E.; Karpinska, M.; Hermanowicz, A.; Debek, W.; Ostrowska, H. Proteasome activity and C-reactive protein concentration in the course of inflammatory reaction in relation to the type of abdominal operation and the surgical technique used. Mediators Inflamm. 2018, 2018, 2469098. [Google Scholar] [CrossRef] [PubMed]
- Dixon, C.L.; Sheller-Miller, S.; Saade, G.R.; Fortunato, S.J.; Lai, A.; Palma, C.; Guanzon, D.; Salomon, C.; Menon, R. Amniotic fluid exosome proteomic profile exhibits unique pathways of term and preterm labor. Endocrinology 2018, 159, 2229–2240. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Silva, S.; Benito-Martin, A.; Sanchez-Redondo, S.; Hernandez-Barranco, A.; Ximenez-Embun, P.; Nogues, L.; Mazariegos, M.S.; Brinkmann, K.; Amor Lopez, A.; Meyer, L.; et al. Use of extracellular vesicles from lymphatic drainage as surrogate markers of melanoma progression and BRAF (V600E) mutation. J. Exp. Med. 2019, 216, 1061–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guha, D.; Lorenz, D.R.; Misra, V.; Chettimada, S.; Morgello, S.; Gabuzda, D. Proteomic analysis of cerebrospinal fluid extracellular vesicles reveals synaptic injury, inflammation, and stress response markers in HIV patients with cognitive impairment. J. Neuroinflammation 2019, 16, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, S.Y.; Choi, Y.R.; Lee, J.H.; Lim, J.M.; Lee, S.E.; Kim, K.P.; Kim, J.Y.; Lee, S.H.; Kim, M.S. In-depth proteomic analysis of human bronchoalveolar lavage fluid toward the biomarker discovery for lung cancers. Proteom. Clin. Appl. 2019, 13, e1900028. [Google Scholar] [CrossRef]
- Sun, Y.; Huo, C.; Qiao, Z.; Shang, Z.; Uzzaman, A.; Liu, S.; Jiang, X.; Fan, L.Y.; Ji, L.; Guan, X.; et al. Comparative proteomic analysis of exosomes and microvesicles in human saliva for lung cancer. J. Proteome Res. 2018, 17, 1101–1107. [Google Scholar] [CrossRef]
- van Herwijnen, M.J.; Zonneveld, M.I.; Goerdayal, S.; Nolte-’t Hoen, E.N.; Garssen, J.; Stahl, B.; Maarten Altelaar, A.F.; Redegeld, F.A.; Wauben, M.H. Comprehensive proteomic analysis of human milk-derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol. Cell Proteoml. 2016, 15, 3412–3423. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Guo, W.B.; Zhang, W.S.; Bian, J.; Yang, J.K.; Zhou, Q.Z.; Chen, M.K.; Peng, W.; Qi, T.; Wang, C.Y.; et al. Comprehensive proteomics analysis of exosomes derived from human seminal plasma. Andrology 2017, 5, 1007–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Zhao, S.Z.; Koh, S.K.; Chen, L.; Vaz, C.; Tanavde, V.; Li, X.R.; Beuerman, R.W. In-depth analysis of the human tear proteome. J. Proteom. 2012, 75, 3877–3885. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, L.; Yu, H.; McKenzie, A.J.; Franklin, J.L.; Patton, J.G.; Liu, Q.; Weaver, A.M. Quantitative proteomic analysis of small and large extracellular vesicles (evs) reveals enrichment of adhesion proteins in small EVs. J. Proteome Res. 2019, 18, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Thery, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tucher, C.; Bode, K.; Schiller, P.; Classen, L.; Birr, C.; Souto-Carneiro, M.M.; Blank, N.; Lorenz, H.M.; Schiller, M. Extracellular vesicle subtypes released from activated or apoptotic T-lymphocytes carry a specific and stimulus-dependent protein cargo. Front. Immunol. 2018, 9, 534. [Google Scholar] [CrossRef] [Green Version]
- Andre-Gregoire, G.; Bidere, N.; Gavard, J. Temozolomide affects extracellular vesicles released by glioblastoma cells. Biochimie 2018, 155, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.C.; Tan, S.S.; Teh, B.J.; Sze, S.K.; Arslan, F.; de Kleijn, D.P.; Choo, A.; Lim, S.K. Proteolytic potential of the MSC exosome proteome: Implications for an exosome-mediated delivery of therapeutic proteasome. Int J. Proteom. 2012, 2012, 971907. [Google Scholar] [CrossRef] [Green Version]
- Kulichkova, V.A.; Artamonova, T.O.; Lyublinskaya, O.G.; Khodorkovskii, M.A.; Tomilin, A.N.; Tsimokha, A.S. Proteomic analysis of affinity-purified extracellular proteasomes reveals exclusively 20S complexes. Oncotarget 2017, 8, 102134–102149. [Google Scholar] [CrossRef] [Green Version]
- Marcoux, G.; Laroche, A.; Hasse, S.; Bellio, M.; Mbarik, M.; Tamagne, M.; Allaeys, I.; Zufferey, A.; Levesque, T.; Rebetz, J.; et al. Platelet EVs contain an active proteasome involved in protein processing for antigen presentation via MHC-I molecules. Blood 2021, 138, 2607–2620. [Google Scholar] [CrossRef]
- Dianzani, C.; Bellavista, E.; Liepe, J.; Verderio, C.; Martucci, M.; Santoro, A.; Chiocchetti, A.; Gigliotti, C.L.; Boggio, E.; Ferrara, B.; et al. Extracellular proteasome-osteopontin circuit regulates cell migration with implications in multiple sclerosis. Sci. Rep. 2017, 7, 43718. [Google Scholar] [CrossRef]
- Ding, X.Q.; Wang, Z.Y.; Xia, D.; Wang, R.X.; Pan, X.R.; Tong, J.H. Proteomic profiling of serum exosomes from patients with metastatic gastric cancer. Front. Oncol. 2020, 10, 1113. [Google Scholar] [CrossRef] [PubMed]
- Yunusova, N.V.; Zambalova, E.A.; Patysheva, M.R.; Kolegova, E.S.; Afanas’ev, S.G.; Cheremisina, O.V.; Grigor’eva, A.E.; Tamkovich, S.N.; Kondakova, I.V. Exosomal protease cargo as prognostic biomarker in colorectal cancer. Asian Pac. J. Cancer Prev. 2021, 22, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Tamkovich, S.N.; Yunusova, N.V.; Tugutova, E.; Somov, A.K.; Proskura, K.V.; Kolomiets, L.A.; Stakheeva, M.N.; Grigor’eva, A.E.; Laktionov, P.P.; Kondakova, I.V. Protease cargo in circulating exosomes of breast cancer and ovarian cancer patients. Asian Pac. J. Cancer Prev. 2019, 20, 255–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Kojima, K.; Mobley, J.A.; West, A.B. Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine 2019, 45, 351–361. [Google Scholar] [CrossRef] [Green Version]
- Thompson, A.G.; Gray, E.; Mager, I.; Thezenas, M.L.; Charles, P.D.; Talbot, K.; Fischer, R.; Kessler, B.M.; Wood, M.; Turner, M.R. CSF extracellular vesicle proteomics demonstrates altered protein homeostasis in amyotrophic lateral sclerosis. Clin. Proteom. 2020, 17, 31. [Google Scholar] [CrossRef]
- Holtzman, J.; Lee, H. Emerging role of extracellular vesicles in the respiratory system. Exp. Mol. Med. 2020, 52, 887–895. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [Green Version]
- Lambeau, G.; Gelb, M.H. Biochemistry and physiology of mammalian secreted phospholipases. Annu. Rev. Biochem. 2008, 77, 495–520. [Google Scholar] [CrossRef] [Green Version]
- Vadas, P.; Pruzanski, W.; Stefanski, E.; Ellies, L.G.; Aubin, J.E.; Sos, A.; Melcher, A. Extracellular phospholipase A2 secretion is a common effector pathway of interleukin-1 and tumour necrosis factor action. Immunol. Lett. 1991, 28, 187–193. [Google Scholar] [CrossRef]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef] [Green Version]
- Sharon, M.; Regev-Rudzki, N. Cell communication and protein degradation: All in one parasitic package. J. Extracell. Vesicles 2021, 10, e12116. [Google Scholar] [CrossRef] [PubMed]
- Jahan, S.; Mukherjee, S.; Ali, S.; Bhardwaj, U.; Choudhary, R.K.; Balakrishnan, S.; Naseem, A.; Mir, S.A.; Banawas, S.; Alaidarous, M.; et al. Pioneer role of extracellular vesicles as modulators of cancer initiation in progression, drug therapy, and vaccine prospects. Cells 2022, 11, 490. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, M.G.; Lee, S.H.; Abdelazim, A.M.; Saadeldin, I.M.; Abomughaid, M.M. Role of extracellular vesicles in compromising cellular resilience to environmental stressors. Biomed. Res. Int. 2021, 2021, 9912281. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.P.R.; Caires, H.R.; Barbosa, M.A.G.; Bergantim, R.; Guimaraes, J.E.; Vasconcelos, M.H. The role of extracellular vesicles in the hallmarks of cancer and drug resistance. Cells 2020, 9, 1141. [Google Scholar] [CrossRef]
- Chang, W.H.; Cerione, R.A.; Antonyak, M.A. Extracellular vesicles and their roles in cancer progression. Methods Mol. Biol. 2021, 2174, 143–170. [Google Scholar]
- Arendt, B.K.; Walters, D.K.; Wu, X.; Tschumper, R.C.; Jelinek, D.F. Multiple myeloma dell-derived microvesicles are enriched in CD147 expression and enhance tumor cell proliferation. Oncotarget 2014, 5, 5686–5699. [Google Scholar] [CrossRef] [Green Version]
- Zarfati, M.; Avivi, I.; Brenner, B.; Katz, T.; Aharon, A. Extracellular vesicles of multiple myeloma cells utilize the proteasome inhibitor mechanism to moderate endothelial angiogenesis. Angiogenesis 2019, 22, 185–196. [Google Scholar] [CrossRef]
- Jordan, K.R.; Hall, J.K.; Schedin, T.; Borakove, M.; Xian, J.J.; Dzieciatkowska, M.; Lyons, T.R.; Schedin, P.; Hansen, K.C.; Borges, V.F. Extracellular vesicles from young women’s breast cancer patients drive increased invasion of non-malignant cells via the Focal Adhesion Kinase pathway: A proteomic appproach. Breast Cancer Res. 2020, 22, 128. [Google Scholar] [CrossRef]
- Park, K.C.; Dharmasivam, M.; Richardson, D.R. The role of extracellular proteases in tumor progression and the development of innovative metal ion chelators that inhibit their activity. Int. J. Mol. Sci. 2020, 21, 6805. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative stress in cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Sijen, T.; Harbison, S. On the identification of body fluids and tissues: A crucial link in the investigation and solution of crime. Genes 2021, 12, 1728. [Google Scholar] [CrossRef] [PubMed]
- Baskoro, B.D.; Nugraha, R.A.; Puspitawati, R.; Redjeki, S. Effect of centrifugation at 7,000 g, 8,000 g, and 9,000 g on the salivary protein profile ≥30 kDa. J. Phys. Conf. Ser. 2017, 884, 012013. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Ben-Nissan, G.; Vimer, S.; Tarnavsky, M.; Sharon, M. Structural mass spectrometry approaches to study the 20S proteasome. Methods Enzymol. 2019, 619, 179–223. [Google Scholar]
- Cvjetkovic, A.; Lotvall, J.; Lasser, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles 2014, 3, 23111. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on strategies and technologies for exosome isolation and purification. Front. Bioengin. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef]
- Benedikter, B.J.; Bouwman, F.G.; Vajen, T.; Heinzmann, A.C.A.; Grauls, G.; Mariman, E.C.; Wouters, E.F.M.; Savelkoul, P.H.; Lopez-Iglesias, C.; Koenen, R.R.; et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci. Rep. 2017, 7, 15297. [Google Scholar] [CrossRef] [PubMed]
- Schipper-Krom, S.; Sanz, A.S.; van Bodegraven, E.J.; Speijer, D.; Florea, B.I.; Ovaa, H.; Reits, E.A. Visualizing proteasome activity and intracellular localization using fluorescent proteins and activity-based probes. Front. Mol. Biosci. 2019, 6, 56. [Google Scholar] [CrossRef] [Green Version]
- Moscovitz, O.; Ben-Nissan, G.; Fainer, I.; Pollack, D.; Mizrachi, L.; Sharon, M. The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015, 6, 6609. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.L.; Njomen, E.; Sjogren, B.; Dexheimer, T.S.; Tepe, J.J. Small molecule enhancement of 20S proteasome activity targets intrinsically disordered proteins. ACS Chem. Biol. 2017, 12, 2240–2247. [Google Scholar] [CrossRef] [Green Version]
- Trader, D.J.; Simanski, S.; Dickson, P.; Kodadek, T. Establishment of a suite of assays that support the discovery of proteasome stimulators. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 892–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Fonts, K.; Matouschek, A. A rapid and versatile method for generating proteins with defined ubiquitin chains. Biochemistry 2016, 55, 1898–1908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharyya, S.; Renn, J.P.; Yu, H.; Marko, J.F.; Matouschek, A. An assay for 26S proteasome activity based on fluorescence anisotropy measurements of dye-labeled protein substrates. Anal. Biochem. 2016, 509, 50–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fricker, L.D. Proteasome inhibitor drugs. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 457–476. [Google Scholar] [CrossRef] [Green Version]
- Gan, J.; Leestemaker, Y.; Sapmaz, A.; Ovaa, H. Highlighting the Proteasome: Using Fluorescence to Visualize Proteasome Activity and Distribution. Front. Mol. Biosci. 2019, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Maher, P. Proteasome assay in cell lysates. Bio. Protoc. 2014, 4, e1028. [Google Scholar] [CrossRef] [Green Version]
- Roelofs, J.; Suppahia, A.; Waite, K.A.; Park, S. Native gel approaches in studying proteasome assembly and chaperones. Methods Mol. Biol. 2018, 1844, 237–260. [Google Scholar]
- Babbitt, S.E.; Kiss, A.; Deffenbaugh, A.E.; Chang, Y.H.; Bailly, E.; Erdjument-Bromage, H.; Tempst, P.; Buranda, T.; Sklar, L.A.; Baumler, J.; et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell 2005, 121, 553–565. [Google Scholar] [CrossRef] [Green Version]
- Solini, A.; Usuelli, V.; Fiorina, P. The dark side of extracellular ATP in kidney diseases. J. Am. Soc. Nephrol. 2015, 26, 1007–1016. [Google Scholar] [CrossRef] [Green Version]
- Trautmann, A. Extracellular ATP in the immune system: More than just a “danger signal”. Sci. Signal 2009, 2, e6. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Xu, C.; Chen, K.; Zhao, Q.; Wang, S.; Yin, Y.; Peng, C.; Ding, Z.; Cong, Y. Cryo-EM of mammalian PA28ab-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28ab. Nat. Commun. 2021, 12, 739. [Google Scholar] [CrossRef] [PubMed]
- Realini, C.; Dubiel, W.; Pratt, G.; Ferrell, K.; Rechsteiner, M. Molecular cloning and expression of a gamma-interferon-inducible activator of the multicatalytic protease. J. Biol. Chem. 1994, 269, 20727–20732. [Google Scholar] [CrossRef]
- Fabre, B.; Lambour, T.; Garrigues, L.; Amalric, F.; Vigneron, N.; Menneteau, T.; Stella, A.; Monsarrat, B.; Van den Eynde, B.; Burlet-Schiltz, O.; et al. Deciphering preferential interactions within supramolecular protein complexes: The proteasome case. Mol. Syst. Biol. 2015, 11, 771. [Google Scholar] [CrossRef] [PubMed]
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28ab reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chem. Biol. 2014, 21, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Castell-Rodríguez, A.P.-Z.G.; Herrera-Enríquez, M.; Jarquín-Yáñez, K.; Medina-Solares, I. Dendritic Cells: Location, Function, and Clinical Implications; Biology of Myelomonocytic Cells; IntechOpen: London, UK, 2017. [Google Scholar]
- Liu, K. Dendritic cells. In Encyclopedia of Cell Biology; Elsevier Public Health Emergency Collection: Amsterdam, The Netherlands, 2016; pp. 741–749. [Google Scholar]
- Embgenbroich, M.; Burgdorf, S. Current concepts of antigen cross-presentation. Front. Immunol. 2018, 9, 1643. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Prassas, I.; Muytjens, C.M.; Diamandis, E.P. Proteomic and peptidomic analysis of human sweat with emphasis on proteolysis. J. Proteom. 2017, 155, 40–48. [Google Scholar] [CrossRef]
- Verma, K.; Verma, M.; Chaphalkar, A.; Chakraborty, K. Recent advances in understanding the role of proteostasis. Fac. Rev. 2021, 10, 72. [Google Scholar] [CrossRef]
- Wyatt, A.R.; Yerbury, J.J.; Ecroyd, H.; Wilson, M.R. Extracellular chaperones and proteostasis. Annu. Rev. Biochem. 2013, 82, 295–322. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Karlsson, M.J.; Hober, A.; Svensson, A.S.; Scheffel, J.; Kotol, D.; Zhong, W.; Tebani, A.; Strandberg, L.; Edfors, F.; et al. The human secretome. Sci. Signal 2019, 12, eaaz0274. [Google Scholar] [CrossRef] [Green Version]
- Tsimokha, A.S.; Artamonova, T.O.; Diakonov, E.E.; Khodorkovskii, M.A.; Tomilin, A.N. Post-translational modifications of extracellular proteasome. Molecules 2020, 25, 3504. [Google Scholar] [CrossRef]
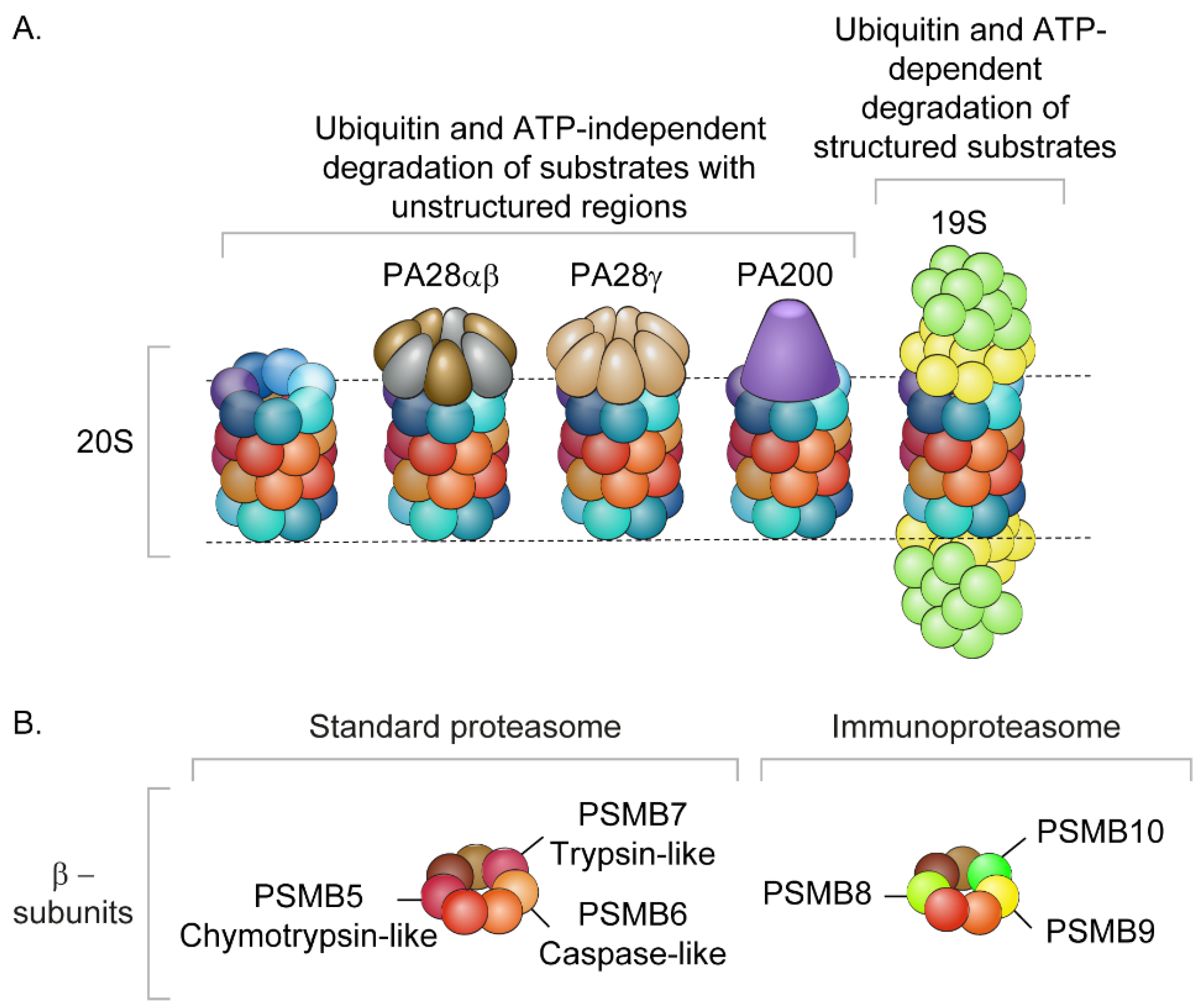
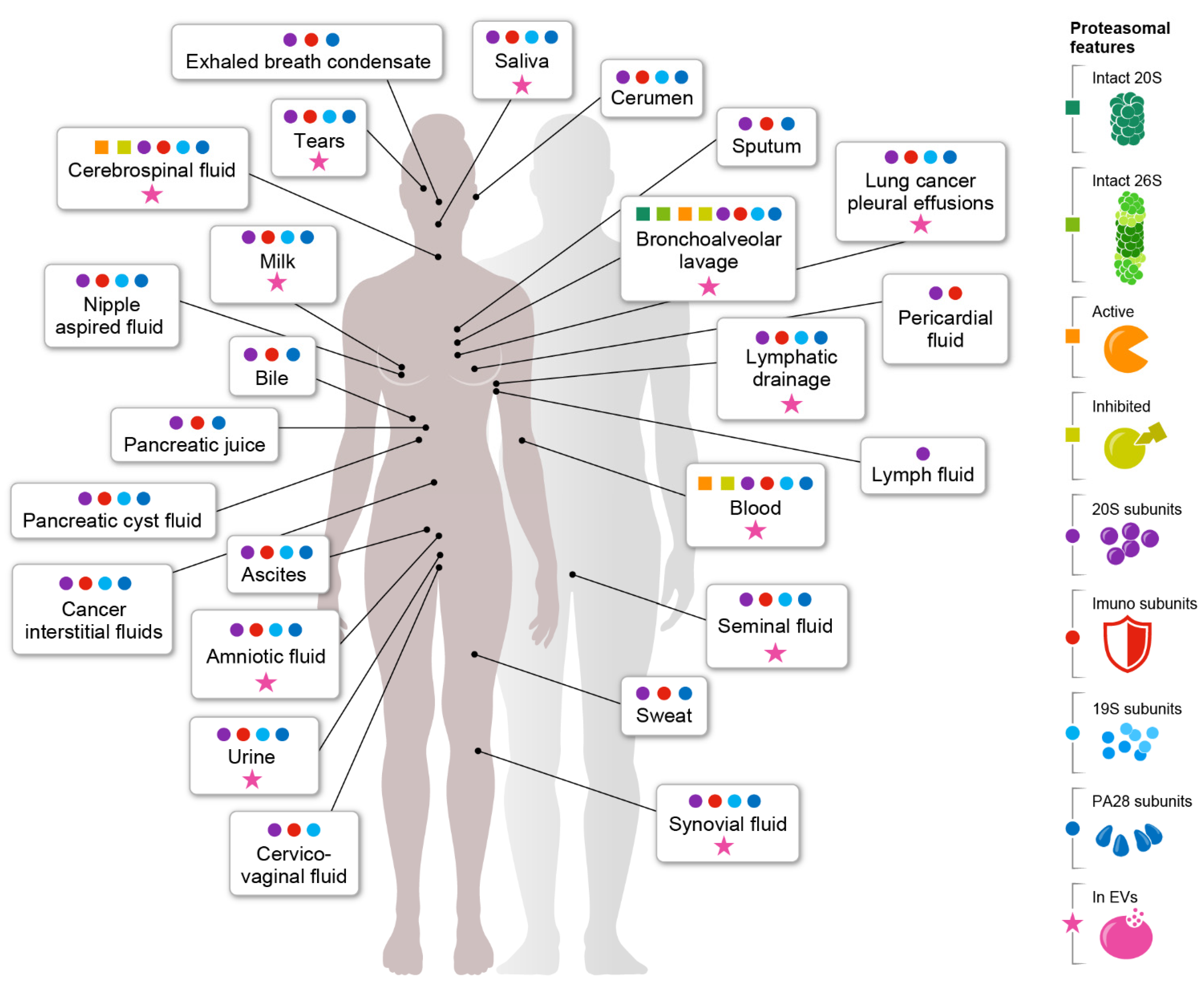

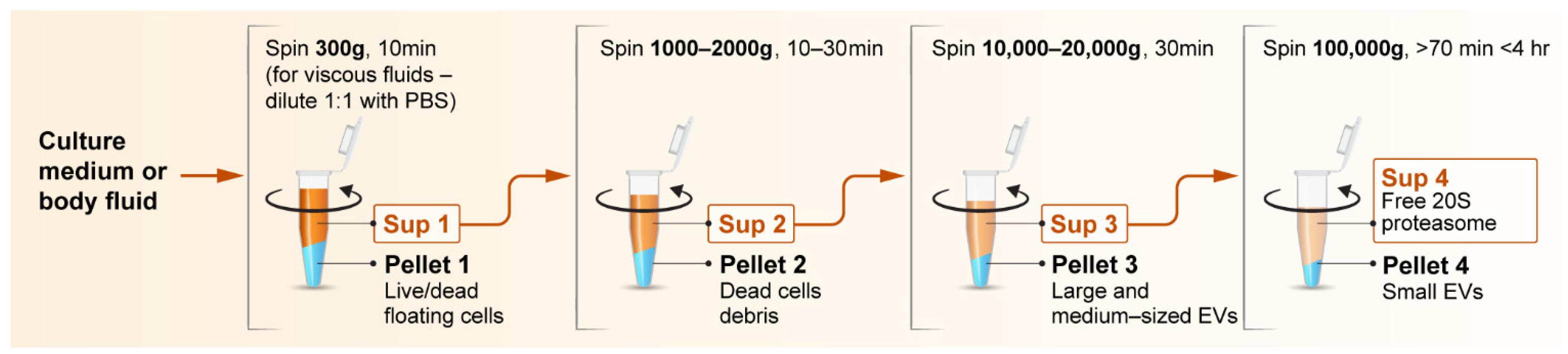
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben-Nissan, G.; Katzir, N.; Füzesi-Levi, M.G.; Sharon, M. Biology of the Extracellular Proteasome. Biomolecules 2022, 12, 619. https://doi.org/10.3390/biom12050619
Ben-Nissan G, Katzir N, Füzesi-Levi MG, Sharon M. Biology of the Extracellular Proteasome. Biomolecules. 2022; 12(5):619. https://doi.org/10.3390/biom12050619
Chicago/Turabian StyleBen-Nissan, Gili, Naama Katzir, Maria Gabriella Füzesi-Levi, and Michal Sharon. 2022. "Biology of the Extracellular Proteasome" Biomolecules 12, no. 5: 619. https://doi.org/10.3390/biom12050619
APA StyleBen-Nissan, G., Katzir, N., Füzesi-Levi, M. G., & Sharon, M. (2022). Biology of the Extracellular Proteasome. Biomolecules, 12(5), 619. https://doi.org/10.3390/biom12050619





