Differentiating Inhibitors of Closely Related Protein Kinases with Single- or Multi-Target Activity via Explainable Machine Learning and Feature Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Compounds and Activity Data
2.2. Target Selection
2.3. Molecular Representation
2.4. Machine Learning
2.5. SHAP Analysis and Feature Extraction
- (i)
- For each correctly predicted MT-CPD, the top-ranked N features with the highest SHAP values were pre-selected and these features were pooled.
- (ii)
- The pool of the top-ranked N features was re-ranked by the feature frequency of occurrence in correctly predicted MT-CPDs, and the top M most frequent features were selected.
3. Results and Discussion
3.1. Study Design
3.2. Systematic Analysis of Kinase Triplets
3.3. Compound Classification
3.4. Representation Features Determining Predictions
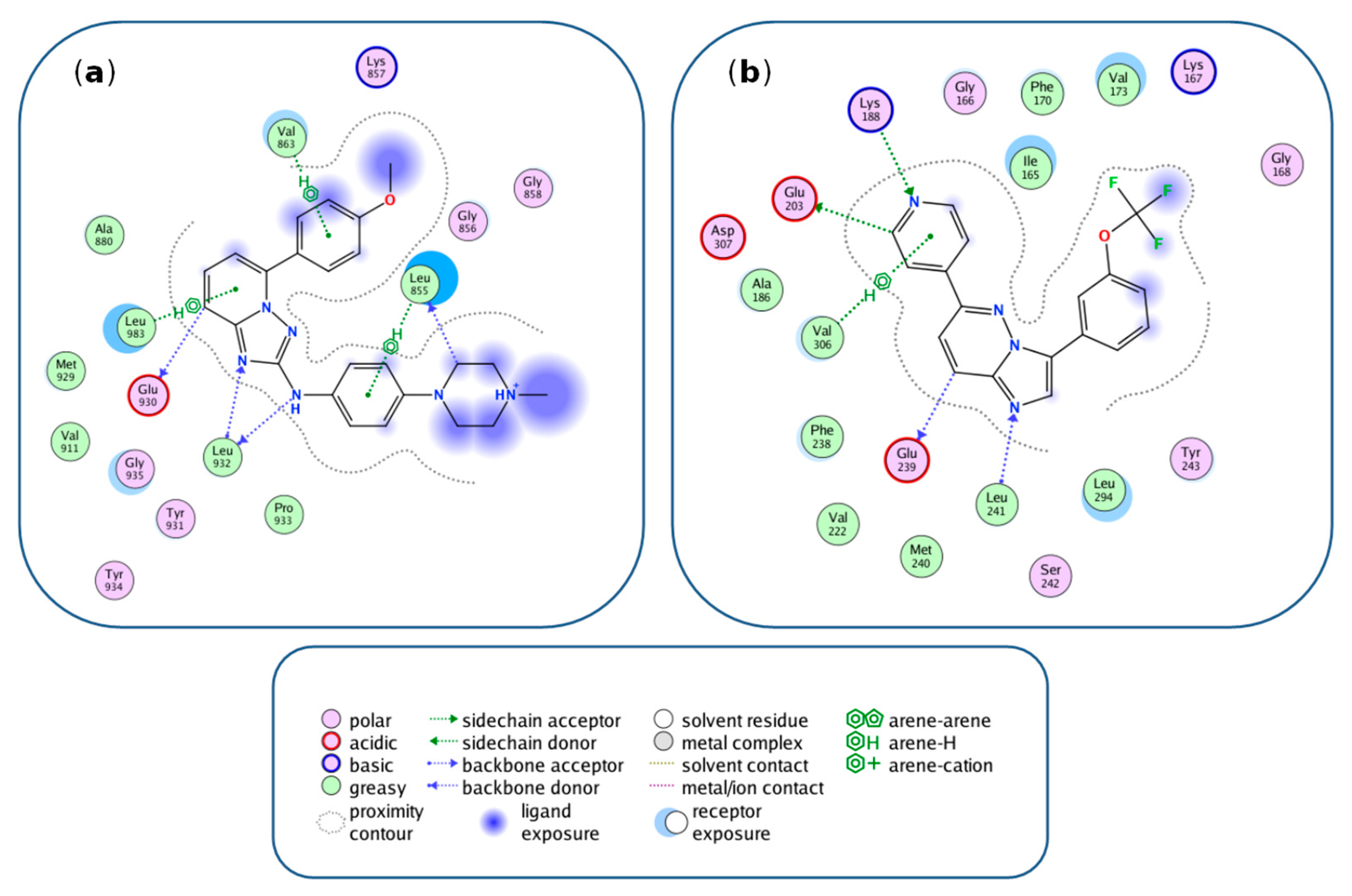
3.5. Feature Mapping and Rationalization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Overington, J.P.; Al-Lazikani, B.; Hopkins, A.L. How Many Drug Targets Are There? Nat. Rev. Drug Discov. 2006, 5, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, M.L.; Cavalli, A. Multitarget Drug Discovery and Polypharmacology. ChemMedChem 2006, 11, 1190–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Pei, J.; Lai, L. Computational Multitarget Drug Design. J. Chem. Inf. Model. 2017, 57, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Proschak, E.; Stark, H.; Merk, D. Polypharmacology by Design: A Medicinal Chemist’s Perspective on Multitargeting Compounds. J. Med. Chem. 2019, 62, 420–444. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, X.; He, S.; Jiang, H.; Feng, F.; Liu, W.; Qu, W.; Sun, H. Rational Design of Multitarget-Directed Ligands: Strategies and Emerging Paradigms. J. Med. Chem. 2019, 62, 8881–8914. [Google Scholar] [CrossRef]
- Rastelli, G.; Pinzi, L. Computational Polypharmacology Comes of Age. Front. Pharmacol. 2015, 6, 157. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Bajorath, J. Entering the ‘Big Data’ Era in medicinal Chemistry: Molecular Promiscuity Analysis Revisited. Future Sci. OA 2017, 3, FSO179. [Google Scholar] [CrossRef] [Green Version]
- Chaudhari, R.; Fong, L.W.; Tan, Z.; Huang, B.; Zhang, S. An Up-To-Date Overview of Computational Polypharmacology in Modern Drug Discovery. Expert Opin. Drug Discov. 2020, 15, 1025–1044. [Google Scholar] [CrossRef]
- Miljković, F.; Bajorath, J. Data Structures for Computational Compound Promiscuity Analysis and Exemplary Applications to Inhibitors of the Human Kinome. J. Comp.-Aided Mol. Des. 2020, 34, 1–10. [Google Scholar] [CrossRef]
- Fabian, M.A.; Biggs, W.H., 3rd; Treiber, D.K.; Atteridge, C.E.; Azimioara, M.D.; Benedetti, M.G.; Carter, T.A.; Ciceri, P.; Edeen, P.T.; Floyd, M.; et al. A Small Molecule-Kinase Interaction Map for Clinical Kinase Inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [Google Scholar] [CrossRef]
- Karaman, M.W.; Herrgard, S.; Treiber, D.K.; Gallant, P.; Atteridge, C.E.; Campbell, B.T.; Chan, K.W.; Ciceri, P.; Davis, M.I.; Edeen, P.T.; et al. A Quantitative Analysis of Kinase Inhibitor Selectivity. Nat. Biotechnol. 2008, 26, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Klaeger, S.; Heinzlmeir, S.; Wilhelm, M.; Polzer, H.; Vick, B.; Koenig, P.A.; Reinecke, M.; Ruprecht, B.; Petzoldt, S.; Meng, C.; et al. The Target Landscape of Clinical Kinase Inhibitors. Science 2017, 358, eaan4368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorne, N.; Auld, D.S.; Inglese, J. Apparent Activity in High-Throughput Screening: Origins of Compound-Dependent Assay Interference. Curr. Opin. Chem. Biol. 2010, 14, 315–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baell, J.B.; Walters, M.A. Chemistry: Chemical Con Artists Foil Drug Discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Bajorath, J. Activity Artifacts in Drug Discovery and Different Facets of Compound Promiscuity. F1000Research 2014, 3, 233. [Google Scholar] [CrossRef] [Green Version]
- Irwin, J.J.; Duan, D.; Torosyan, H.; Doak, A.K.; Ziebart, K.T.; Sterling, T.; Tumanian, G.; Shoichet, B.K. An Aggregation Advisor for Ligand Discovery. J. Med. Chem. 2015, 58, 1712–1722. [Google Scholar] [CrossRef] [Green Version]
- Gilberg, E.; Bajorath, J. Recent Progress in Structure-Based Evaluation of Compound Promiscuity. ACS Omega 2019, 4, 2758–2765. [Google Scholar] [CrossRef] [Green Version]
- Feldmann, C.; Yonchev, D.; Stumpfe, D.; Bajorath, J. Systematic Data Analysis and Diagnostic Machine Learning Reveal differences between Compounds with Single- and Multitarget Activity. Mol. Pharm. 2020, 17, 4652–4666. [Google Scholar] [CrossRef]
- Feldmann, C.; Yonchev, D.; Bajorath, J. Analysis of Biological Screening Compounds with Single- or Multi-Target Activity via Diagnostic Machine Learning. Biomolecules 2020, 10, 1605. [Google Scholar] [CrossRef]
- Feldmann, C.; Bajorath, J. Machine Learning Reveals that Structural Features Distinguishing Promiscuous and Non-Promiscuous Compounds Depend on Target Combinations. Sci. Rep. 2021, 11, 7863. [Google Scholar] [CrossRef]
- Castelvecchi, D. Can We Open the Black Box of AI? Nature 2015, 538, 20–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dybowski, R. Interpretable Machine Learning as a Tool for Scientific Discovery in Chemistry. New J. Chem. 2020, 44, 20914–20920. [Google Scholar] [CrossRef]
- Feng, J.; Lansford, J.L.; Katsoulakis, M.A.; Vlachos, D.G. Explainable and Trustworthy Artificial Intelligence for Correctable Modeling in Chemical Sciences. Sci. Adv. 2020, 6, eabc3204. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pérez, R.; Bajorath, J. Chemistry-Centric Explanation of Machine Learning Models. Artif. Intell. Life Sci. 2021, 1, 100009. [Google Scholar] [CrossRef]
- Stepin, I.; Alonso, J.M.; Catala, A.; Pereira-Fariña, M. A Survey of Contrastive and Counterfactual Explanation Generation Methods for Explainable Artificial Intelligence. IEEE Access 2021, 9, 11974–12001. [Google Scholar] [CrossRef]
- Shapley, L.S. A Value for N-Person Games. In Contributions to the Theory of Games; Kuhn, H.W., Tucker, A.W., Eds.; Annals of Mathematical Studies; Princeton University Press: Princeton, NJ, USA, 1953; pp. 307–317. [Google Scholar]
- Young, H.P. Monotonic Solutions of Cooperative Games. Int. J. Game Theory 1985, 14, 65–72. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S. A Unified Approach to Interpreting Model Predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Rodríguez-Pérez, R.; Bajorath, J. Interpretation of Compound Activity Predictions from Complex Machine Learning Models Using Local Approximations and Shapley Values. J. Med. Chem. 2019, 63, 8761–8777. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Feldmann, C.; Philipps, M.; Bajorath, J. Explainable Machine Learning Predictions of Dual-Target Compounds Reveal Characteristic Structural Features. Sci. Rep. 2021, 11, 21594. [Google Scholar] [CrossRef]
- Ferguson, F.M.; Gray, N.S. Kinase Inhibitors: The Road Ahead. Nat. Rev. Drug Discov. 2018, 17, 353–376. [Google Scholar] [CrossRef]
- Knight, Z.A.; Lin, H.; Shokat, K.M. Targeting the Cancer Kinome through Polypharmacology. Nat. Rev. Cancer 2010, 10, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Gavrin, L.K.; Saiah, E. Approaches to Discover Non-ATP Site Kinase Inhibitors. Med. Chem. Commun. 2013, 4, 41–51. [Google Scholar] [CrossRef]
- Hu, Y.; Furtmann, N.; Bajorath, J. Current Compound Coverage of the Kinome. J. Med. Chem. 2015, 58, 30–40. [Google Scholar] [CrossRef]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Krüger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL Bioactivity Database: An Update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef] [Green Version]
- RDKit: Cheminformatics and Machine Learning Software (2013). Available online: http://www.rdkit.org (accessed on 1 October 2021).
- Bruns, R.F.; Watson, I.A. Rules for Identifying Potentially Reactive or Promiscuous Compounds. J. Med. Chem. 2012, 55, 9763–9772. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lemaître, G.; Nogueira, F.; Aridas, C.K. Imbalanced-learn: A Python Toolbox to Tackle the Curse of Imbalanced Data Sets in machine Learning. J. Mach. Learn. Res. 2017, 18, 1–5. [Google Scholar]
- Brodersen, K.H.; Ong, C.S.; Stephan, K.E.; Buhmann, J.M. The Balanced Accuracy and Its Posterior Distribution. In Proceedings of the 20th International Conference on Pattern Recognition (ICPR), Istanbul, Turkey, 23–26 August 2010; pp. 3121–3124. [Google Scholar]
- Van Rijsbergen, C.J. Information Retrieval, 2nd ed.; Butterworth-Heinemann: Oxford, UK, 1979. [Google Scholar]
- Matthews, B. Comparison of the Predicted and Observed Secondary Structure of T4 phage Lysozyme. Biochim. Biophys. Acta 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Curry, M.A.; Dorsey, B.D.; Dugan, B.D.; Gingrich, D.E.; Mesaros, E.F.; Milkiewicz, K.L. Preparation and Uses of 1,2,4-Triazolo [1,5a] Pyridine. Derivatives. Patent US-8501936-B2, 2013. [Google Scholar]
- Bendjeddou, L.Z.; Loaëc, N.; Villiers, B.; Prina, E.; Späth, G.F.; Galons, H.; Meijer, L.; Oumata, N. Exploration of the Imidazo[1,2-b]Pyridazine Scaffold as a Protein Kinase Inhibitor. Eur. J. Med. Chem. 2017, 125, 696–709. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?” Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
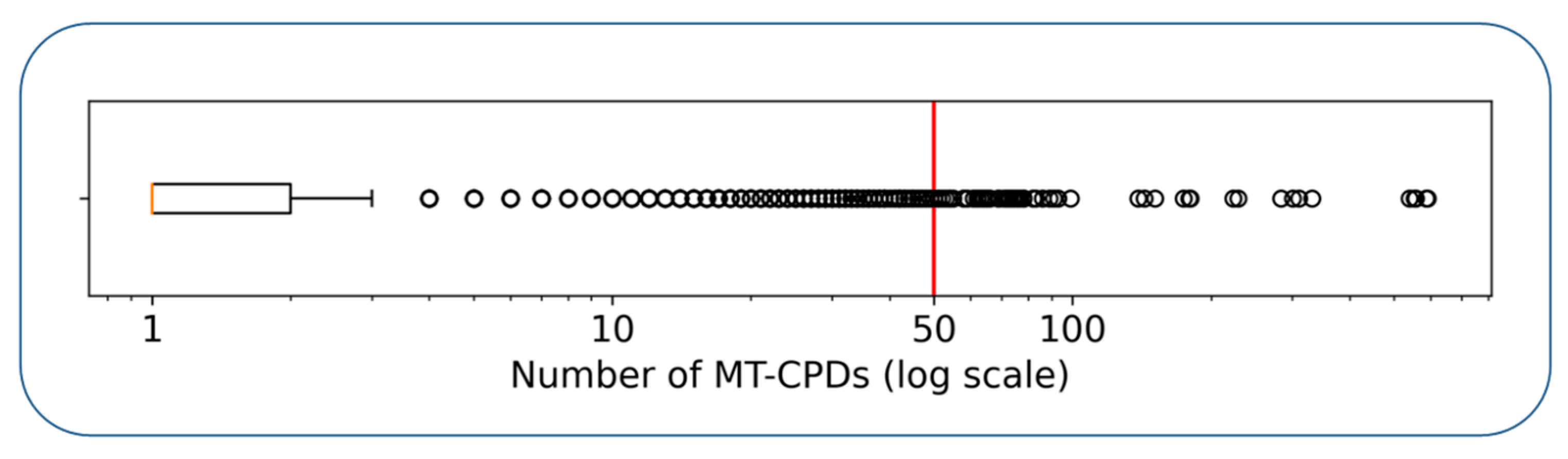




| Annotation | Number of Inhibitors | |
|---|---|---|
| Triplet 1 | (Triple-target) MT-CPDs | 223 |
| ST-CPDs Tyrosine-protein kinase JAK2 | 1225 | |
| Tyrosine-protein kinase JAK3 | 724 | |
| Adhesion kinase 1 | 505 | |
| Triplet 2 | (Triple-target) MT-CPDs | 74 |
| ST-CPDs Dual specificity tyrosine-phosphorylation-regulated kinase 1A | 343 | |
| Dual specificity tyrosine-phosphorylation-regulated kinase 1B | 19 | |
| Dual specificity tyrosine-phosphorylation-regulated kinase 2 | 51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feldmann, C.; Bajorath, J. Differentiating Inhibitors of Closely Related Protein Kinases with Single- or Multi-Target Activity via Explainable Machine Learning and Feature Analysis. Biomolecules 2022, 12, 557. https://doi.org/10.3390/biom12040557
Feldmann C, Bajorath J. Differentiating Inhibitors of Closely Related Protein Kinases with Single- or Multi-Target Activity via Explainable Machine Learning and Feature Analysis. Biomolecules. 2022; 12(4):557. https://doi.org/10.3390/biom12040557
Chicago/Turabian StyleFeldmann, Christian, and Jürgen Bajorath. 2022. "Differentiating Inhibitors of Closely Related Protein Kinases with Single- or Multi-Target Activity via Explainable Machine Learning and Feature Analysis" Biomolecules 12, no. 4: 557. https://doi.org/10.3390/biom12040557
APA StyleFeldmann, C., & Bajorath, J. (2022). Differentiating Inhibitors of Closely Related Protein Kinases with Single- or Multi-Target Activity via Explainable Machine Learning and Feature Analysis. Biomolecules, 12(4), 557. https://doi.org/10.3390/biom12040557







