GroEL—A Versatile Chaperone for Engineering and a Plethora of Applications
Abstract
:1. Introduction
2. Direct Use of Unmodified Chaperones
3. Engineered GroEL Forms
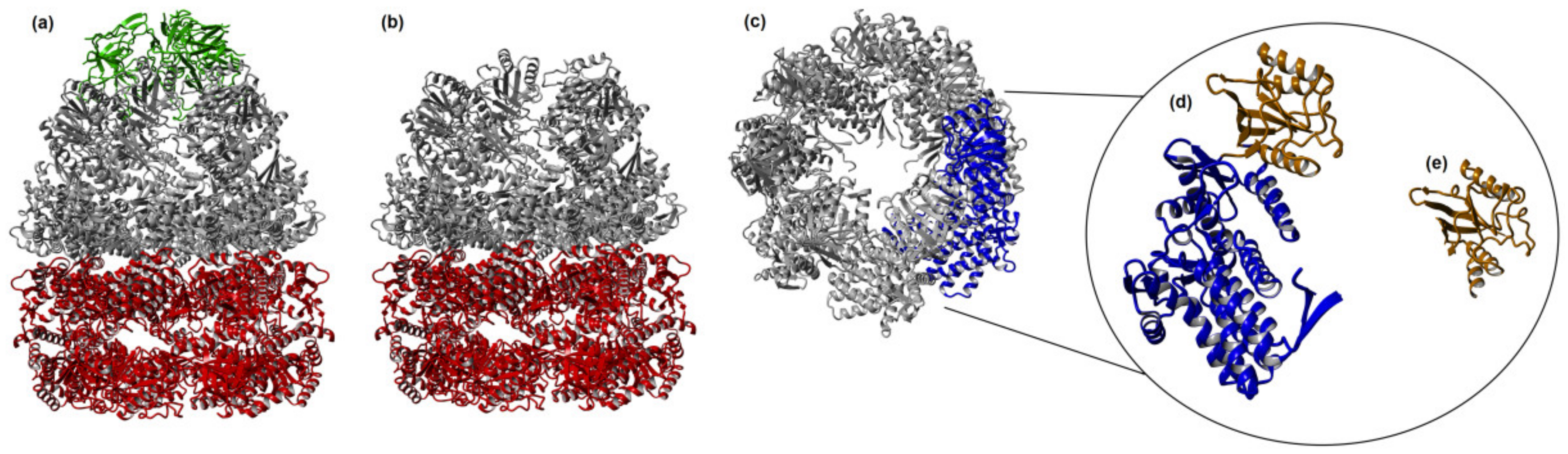
3.1. GroE Complex
3.2. Monomers
3.3. Minichaperone
4. Refolding In Vitro
5. GroEL as a Scaffold
6. GroEL as an Adjuvant and Antigen
7. Beyond GroEL
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Fedorov, A.N. Biosynthetic Protein Folding and Molecular Chaperons. Biochemistry 2022, 87, S128–S145. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Thirumalai, D.; Lorimer, G.H. Chaperonin-Mediated Protein Folding. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 245–269. [Google Scholar] [CrossRef]
- Horwich, A.L.; Farr, G.W.; Fenton, W.A. GroEL-GroES-Mediated Protein Folding. Chem. Rev. 2006, 106, 1917–1930. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone Machines for Protein Folding, Unfolding and Disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [Green Version]
- Horwich, A.L.; Fenton, W.A.; Chapman, E.; Farr, G.W. Two Families of Chaperonin: Physiology and Mechanism. Annu. Rev. Cell Dev. Biol. 2007, 23, 115–145. [Google Scholar] [CrossRef]
- Chaudhuri, T.K.; Verma, V.K.; Maheshwari, A. GroEL Assisted Folding of Large Polypeptide Substrates in Escherichia coli: Present Scenario and Assignments for the Future. Prog. Biophys. Mol. Biol. 2009, 99, 42–50. [Google Scholar] [CrossRef]
- Gray, T.E.; Fersht, A.R. Cooperativity in ATP Hydrolysis by GroEL Is Increased by GroES. FEBS Lett. 1991, 292, 254–258. [Google Scholar] [CrossRef] [Green Version]
- Yifrach, O.; Horovitz, A. Nested Cooperativity in the ATPase Activity of the Oligomeric Chaperonin GroEL. Biochemistry 1995, 34, 5303–5308. [Google Scholar] [CrossRef]
- Weissman, J.S.; Hohl, C.M.; Kovalenko, O.; Kashi, Y.; Chen, S.; Braig, K.; Saibil, H.R.; Fenton, W.A.; Horwich, A.L. Mechanism of GroEL Action: Productive Release of Polypeptide from a Sequestered Position under GroES. Cell 1995, 83, 577–587. [Google Scholar] [CrossRef] [Green Version]
- Chatellier, J.; Hill, F.; Foster, N.W.; Goloubinoff, P.; Fersht, A.R. From Minichaperone to GroEL 3: Properties of an Active Single-Ring Mutant of GroEL. J. Mol. Biol. 2000, 304, 897–910. [Google Scholar] [CrossRef]
- Kovács, E.; Sun, Z.; Liu, H.; Scott, D.J.; Karsisiotis, A.I.; Clarke, A.R.; Burston, S.G.; Lund, P.A. Characterisation of a GroEL Single-Ring Mutant That Supports Growth of Escherichia coli and Has GroES-Dependent ATPase Activity. J. Mol. Biol. 2010, 396, 1271–1283. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding Non-Homologous Proteins by Coupling Deep-Learning Contact Maps with I-TASSER Assembly Simulations. Cell Rep. Methods 2021, 1, 100014. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Tong, Y.; Feng, S.; Xin, Y.; Yang, H.; Zhang, L.; Wang, W.; Chen, W. Enhancement of Soluble Expression of Codon-Optimized Thermomicrobium Roseum Sarcosine Oxidase in Escherichia coli via Chaperone Co-Expression. J. Biotechnol. 2016, 218, 75–84. [Google Scholar] [CrossRef]
- Kim, S.-G.; Kweon, D.-H.; Lee, D.-H.; Park, Y.-C.; Seo, J.-H. Coexpression of Folding Accessory Proteins for Production of Active Cyclodextrin Glycosyltransferase of Bacillus Macerans in Recombinant Escherichia coli. Protein Expr. Purif. 2005, 41, 426–432. [Google Scholar] [CrossRef]
- Veisi, K.; Farajnia, S.; Zarghami, N.; Khorshid, H.R.K.; Samadi, N.; Khosroshahi, S.A.; Jaliani, H.Z. Chaperone-Assisted Soluble Expression of a Humanized Anti-EGFR ScFv Antibody in E. coli. Adv. Pharm. Bull. 2015, 5, 621. [Google Scholar] [CrossRef] [Green Version]
- Temer, B.; Santos, L.V.; Negri, V.A.; Galhardo, J.P.; Magalhães, P.H.M.; José, J.; Marschalk, C.; Corrêa, T.L.R.; Carazzolle, M.F.; Pereira, G.A.G. Conversion of an Inactive Xylose Isomerase into a Functional Enzyme by Co-Expression of GroEL-GroES Chaperonins in Saccharomyces cerevisiae. BMC Biotechnol. 2017, 17, 71. [Google Scholar] [CrossRef]
- Goyal, M.; Chaudhuri, T.K. GroEL–GroES Assisted Folding of Multiple Recombinant Proteins Simultaneously Over-Expressed in Escherichia coli. Int. J. Biochem. Cell Biol. 2015, 64, 277–286. [Google Scholar] [CrossRef]
- De Marco, A.; Deuerling, E.; Mogk, A.; Tomoyasu, T.; Bukau, B. Chaperone-Based Procedure to Increase Yields of Soluble Recombinant Proteins Produced in E. coli. BMC Biotechnol. 2007, 7, 32. [Google Scholar] [CrossRef] [Green Version]
- Yanase, H.; Moriya, K.; Mukai, N.; Kawata, Y.; Okamoto, K.; Kato, N. Effects of GroESL Coexpression on the Folding of Nicotinoprotein Formaldehyde Dismutase from Pseudomonas putida F61. Biosci. Biotechnol. Biochem. 2002, 66, 85–91. [Google Scholar] [CrossRef]
- Xu, Y.; Weng, C.-L.; Narayanan, N.; Hsieh, M.-Y.; Anderson, W.A.; Scharer, J.M.; Moo-Young, M.; Chou, C.P. Chaperone-Mediated Folding and Maturation of the Penicillin Acylase Precursor in the Cytoplasm of Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 6247–6253. [Google Scholar] [CrossRef] [Green Version]
- Vonrhein, C.; Schmidt, U.; Ziegler, G.A.; Schweiger, S.; Hanukoglu, I.; Schulz, G.E. Chaperone-Assisted Expression of Authentic Bovine Adrenodoxin Reductase in Escherichia coli. FEBS Lett. 1999, 443, 167–169. [Google Scholar] [CrossRef] [Green Version]
- Stevens, J.M.; Rao Saroja, N.; Jaouen, M.; Belghazi, M.; Schmitter, J.M.; Mansuy, D.; Artaud, I.; Sari, M.A. Chaperone-Assisted Expression, Purification, and Characterization of Recombinant Nitrile Hydratase NI1 from Comamonas testosteroni. Protein Expr. Purif. 2003, 29, 70–76. [Google Scholar] [CrossRef]
- Nishihara, K.; Kanemori, M.; Kitagawa, M.; Yanagi, H.; Yura, T. Chaperone Coexpression Plasmids: Differential and Synergistic Roles of DnaK-DnaJ-GrpE and GroEL-GroES in Assisting Folding of an Allergen of Japanese Cedar Pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 1998, 64, 1694–1699. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhang, M.; Lv, X.; Fan, J.; Zhang, J.; Sun, J.; Shen, Y. GroEL/ES Mediated the in Vivo Recovery of TRAIL Inclusion Bodies in Escherichia coli. Sci. Rep. 2018, 8, 15766. [Google Scholar] [CrossRef]
- Kyratsous, C.A.; Silverstein, S.J.; DeLong, C.R.; Panagiotidis, C.A. Chaperone-Fusion Expression Plasmid Vectors for Improved Solubility of Recombinant Proteins in Escherichia coli. Gene 2009, 440, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Kyratsous, C.A.; Panagiotidis, C.A. Heat-Shock Protein Fusion Vectors for Improved Expression of Soluble Recombinant Proteins in Escherichia coli. Methods Mol. Biol. 2012, 824, 109–129. [Google Scholar] [CrossRef]
- Chitradevi, S.T.S.; Kaur, G.; Sivaramakrishna, U.; Pratap Singh, D.; Bansal, A. Development of Recombinant Vaccine Candidate Molecule against Shigella Infection. Vaccine 2016, 34, 5376–5383. [Google Scholar] [CrossRef]
- Amatore, D.; Baneyx, F. Insertion Mutagenesis of Escherichia coli GroEL. Biochem. Biophys. Res. Commun. 2003, 302, 246–252. [Google Scholar] [CrossRef]
- Furutani, M. An Engineered Chaperonin Caging a Guest Protein: Structural Insights and Potential as a Protein Expression Tool. Protein Sci. 2005, 14, 341–350. [Google Scholar] [CrossRef]
- Ayling, A.; Baneyx, F. Influence of the GroE Molecular Chaperone Machine on the in Vitro Refolding of Escherichia coli Beta-Galactosidase. Protein Sci. Publ. Protein Soc. 1996, 5, 478–487. [Google Scholar] [CrossRef]
- Yurkova, M.S.; Savvin, O.I.; Zenin, V.A.; Fedorov, A.N. Design and Characterization of a Methionineless Variant of Thermostable Chaperon GroEL from Thermus thermophilus. Appl. Biochem. Microbiol. 2019, 55, 112–116. [Google Scholar] [CrossRef]
- Yurkova, M.S.; Sadykhov, E.G.; Fedorov, A.N. Production of a Toxic Polypeptide as a Fusion inside GroEL Cavity. Sci. Rep. 2020, 10, 21024. [Google Scholar] [CrossRef]
- Powers, J.-P.S.; Rozek, A.; Hancock, R.E.W. Structure-Activity Relationships for the Beta-Hairpin Cationic Antimicrobial Peptide Polyphemusin I. Biochim. Biophys. Acta 2004, 1698, 239–250. [Google Scholar] [CrossRef]
- Tormay, P.; Coates, A.R.M.; Henderson, B. The Intercellular Signaling Activity of the Mycobacterium Tuberculosis Chaperonin 60.1 Protein Resides in the Equatorial Domain. J. Biol. Chem. 2005, 280, 14272–14277. [Google Scholar] [CrossRef] [Green Version]
- Qamra, R.; Mande, S.C. Crystal Structure of the 65-Kilodalton Heat Shock Protein, Chaperonin 60.2, of Mycobacterium tuberculosis. J. Bacteriol. 2004, 186, 8105–8113. [Google Scholar] [CrossRef] [Green Version]
- Qamra, R.; Srinivas, V.; Mande, S.C. Mycobacterium tuberculosis GroEL Homologues Unusually Exist as Lower Oligomers and Retain the Ability to Suppress Aggregation of Substrate Proteins. J. Mol. Biol. 2004, 342, 605–617. [Google Scholar] [CrossRef] [Green Version]
- Safaeian, M.; Porras, C.; Schiffman, M.; Rodriguez, A.C.; Wacholder, S.; Gonzalez, P.; Quint, W.; van Doorn, L.-J.; Sherman, M.E.; Xhenseval, V.; et al. Epidemiological Study of Anti-HPV16/18 Seropositivity and Subsequent Risk of HPV16 and -18 Infections. JNCI J. Natl. Cancer Inst. 2010, 102, 1653–1662. [Google Scholar] [CrossRef]
- McLemore, M. Gardasil®: Introducing the New Human Papillomavirus Vaccine. Clin. J. Oncol. Nurs. 2006, 10, 559–560. [Google Scholar] [CrossRef]
- Holzinger, D.; Wichmann, G.; Baboci, L.; Michel, A.; Höfler, D.; Wiesenfarth, M.; Schroeder, L.; Boscolo-Rizzo, P.; Herold-Mende, C.; Dyckhoff, G.; et al. Sensitivity and Specificity of Antibodies against HPV16 E6 and Other Early Proteins for the Detection of HPV16-Driven Oropharyngeal Squamous Cell Carcinoma: Sensitivity of HPV16 Early Antigen Serology. Int. J. Cancer 2017, 140, 2748–2757. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Zhu, T.; Ye, X.; Yang, L.; Wang, B.; Liang, X.; Lu, L.; Tsao, Y.-P.; Chen, S.-L.; Le, C.; et al. Long-Term Protection Against Human Papillomavirus E7-Positive Tumor by a Single Vaccination of Adeno-Associated Virus Vectors Encoding a Fusion Protein of Inactivated E7 of Human Papillomavirus 16/18 and Heat Shock Protein 70. Hum. Gene Ther. 2009, 21, 109–119. [Google Scholar] [CrossRef]
- Zhou, C.-M.; Zhang, G.-X.; Ma, X.-X. Characterization and Evaluation of the Immune Responses Elicited by a Novel Human Papillomavirus (HPV) Therapeutic Vaccine: HPV 16E7-HBcAg-Hsp65 Fusion Protein. J. Virol. Methods 2014, 197, 1–6. [Google Scholar] [CrossRef]
- Sharapova, O.A.; Yurkova, M.S.; Fedorov, A.N. A Minichaperone-Based Fusion System for Producing Insoluble Proteins in Soluble Stable Forms. Protein Eng. Des. Sel. PEDS 2016, 29, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Zahn, R.; Buckle, A.M.; Perrett, S.; Johnson, C.M.; Corrales, F.J.; Golbik, R.; Fersht, A.R. Chaperone Activity and Structure of Monomeric Polypeptide Binding Domains of GroEL. Proc. Natl. Acad. Sci. USA 1996, 93, 15024–15029. [Google Scholar] [CrossRef] [Green Version]
- Buckle, A.M.; Zahn, R.; Fersht, A.R. A Structural Model for GroEL–polypeptide Recognition. Proc. Natl. Acad. Sci. USA 1997, 94, 3571–3575. [Google Scholar] [CrossRef] [Green Version]
- Chatellier, J.; Hill, F.; Fersht, A.R. From Minichaperone to GroEL 2: Importance of Avidity of the Multisite Ring Structure. J. Mol. Biol. 2000, 304, 883–896. [Google Scholar] [CrossRef]
- Wang, Q.; Buckle, A.M.; Foster, N.W.; Johnson, C.M.; Fersht, A.R. Design of Highly Stable Functional GroEL Minichaperones. Protein Sci. 1999, 8, 2186–2193. [Google Scholar] [CrossRef]
- Hua, Q.; Dementieva, I.S.; Walsh, M.A.; Hallenga, K.; Weiss, M.A.; Joachimiak, A. A Thermophilic Mini-Chaperonin Contains a Conserved Polypeptide-Binding Surface: Combined Crystallographic and NMR Studies of the GroEL Apical Domain with Implications for Substrate Interactions. J. Mol. Biol. 2001, 306, 513–525. [Google Scholar] [CrossRef]
- Yurkova, M.S.; Sharapova, O.A.; Zenin, V.A.; Fedorov, A.N. Versatile Format of Minichaperone-Based Protein Fusion System. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.-H.; Kim, S.-G.; Kweon, D.-H.; Seo, J.-H. Folding Machineries Displayed on a Cation-Exchanger for the Concerted Refolding of Cysteine- or Proline-Rich Proteins. BMC Biotechnol. 2009, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Antonio-Pérez, A.; Ramón-Luing, L.A.; Ortega-López, J. Chromatographic Refolding of Rhodanese and Lysozyme Assisted by the GroEL Apical Domain, DsbA and DsbC Immobilized on Cellulose. J. Chromatogr. A 2012, 1248, 122–129. [Google Scholar] [CrossRef]
- Koculi, E.; Horst, R.; Horwich, A.L.; Wüthrich, K. Nuclear Magnetic Resonance Spectroscopy with the Stringent Substrate Rhodanese Bound to the Single-Ring Variant SR1 of the E. coli Chaperonin GroEL. Protein Sci. Publ. Protein Soc. 2011, 20, 1380–1386. [Google Scholar] [CrossRef] [Green Version]
- Elad, N.; Farr, G.W.; Clare, D.K.; Orlova, E.V.; Horwich, A.L.; Saibil, H.R. Topologies of a Substrate Protein Bound to the Chaperonin GroEL. Mol. Cell 2007, 26, 415–426. [Google Scholar] [CrossRef]
- Horst, R.; Bertelsen, E.B.; Fiaux, J.; Wider, G.; Horwich, A.L.; Wüthrich, K. Direct NMR Observation of a Substrate Protein Bound to the Chaperonin GroEL. Proc. Natl. Acad. Sci. USA 2005, 102, 12748–12753. [Google Scholar] [CrossRef] [Green Version]
- Bracher, A.; Starling-Windhof, A.; Hartl, F.U.; Hayer-Hartl, M. Crystal Structure of a Chaperone-Bound Assembly Intermediate of Form I Rubisco. Nat. Struct. Mol. Biol. 2011, 18, 875–880. [Google Scholar] [CrossRef]
- Chen, L.; Sigler, P.B. The Crystal Structure of a GroEL/Peptide Complex: Plasticity as a Basis for Substrate Diversity. Cell 1999, 99, 757–768. [Google Scholar] [CrossRef] [Green Version]
- O’Neil, P.T.; Machen, A.J.; Deatherage, B.C.; Trecazzi, C.; Tischer, A.; Machha, V.R.; Auton, M.T.; Baldwin, M.R.; White, T.A.; Fisher, M.T. The Chaperonin GroEL: A Versatile Tool for Applied Biotechnology Platforms. Front. Mol. Biosci. 2018, 5, 46. [Google Scholar] [CrossRef] [Green Version]
- Machida, K.; Fujiwara, R.; Tanaka, T.; Sakane, I.; Hongo, K.; Mizobata, T.; Kawata, Y. Gly192 at Hinge 2 Site in the Chaperonin GroEL Plays a Pivotal Role in the Dynamic Apical Domain Movement That Leads to GroES Binding and Efficient Encapsulation of Substrate Proteins. Biochim. Biophys. Acta 2009, 1794, 1344–1354. [Google Scholar] [CrossRef]
- Ojha, B.; Fukui, N.; Hongo, K.; Mizobata, T.; Kawata, Y. Suppression of Amyloid Fibrils Using the GroEL Apical Domain. Sci. Rep. 2016, 6, 31041. [Google Scholar] [CrossRef] [Green Version]
- Fukui, N.; Araki, K.; Hongo, K.; Mizobata, T.; Kawata, Y. Modulating the Effects of the Bacterial Chaperonin GroEL on Fibrillogenic Polypeptides through Modification of Domain Hinge Architecture. J. Biol. Chem. 2016, 291, 25217–25226. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Yagi, H.; Sormanni, P.; Vendruscolo, M.; Makabe, K.; Nakamura, T.; Goto, Y.; Kuwajima, K. Fibrillogenic Propensity of the GroEL Apical Domain: A Janus-faced Minichaperone. FEBS Lett. 2012, 586, 1120–1127. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Yagi, H.; Furutani, Y.; Nakamura, T.; Inaguma, A.; Guo, H.; Kong, Y.; Goto, Y. Self-Assembly of the Chaperonin GroEL Nanocage Induced at Submicellar Detergent. Sci. Rep. 2014, 4, 5614. [Google Scholar] [CrossRef] [Green Version]
- Hayer-Hartl, M.; Bracher, A.; Hartl, F.U. The GroEL–GroES Chaperonin Machine: A Nano-Cage for Protein Folding. Trends Biochem. Sci. 2016, 41, 62–76. [Google Scholar] [CrossRef]
- Fedorov, A.N.; Yurkova, S.M. Molecular Chaperone GroEL—Toward a Nano Toolkit in Protein Engineering, Production and Pharmacy. NanoWorld J. 2018, 4, 8–15. [Google Scholar]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK Stress Response Proteins as Antigens for Bacterial Diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef]
- Milani, A.; Basirnejad, M.; Bolhassani, A. Heat-Shock Proteins in Diagnosis and Treatment: An Overview of Different Biochemical and Immunological Functions. Immunotherapy 2019, 11, 215–239. [Google Scholar] [CrossRef]
- Wang, C.-H.; Hsieh, Y.-H.; Powers, Z.M.; Kao, C.-Y. Defeating Antibiotic-Resistant Bacteria: Exploring Alternative Therapies for a Post-Antibiotic Era. Int. J. Mol. Sci. 2020, 21, 1061. [Google Scholar] [CrossRef] [Green Version]
- Obuchowski, I.; Liberek, K. Small but Mighty: A Functional Look at Bacterial SHSPs. Cell Stress Chaperones 2020, 25, 593–600. [Google Scholar] [CrossRef]
- Bajzert, J.; Gorczykowski, M.; Galli, J.; Stefaniak, T. The Evaluation of Immunogenic Impact of Selected Bacterial, Recombinant Hsp60 Antigens in DBA/2J Mice. Microb. Pathog. 2017, 115, 100–111. [Google Scholar] [CrossRef]
- Khan, M.; Khan, S.; Ali, A.; Akbar, H.; Sayaf, A.M.; Khan, A.; Wei, D.-Q. Immunoinformatics Approaches to Explore Helicobacter Pylori Proteome (Virulence Factors) to Design B and T Cell Multi-Epitope Subunit Vaccine. Sci. Rep. 2019, 9, 13321. [Google Scholar] [CrossRef]
- Verma, S.; Singh, K.; Bansal, A. Multi-Epitope DnaK Peptide Vaccine Accords Protection against Lethal S. typhimurium Challenge: Elicits Both Cell Mediated Immunity and Long-Lasting Serum-Neutralizing Antibody Titers. Pharmacol. Res. 2021, 169, 105652. [Google Scholar] [CrossRef]
- Liu, F.; Tang, X.; Sheng, X.; Xing, J.; Zhan, W. DNA Vaccine Encoding Molecular Chaperone GroEL of Edwardsiella tarda Confers Protective Efficacy against Edwardsiellosis. Mol. Immunol. 2016, 79, 55–65. [Google Scholar] [CrossRef]
- Silvaraj, S.; Yasin, I.S.M.; Karim, M.M.A.; Saad, M.Z. Elucidating the Efficacy of Vaccination against Vibriosis in Lates Calcarifer Using Two Recombinant Protein Vaccines Containing the Outer Membrane Protein K (r-OmpK) of Vibrio Alginolyticus and the DNA Chaperone J (r-DnaJ) of Vibrio harveyi. Vaccines 2020, 8, 660. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Hou, S.; Fu, W.; Cai, J.; Xia, L.; Lu, Y. Comparison of Protective Efficacy between Two DNA Vaccines Encoding DnaK and GroEL against Fish Nocardiosis. Fish Shellfish Immunol. 2019, 95, 128–139. [Google Scholar] [CrossRef]
- Kardani, K.; Sadat, S.M.; Kardani, M.; Bolhassani, A. The next Generation of HCV Vaccines: A Focus on Novel Adjuvant Development. Expert Rev. Vaccines 2021, 20, 839–855. [Google Scholar] [CrossRef]
- Das, J.K.; Xiong, X.; Ren, X.; Yang, J.-M.; Song, J. Heat Shock Proteins in Cancer Immunotherapy. J. Oncol. 2019, 2019, 3267207. [Google Scholar] [CrossRef] [Green Version]
- Calderwood, S.K.; Gong, J.; Stevenson, M.A.; Murshid, A. Cellular and Molecular Chaperone Fusion Vaccines: Targeting Resistant Cancer Cell Populations. Int. J. Hyperth. Off. J. Eur. Soc. Hyperthermic Oncol. N. Am. Hyperth. Group 2013, 29, 376–379. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Li, S.; Zhu, X. The Mechanism of Stimulating and Mobilizing the Immune System Enhancing the Anti-Tumor Immunity. Front. Immunol. 2021, 12, 2243. [Google Scholar] [CrossRef]
- Malys, M.K.; Campbell, L.; Malys, N. Symbiotic and Antibiotic Interactions between Gut Commensal Microbiota and Host Immune System. Medicina 2015, 51, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Som Chaudhury, S.; Das Mukhopadhyay, C. Functional Amyloids: Interrelationship with Other Amyloids and Therapeutic Assessment to Treat Neurodegenerative Diseases. Int. J. Neurosci. 2018, 128, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Convertino, M.; Das, J.; Dokholyan, N. Pharmacological Chaperones: Design and Development of New Therapeutic Strategies for the Treatment of Conformational Diseases. ACS Chem. Biol. 2016, 11, 1471–1489. [Google Scholar] [CrossRef] [PubMed]
- Bongiovanni, M.N.; Aprile, F.A.; Sormanni, P.; Vendruscolo, M. A Rationally Designed Hsp70 Variant Rescues the Aggregation-Associated Toxicity of Human IAPP in Cultured Pancreatic Islet β-Cells. Int. J. Mol. Sci. 2018, 19, 1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newby, G.; Kiriakov, S.; Hallacli, E.; Kayatekin, C.; Tsvetkov, P.; Mancuso, C.; Bonner, J.; Hesse, W.; Chakrabortee, S.; Manogaran, A.; et al. A Genetic Tool to Track Protein Aggregates and Control Prion Inheritance. Cell 2017, 171, 966–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindberg, I.; Shorter, J.; Wiseman, R.L.; Chiti, F.; Dickey, C.A.; McLean, P.J. Chaperones in Neurodegeneration. J. Neurosci. 2015, 35, 13853–13859. [Google Scholar] [CrossRef] [Green Version]
- Mack, K.L.; Shorter, J. Engineering and Evolution of Molecular Chaperones and Protein Disaggregases with Enhanced Activity. Front. Mol. Biosci. 2016, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.D.; Herman, C.; Tipton, K.A.; Gross, C.A.; Weissman, J.S. Directed Evolution of Substrate-Optimized GroEL/S Chaperonins. Cell 2002, 111, 1027–1039. [Google Scholar] [CrossRef] [Green Version]
- Quan, S.; Koldewey, P.; Tapley, T.; Kirsch, N.; Ruane, K.M.; Pfizenmaier, J.; Shi, R.; Hofmann, S.; Foit, L.; Ren, G.; et al. Genetic Selection Designed to Stabilize Proteins Uncovers a Chaperone Called Spy. Nat. Struct. Mol. Biol. 2011, 18, 262–269. [Google Scholar] [CrossRef]
- Quan, S.; Wang, L.; Petrotchenko, E.V.; Makepeace, K.A.; Horowitz, S.; Yang, J.; Zhang, Y.; Borchers, C.H.; Bardwell, J.C. Super Spy Variants Implicate Flexibility in Chaperone Action. eLife 2014, 3, e01584. [Google Scholar] [CrossRef]
- Aponte, R.A.; Zimmermann, S.; Reinstein, J. Directed Evolution of the DnaK Chaperone: Mutations in the Lid Domain Result in Enhanced Chaperone Activity. J. Mol. Biol. 2010, 399, 154–167. [Google Scholar] [CrossRef]
- Jackrel, M.E.; Shorter, J. Engineering Enhanced Protein Disaggregases for Neurodegenerative Disease. Prion 2015, 9, 90–109. [Google Scholar] [CrossRef] [Green Version]
- Jackrel, M.E.; Shorter, J. Potentiated Hsp104 Variants Suppress Toxicity of Diverse Neurodegenerative Disease-Linked Proteins. Dis. Model. Mech. 2014, 7, 1175–1184. [Google Scholar] [CrossRef] [Green Version]
- Julian, M.C.; Lee, C.C.; Tiller, K.E.; Rabia, L.A.; Day, E.K.; Schick, A.J.; Tessier, P.M. Co-Evolution of Affinity and Stability of Grafted Amyloid-Motif Domain Antibodies. Protein Eng. Des. Sel. PEDS 2015, 28, 339–350. [Google Scholar] [CrossRef] [Green Version]
- Aprile, F.A.; Sormanni, P.; Vendruscolo, M. A Rational Design Strategy for the Selective Activity Enhancement of a Molecular Chaperone toward a Target Substrate. Biochemistry 2015, 54, 5103–5112. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yurkova, M.S.; Fedorov, A.N. GroEL—A Versatile Chaperone for Engineering and a Plethora of Applications. Biomolecules 2022, 12, 607. https://doi.org/10.3390/biom12050607
Yurkova MS, Fedorov AN. GroEL—A Versatile Chaperone for Engineering and a Plethora of Applications. Biomolecules. 2022; 12(5):607. https://doi.org/10.3390/biom12050607
Chicago/Turabian StyleYurkova, Maria S., and Alexey N. Fedorov. 2022. "GroEL—A Versatile Chaperone for Engineering and a Plethora of Applications" Biomolecules 12, no. 5: 607. https://doi.org/10.3390/biom12050607
APA StyleYurkova, M. S., & Fedorov, A. N. (2022). GroEL—A Versatile Chaperone for Engineering and a Plethora of Applications. Biomolecules, 12(5), 607. https://doi.org/10.3390/biom12050607




