Multiple Mutations in the Non-Ordered Red Ω-Loop Enhance the Membrane-Permeabilizing and Peroxidase-like Activity of Cytochrome c
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction of the Mutant Genes of CytC
2.2. Expression of the Mutant Genes of CytC, Protein Isolation, and Purification
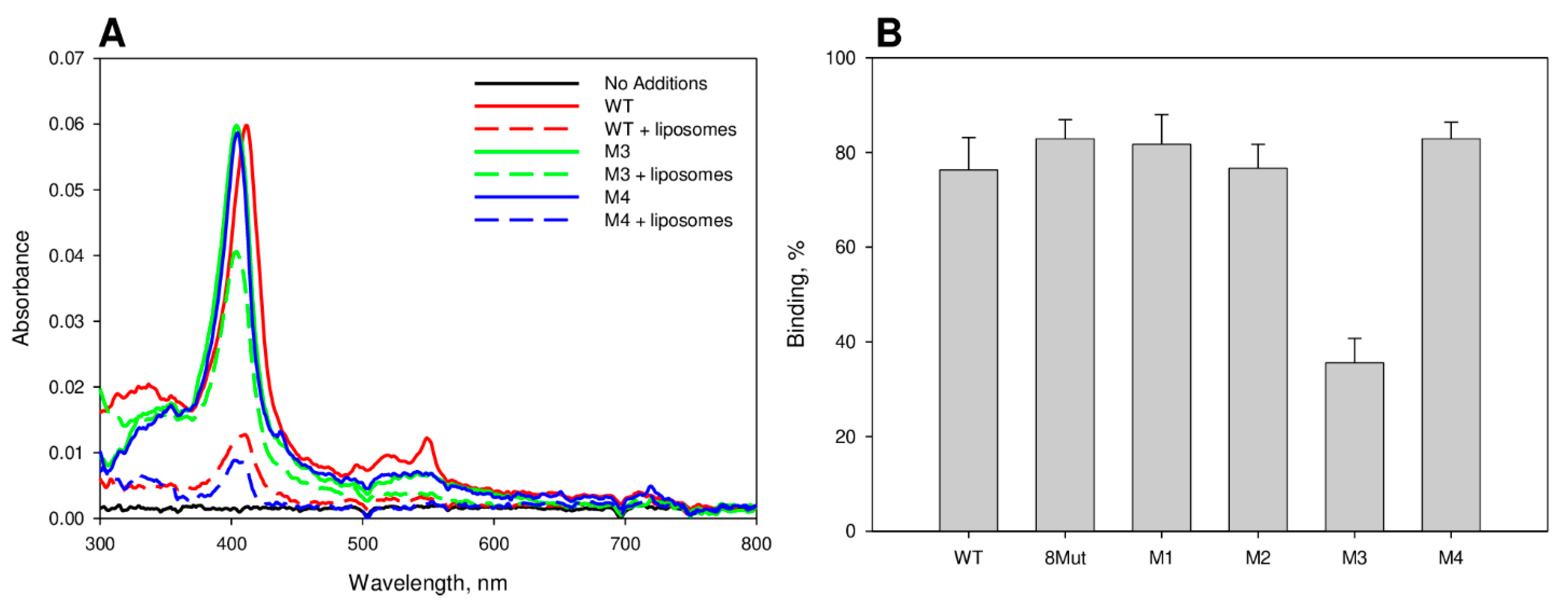
2.3. Centrifugation Binding Assay
2.4. Estimation of the CytC Membrane-Permeabilization Activity
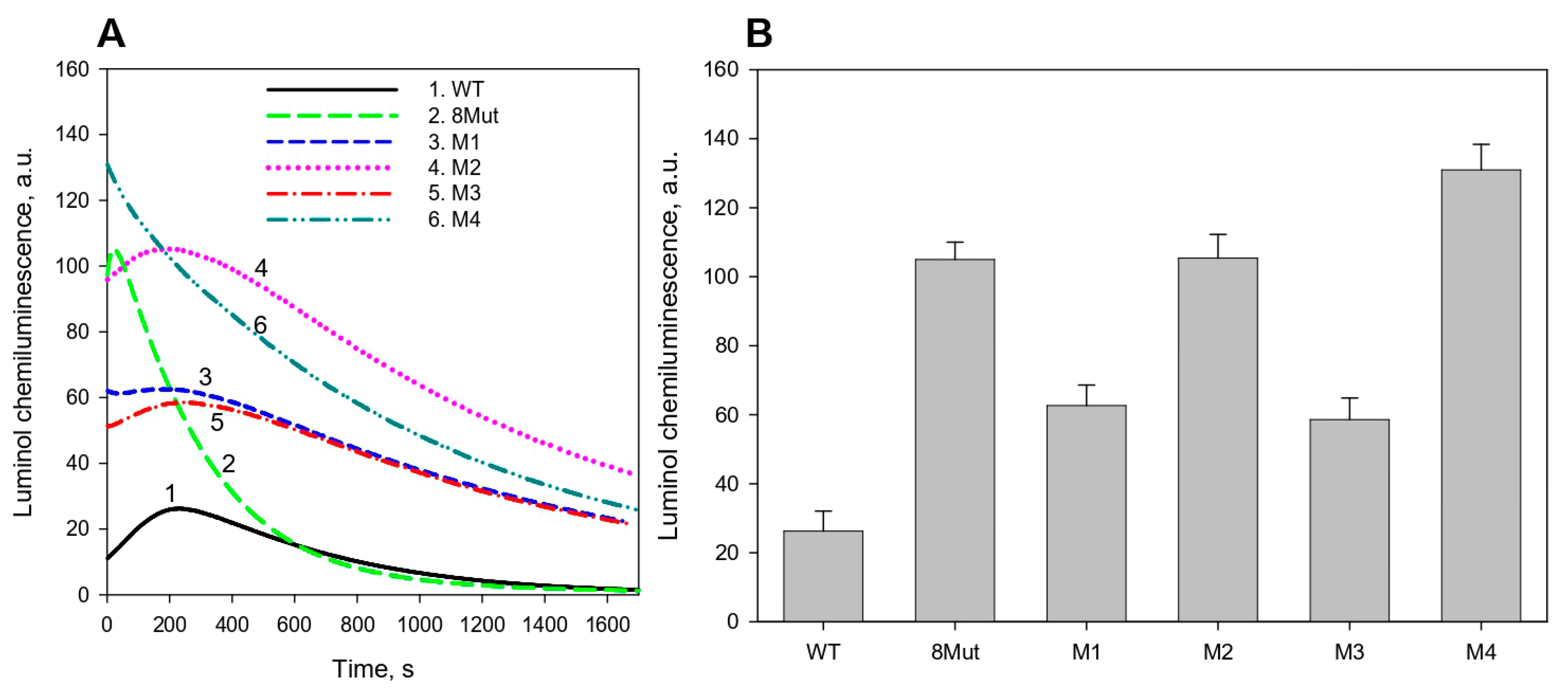
2.5. Peroxidase Activity Assay by Luminol Chemiluminescence
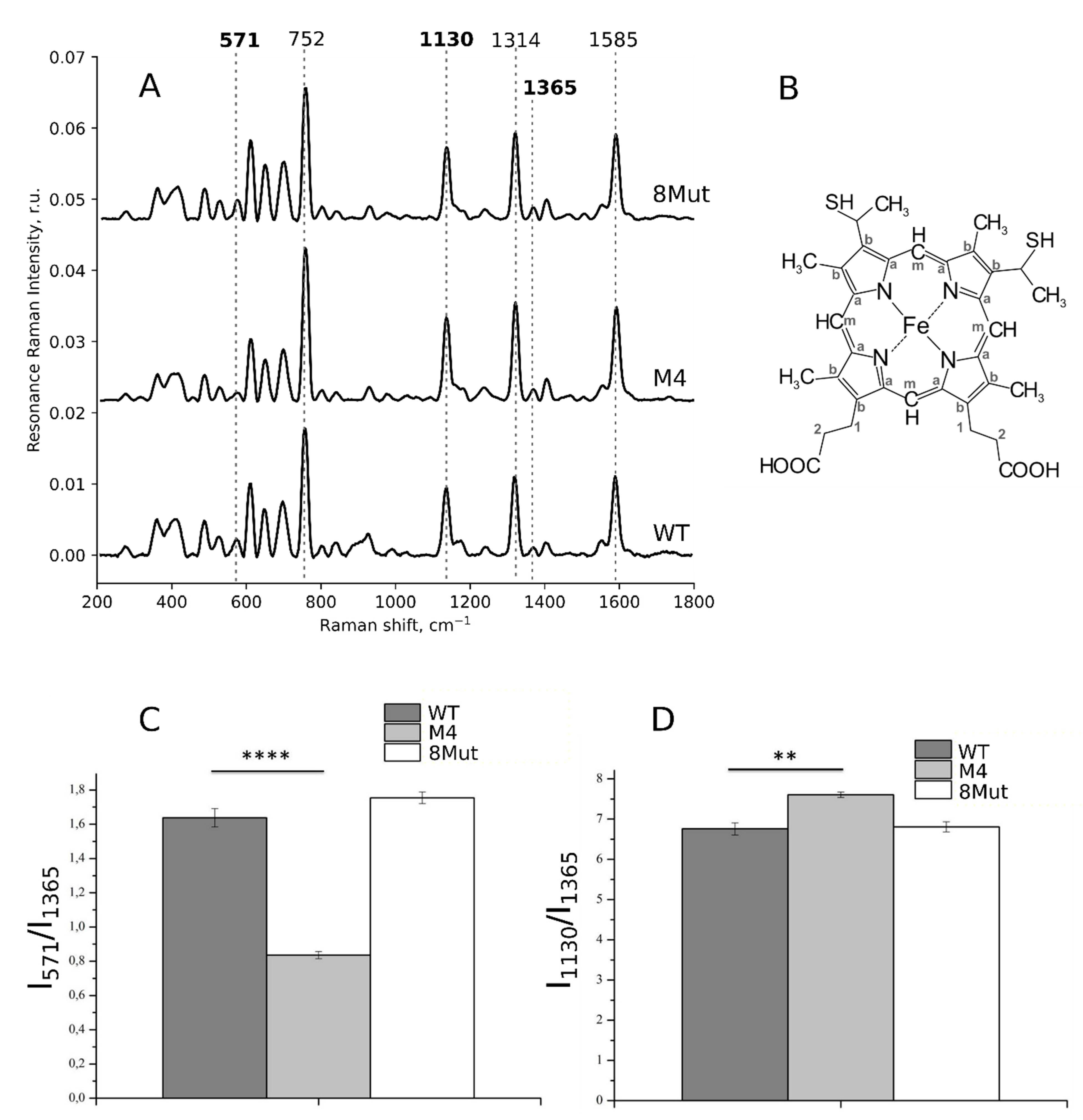
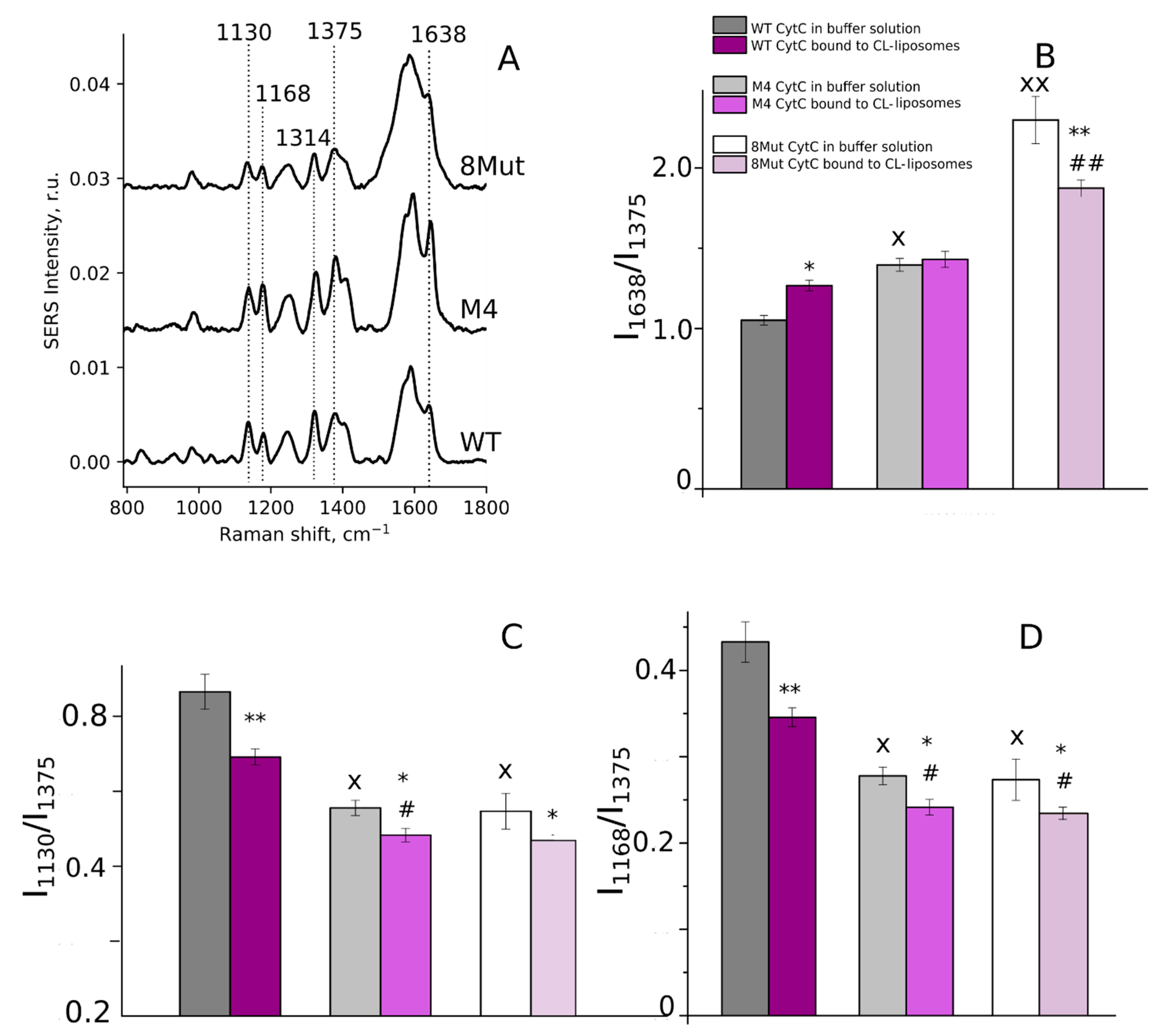
2.6. Resonance Raman and Surface-Enhanced Raman Spectroscopy of CytC
2.7. Liposome Preparation
3. Results and Discussion
3.1. CytC Binding to Cardiolipin-Containing Liposomes and the Peroxidative Permeabilization of Liposomes Induced by CytC Mutants
3.2. Conformation of Heme and Its Local Protein Environment in Cytochrome c Are Affected by the Mutations in the Ω-Loop and in the Universal Binding Site
3.2.1. Conformation of Heme in Unbound Reduced and Oxidized Cytochrome c Molecules
3.2.2. Ordering of Lipids in CL-Containing Liposomes upon CytC Binding
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kulikov, A.V.; Shilov, E.S.; Mufazalov, I.A.; Gogvadze, V.; Nedospasov, S.A.; Zhivotovsky, B. Cytochrome c: The Achilles’ heel in apoptosis. Cell. Mol. Life Sci. 2012, 69, 1787–1797. [Google Scholar] [CrossRef] [PubMed]
- Ow, Y.P.; Green, D.R.; Hao, Z.; Mak, T.W. Cytochrome c: Functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008, 9, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Chipuk, J.E.; Moldoveanu, T.; Llambi, F.; Parsons, M.J.; Green, D.R. The BCL-2 family reunion. Mol. Cell. 2010, 37, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Strasser, A.; McDunn, J.E.; Swanson, P.E. Cell death. N. Engl. J. Med. 2009, 361, 1570–1583. [Google Scholar] [CrossRef] [Green Version]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef]
- Firsov, A.M.; Kotova, E.A.; Korepanova, E.A.; Osipov, A.N.; Antonenko, Y.N. Peroxidative permeabilization of liposomes induced by cytochrome c/cardiolipin complex. Biochim. Biophys. Acta 2015, 1848, 767–774. [Google Scholar] [CrossRef] [Green Version]
- Kagan, V.E.; Tyurin, V.A.; Jiang, J.; Tyurina, Y.Y.; Ritov, V.B.; Amoscato, A.A.; Osipov, A.N.; Belikova, N.A.; Kapralov, A.A.; Kini, V.; et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005, 1, 223–232. [Google Scholar] [CrossRef]
- Kriska, T.; Korytowski, W.; Girotti, A.W. Role of mitochondrial cardiolipin peroxidation in apoptotic photokilling of 5-aminolevulinate-treated tumor cells. Arch. Biochem. Biophys. 2005, 433, 435–446. [Google Scholar] [CrossRef]
- Kagan, V.E.; Bayir, H.A.; Belikova, N.A.; Kapralov, O.; Tyurina, Y.Y.; Tyurin, V.A.; Jiang, J.; Stoyanovsky, D.A.; Wipf, P.; Kochanek, P.M.; et al. Cytochrome c/cardiolipin relations in mitochondria: A kiss of death. Free Radic. Biol. Med. 2009, 46, 1439–1453. [Google Scholar] [CrossRef] [Green Version]
- Muenzner, J.; Pletneva, E.V. Structural transformations of cytochrome c upon interaction with cardiolipin. Chem. Phys. Lipids 2014, 179, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Antonov, V.F.; Puchkov, M.N.; Korepanova, E.A.; Nemchenko, O.Y.; Borodulin, V. Soft perforation of cardiolipin-containing planar lipid bilayer membrane by cytochrome c and H2O2. Eur. Biophys. J. 2014, 43, 469–476. [Google Scholar] [CrossRef]
- Puchkov, M.N.; Vassarais, R.A.; Korepanova, E.A.; Osipov, A.N. Cytochrome c produces pores in cardiolipin-containing planar bilayer lipid membranes in the presence of hydrogen peroxide. Biochim. Biophys. Acta 2013, 1828, 208–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergstrom, C.L.; Beales, P.A.; Lv, Y.; Groves, J.T. Cytochrome c causes pore formation in cardiolipin containing membranes. Proc. Natl. Acad. Sci. USA 2013, 110, 6269–6274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gusev, I.D.; Firsov, A.M.; Chertkova, R.V.; Kotova, E.A.; Dolgikh, D.A.; Kirpichnikov, M.P.; Antonenko, Y.N. Study of interaction of fluorescent cytochrome c with liposomes, mitochondria, and mitoplasts by fluorescence correlation spectroscopy. Russ. J. Bioorg. Chem. 2020, 46, 1162–1171. [Google Scholar] [CrossRef]
- Chertkova, R.V.; Firsov, A.M.; Kotova, E.A.; Gusev, I.D.; Dolgikh, D.A.; Kirpichnikov, M.P.; Antonenko, Y.N. Lysine 72 substitutions differently affect lipid membrane permeabilizing and proapoptotic activities of horse heart cytochrome c. Biochem. Biophys. Res. Commun. 2021, 548, 74–77. [Google Scholar] [CrossRef]
- Chertkova, R.V.; Brazhe, N.A.; Bryantseva, T.V.; Nekrasov, A.N.; Dolgikh, D.A.; Yusipovich, A.I.; Maksimov, G.V.; Rubin, A.B.; Kirpichnikov, M.P. New insight into the mechanism of mitochondrial cytochrome c function. PLoS ONE 2017, 12, e0178280. [Google Scholar] [CrossRef]
- Chertkova, R.V.; Bryantseva, T.V.; Brazhe, N.A.; Kudryashova, K.S.; Revin, V.V.; Nekrasov, A.N.; Yusipovich, A.I.; Brazhe, A.R.; Rubin, A.B.; Dolgikh, D.A.; et al. Amino acid substitutions in the non-ordered Ω-loop 70–85 affect electron transfer function and secondary structure of mitochondrial cytochrome c. Crystals 2021, 11, 973. [Google Scholar] [CrossRef]
- Pepelina, T.Y.; Chertkova, R.V.; Ostroverkhova, T.V.; Dolgikh, D.A.; Kirpichnikov, M.P.; Grivennikova, V.G.; Vinogradov, A.D. Site-directed mutagenesis of cytochrome c: Reactions with respiratory chain components and superoxide radical. Biochemistry (Moscow) 2009, 74, 625–632. [Google Scholar] [CrossRef]
- Dolgikh, D.A.; Latypov, R.F.; Abdullaev, Z.K.; Kolov, V.; Roder, H.; Kirpichnikov, M.P. Expression of mutant horse cytochrome c genes in Escherichia coli. Russ. J. Bioorg. Chem. 1998, 24, 756–759. [Google Scholar]
- Pepelina, T.Y.; Chertkova, R.V.; Dolgikh, D.A.; Kirpichnikov, M.P. The role of individual lysine residues of horse cytochrome c in the formation of reactive complexes with components of the respiratory chain. Russ. J. Bioorg. Chem. 2010, 36, 90–96. [Google Scholar] [CrossRef]
- Chertkova, R.V.; Sharonov, G.V.; Feofanov, A.V.; Bocharova, O.V.; Latypov, R.F.; Chernyak, B.V.; Arseniev, A.S.; Dolgikh, D.A.; Kirpichnikov, M.P. Proapoptotic activity of cytochrome c in living cells: Effect of K72 substitutions and species differences. Mol. Cell. Biochem. 2008, 314, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Radi, R.; Turrens, J.F.; Freeman, B.A. Cytochrome c-catalyzed membrane lipid peroxidation by hydrogen peroxide. Arch. Biochem. Biophys. 1991, 288, 118–125. [Google Scholar] [CrossRef]
- Hanske, J.; Toffey, J.R.; Morenz, A.M.; Bonilla, A.J.; Schiavoni, K.H.; Pletneva, E.V. Conformational properties of cardiolipin-bound cytochrome c. Proc. Natl. Acad. Sci. USA 2012, 109, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muenzner, J.; Toffey, J.R.; Hong, Y.; Pletneva, E.V. Becoming a peroxidase: Cardiolipin-induced unfolding of cytochrome c. J. Phys. Chem. B 2013, 117, 12878–12886. [Google Scholar] [CrossRef] [Green Version]
- Firsov, A.M.; Kotova, E.A.; Antonenko, Y.N. Calcein leakage as a robust assay for cytochrome c/H2O2-mediated liposome permeabilization. Anal. Biochem. 2018, 552, 19–23. [Google Scholar] [CrossRef]
- Radi, R.; Thomson, L.; Rubbo, H.; Prodanov, E. Cytochrome c-catalyzed oxidation of organic molecules by hydrogen peroxide. Arch. Biochem. Biophys. 1991, 288, 112–117. [Google Scholar] [CrossRef]
- Vladimirov, Y.A.; Proskurnina, E.V.; Izmailov, D.Y.; Novikov, A.A.; Brusnichkin, A.V.; Osipov, A.N.; Kagan, V.E. Mechanism of activation of cytochrome c peroxidase activity by cardiolipin. Biochemistry (Moscow) 2006, 71, 989–997. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Evlyukhin, A.B.; Goodilin, E.A.; Semenova, A.A.; Novikov, S.M.; Bozhevolnyi, S.I.; Chichkov, B.N.; Sarycheva, A.S.; Baizhumanov, A.A.; Nikelshparg, E.I.; et al. Probing cytochrome c in living mitochondria with surface-enhanced raman spectroscopy. Sci. Rep. 2015, 5, 13793. [Google Scholar] [CrossRef] [Green Version]
- Lei, H.; Nold, S.M.; Motta, L.J.; Bowler, B.E. Effect of V83G and I81A substitutions to human cytochrome c on acid unfolding and peroxidase activity below a neutral pH. Biochemistry 2019, 58, 2921–2933. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, X.-C.; Li, L.; Gao, S.Q.; Wen, G.-B.; Lin, Y.-W. Naturally occurring I81N mutation in human cytochrome c regulates both inherent peroxidase activity and interactions with neuroglobin. ACS Omega 2022, 7, 11510–11518. [Google Scholar] [CrossRef]
- Samsri, S.; Prasertsuk, P.; Nutho, B.; Pornsuwan, S. Molecular insights on the conformational dynamics of a P76C mutant of human cytochrome c and the enhancement on its peroxidase activity. Arch. Biochem. Biophys. 2022, 716, 109112. [Google Scholar] [CrossRef] [PubMed]
- Lalli, D.; Rosa, C.; Allegrozzi, M.; Turano, P. Distal unfolding of ferricytochrome c induced by the F82K mutation. Int. J. Mol. Sci. 2020, 21, 2134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Benabbas, A.; Zeng, W.; Kleingardner, J.G.; Bren, K.L.; Champion, P.M. Investigations of heme distortion, low-frequency vibrational excitations, and electron transfer in cytochrome c. Proc. Natl. Acad. Sci. USA 2014, 111, 6570–6575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, M.; Harada, Y.; Yamaoka, Y.; Fujita, K.; Yaku, H.; Takamatsu, T. Label-free biochemical imaging of heart tissue with high-speed spontaneous Raman microscopy. Biochem Biophys Res Commun. 2009, 382, 370–374. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Treiman, M.; Brazhe, A.R.; Find, N.L.; Maksimov, G.V.; Sosnovtseva, O.V. Mapping of redox state of mitochondrial cytochromes in live cardiomyocytes using Raman microspectroscopy. PLoS ONE 2012, 7, 41990. [Google Scholar] [CrossRef]
- Nikelshparg, E.I.; Grivennikova, V.G.; Baizhumanov, A.A.; Semenova, A.A.; Sosnovtseva, V.; Goodilin, E.A.; Maksimov, G.V.; Brazhe, N.A. Probing lipids in biological membranes using SERS. Mendeleev Commun. 2019, 29, 635–637. [Google Scholar] [CrossRef]
- Sarycheva, A.S.; Brazhe, N.A.; Baizhumanov, A.A.; Nikelshparg, E.I.; Semenova, A.A.; Garshev, A.V.; Baranchikov, A.E.; Ivanov, V.K.; Maksimov, G.V.; Sosnovtseva, O.V.; et al. New nanocomposites for SERS studies of living cells and mitochondria. J. Mater. Chem. B 2016, 3, 539–546. [Google Scholar] [CrossRef]
- Orendorff, C.J.; Ducey, M.W., Jr.; Pemberton, J.E. Quantitative correlation of Raman spectral indicators in determining conformational order in alkyl chains. J. Phys. Chem. A 2002, 106, 6991–6998. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2014, 46, 4–20. [Google Scholar] [CrossRef]

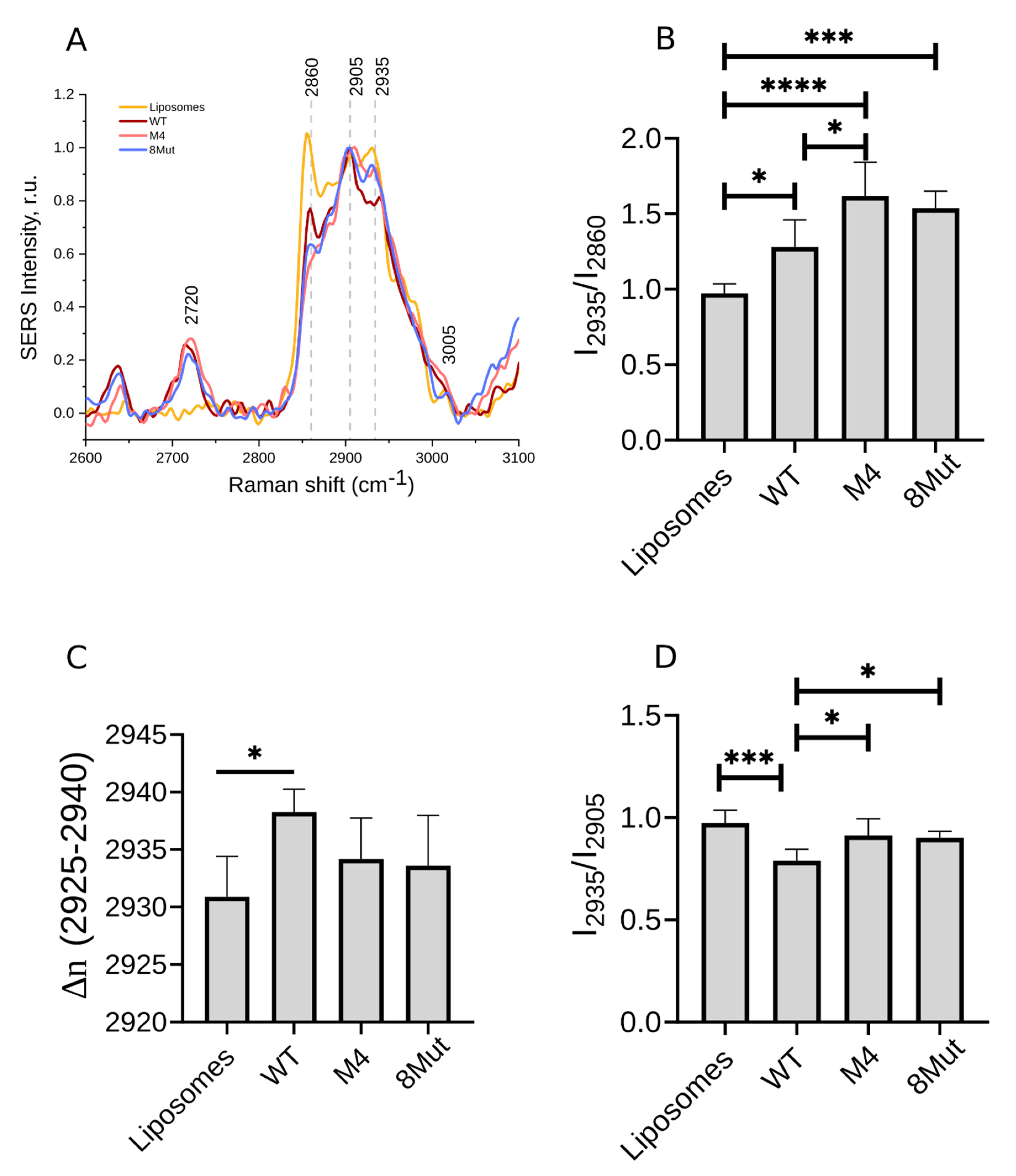
| Position of a Peak Maximum in SERS Spectra CL-Containing Liposomes with Bound CytC | Raman Peak Position in Spectra of CL-Containing Liposomes without CytC | Description | Comments |
|---|---|---|---|
| 2627 | - | ν(C–H) | |
| 2720 | - | ν(C–H) | |
| 2860 | 2854 | νs(-CH2-) | |
| 2905 | 2906 | νas(-CH2-) | |
| 2925–2940 | 2930 | νs(CH3) | |
| 3005 | 3014 | н(=CH-) | Correlates with the number of double C=C bonds in CL [39] and is affected by CytC-specific interaction with CL |
| Parameter, Calculated from Raman Spectra | Meaning | Comments |
|---|---|---|
| I2935/I2860 | The relative input of symmetric vibrations of C-H bonds in amino acids’ CH3-groups compared to the symmetric vibrations of -CH2- backbone bonds | The ratio increase corresponds to the easier -CH3 bond vibrations due to the decrease in the ordering of the membrane lipid tails and due to the appearance of disturbances in the membrane lipid phase |
| I2935/I2905 | The relative input of the symmetric vibrations of C-H bonds in amino acids’ CH3-groups compared to the asymmetric vibrations of -CH2- backbone bonds | The ratio gives similar information as the ratio I2935/I2860. The disordering of lipid tails affects this ratio to a greater degree than the ratio I2935/I2860 |
| ν(2925–2940) | The peak corresponds to the symmetric vibrations of C-H bonds in amino acids’ CH3-groups | Peak maximum shifts under changes in the lipid ordering and membrane microfluidity in the CytC surrounding |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chertkova, R.V.; Firsov, A.M.; Brazhe, N.A.; Nikelshparg, E.I.; Bochkova, Z.V.; Bryantseva, T.V.; Semenova, M.A.; Baizhumanov, A.A.; Kotova, E.A.; Kirpichnikov, M.P.; et al. Multiple Mutations in the Non-Ordered Red Ω-Loop Enhance the Membrane-Permeabilizing and Peroxidase-like Activity of Cytochrome c. Biomolecules 2022, 12, 665. https://doi.org/10.3390/biom12050665
Chertkova RV, Firsov AM, Brazhe NA, Nikelshparg EI, Bochkova ZV, Bryantseva TV, Semenova MA, Baizhumanov AA, Kotova EA, Kirpichnikov MP, et al. Multiple Mutations in the Non-Ordered Red Ω-Loop Enhance the Membrane-Permeabilizing and Peroxidase-like Activity of Cytochrome c. Biomolecules. 2022; 12(5):665. https://doi.org/10.3390/biom12050665
Chicago/Turabian StyleChertkova, Rita V., Alexander M. Firsov, Nadezda A. Brazhe, Evelina I. Nikelshparg, Zhanna V. Bochkova, Tatyana V. Bryantseva, Marina A. Semenova, Adil A. Baizhumanov, Elena A. Kotova, Mikhail P. Kirpichnikov, and et al. 2022. "Multiple Mutations in the Non-Ordered Red Ω-Loop Enhance the Membrane-Permeabilizing and Peroxidase-like Activity of Cytochrome c" Biomolecules 12, no. 5: 665. https://doi.org/10.3390/biom12050665
APA StyleChertkova, R. V., Firsov, A. M., Brazhe, N. A., Nikelshparg, E. I., Bochkova, Z. V., Bryantseva, T. V., Semenova, M. A., Baizhumanov, A. A., Kotova, E. A., Kirpichnikov, M. P., Maksimov, G. V., Antonenko, Y. N., & Dolgikh, D. A. (2022). Multiple Mutations in the Non-Ordered Red Ω-Loop Enhance the Membrane-Permeabilizing and Peroxidase-like Activity of Cytochrome c. Biomolecules, 12(5), 665. https://doi.org/10.3390/biom12050665









