Fine Mapping and Characterization of a Major Gene Responsible for Chlorophyll Biosynthesis in Brassica napus L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Quantitative Trait Loci Sequencing Analysis
2.3. Fine Mapping of Candidate Genes
2.4. RNA Library Construction and RNA-Seq Analysis
2.5. Gene Cloning and Structure Prediction
2.6. Gene Expression Analysis by Quantitative Real-Time PCR (qRT-PCR)
2.7. Developing Molecular Marker
2.8. Statistical Analysis
3. Results
3.1. Phenotypic Characterization and Genetic Analysis
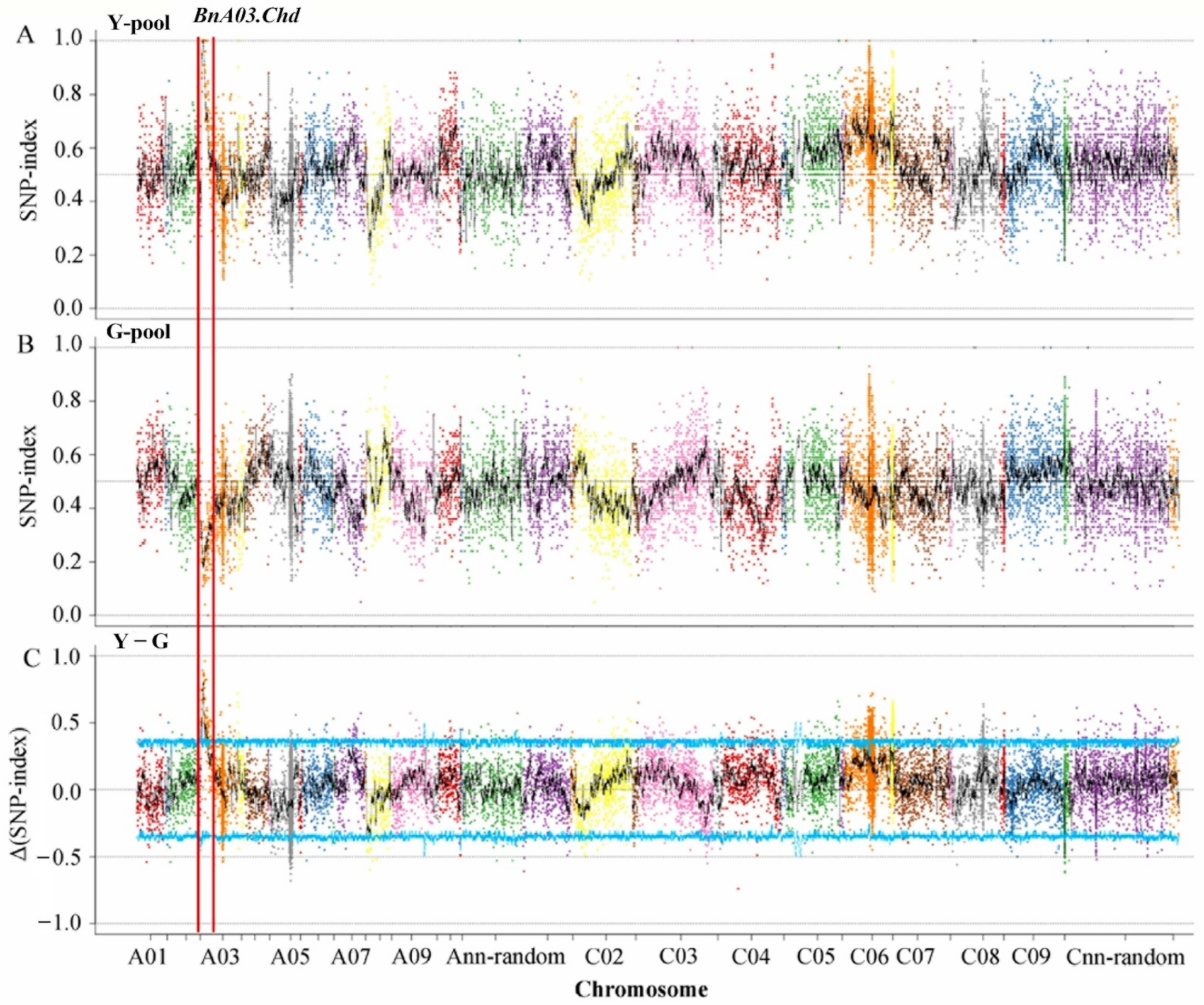
3.2. Initial Localization of Candidate Genes
3.3. Fine Mapping of the BnA03.Chd Gene
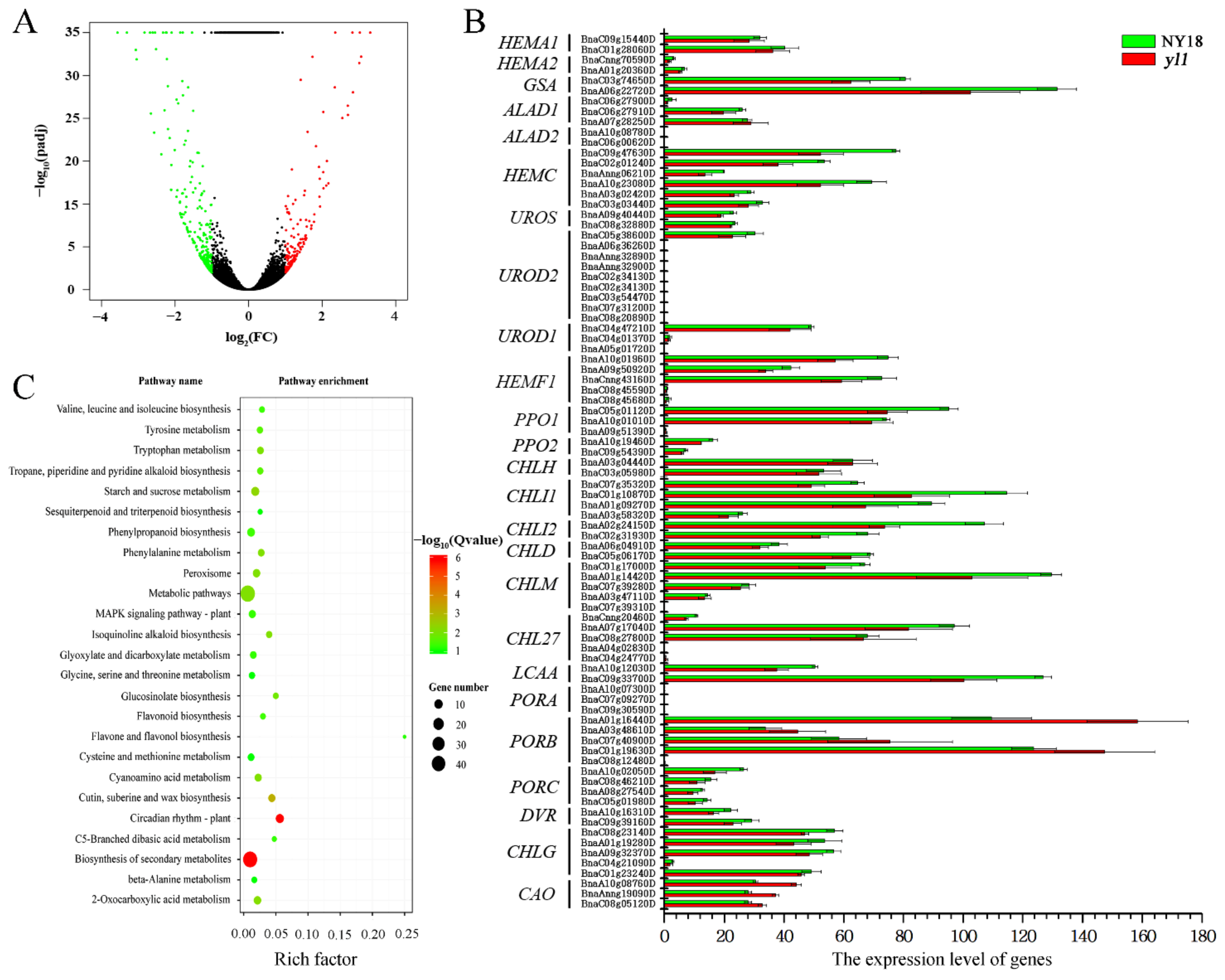
3.4. Genome-Wide Transcriptomic Analyses of NY18 and yl1
3.5. Identification of a Candidate Gene
3.6. Sequence Analysis of the Candidate Gene for BnA03.Chd
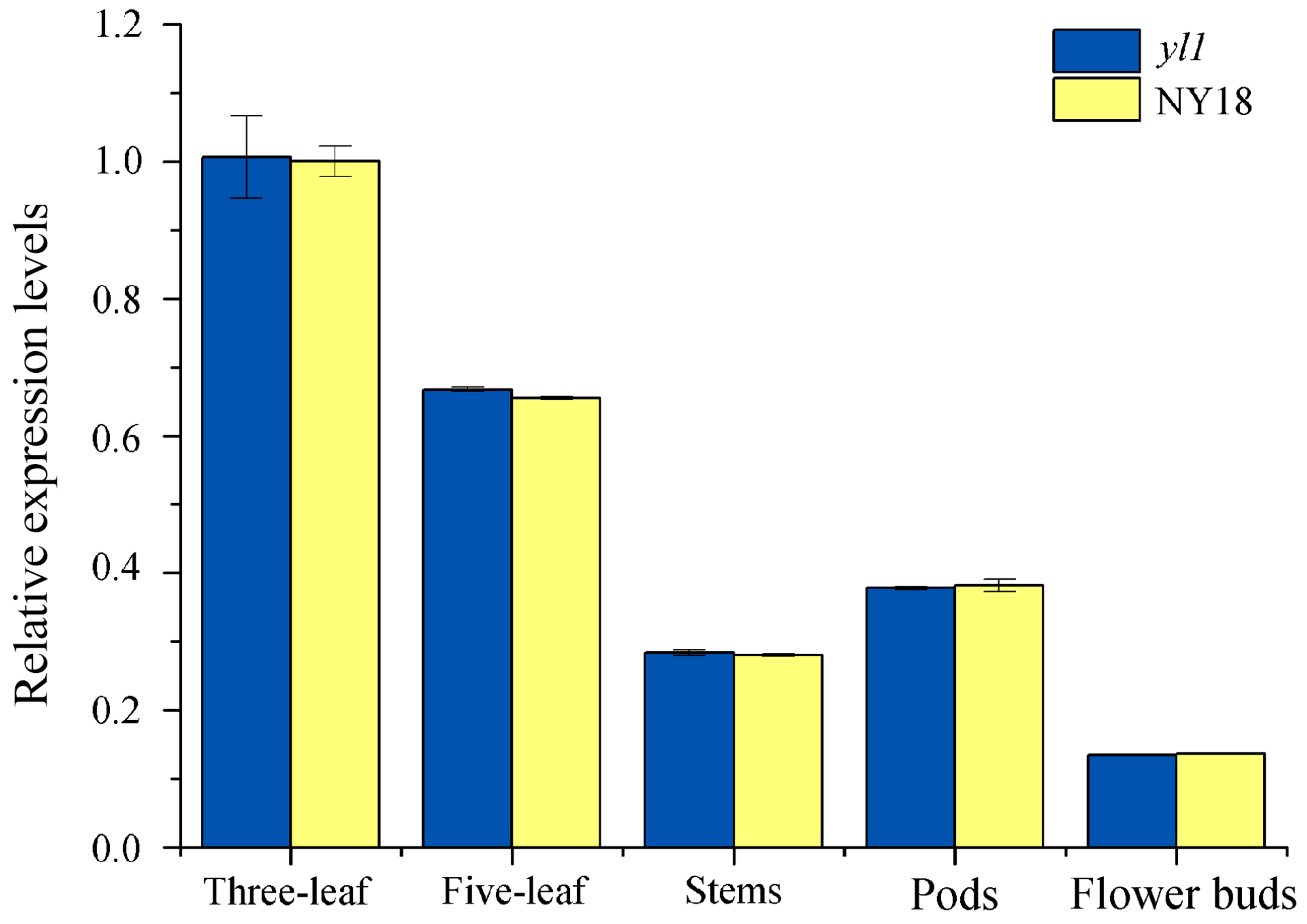
3.7. Expression Analysis of the Candidate Genes
3.8. Development of the PARMS Marker for BnA03.Chd
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Lu, C.; Napier, J.A.; Clemente, T.E.; Cahoon, E.B. New frontiers in oilseed biotechnology: Meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr. Opin. Biotech. 2011, 22, 252–259. [Google Scholar] [CrossRef]
- Durrett, T.P.; Benning, C.; Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008, 54, 593–607. [Google Scholar] [CrossRef]
- An, H.; Qi, X.; Gaynor, M.L.; Hao, Y.; Gebken, S.C.; Mabry, M.E.; McAlvay, A.C.; Teakle, G.R.; Conant, G.C.; Barker, M.S.; et al. Transcriptome and organellar sequencing highlights the complex origin and diversification of allotetraploid Brassica napus. Nat. Commun. 2019, 10, 2878. [Google Scholar] [CrossRef]
- Nagata, N.; Tanaka, R.; Satoh, S.; Tanaka, A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of prochlorococcus species. Plant Cell 2005, 17, 233–240. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, X.; Hu, K.; Wang, Y.; Wang, X.; Du, S.; Li, Y.; Hu, D.; Cheng, K.; An, B.; et al. Impaired magnesium protoporphyrin IX methyltransferase (ChlM) impedes chlorophyll synthesis and plant growth in rice. Front. Plant Sci. 2017, 8, 1694. [Google Scholar] [CrossRef]
- Frick, G.; Su, Q.; Apel, K.; Armstrong, G.A. An Arabidopsis porB porC double mutant lacking light-dependent NADPH: Protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J. 2003, 35, 141–153. [Google Scholar] [CrossRef]
- Runge, S.; van Cleve, B.; Lebedev, N.; Armstrong, G.; Apel, K. Isolation and classification of chlorophyll-deficient xantha mutants of Arabidopsis thaliana. Planta 1995, 197, 490–500. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, Y.; Lv, J.; Zhang, J.; Li, P.; Shi, X.; Wang, Y.; Zhang, H.; He, Z.; Teng, S. Characterization and fine mapping of a novel rice albino mutant low temperature albino 1. J. Genet. Genom. 2012, 39, 385–396. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Hu, Z.; Xia, Y.; Huang, Q.; Yu, T.; Yi, H.; Lu, Y.; Wang, J.; Cao, M. A valine residue deletion in ZmSig2A, a sigma factor, accounts for a revertible leaf-color mutation in maize. Crop J. 2021, 9, 1330–1343. [Google Scholar] [CrossRef]
- Zhu, X.; Pan, Y.; Liu, Z.; Liu, Y.; Zhong, D.; Duan, Z.; Tian, Z.; Zhu, B.; Zhou, G. Mutation of YL results in a yellow leaf with chloroplast RNA editing defect in soybean. Int. J. Mol. Sci. 2020, 21, 4275. [Google Scholar] [CrossRef]
- Liu, C.; Shi, N.; Wu, H.; An, X.; Zheng, J.; Duan, Y.; Sun, D.; Feng, Y.; Zhang, L. Cytogenetic analyses of PSL1 mutant, a novel low-temperature-sensitive purple-striped leaf color mutant in wheat. Crop Sci. 2018, 58, 1919–1931. [Google Scholar] [CrossRef]
- Wang, Y.; He, Y.; Yang, M.; He, J.; Xu, P.; Shao, M.; Chu, P.; Guan, R. Fine mapping of a dominant gene conferring chlorophyll-deficiency in Brassica napus. Sci. Rep. 2016, 6, 31419. [Google Scholar] [CrossRef]
- Zhu, L.; Zeng, X.; Chen, Y.; Yang, Z.; Qi, L.; Pu, Y.; Yi, B.; Wen, J.; Ma, C.; Shen, J.; et al. Genetic characterisation and fine mapping of a chlorophyll-deficient mutant (BnaC.ygl) in Brassica napus. Mol. Breed. 2014, 34, 603–614. [Google Scholar] [CrossRef]
- Tsang, E.W.T.; Yang, J.; Chang, Q.; Nowak, G.; Kolenovsky, A.; McGregor, D.I.; Keller, W.A. Chlorophyll reduction in the seed of Brassica napus with a glutamate 1-semialdehyde aminotransferase antisense gene. Plant Mol. Biol. 2003, 51, 191–201. [Google Scholar] [CrossRef]
- Liu, M.; Lu, Y.; Wang, S.; Wu, F.; Li, J.; Wang, Y.; Zhao, J.; Shen, S. Characterization of the leaf color mutant hy and identification of the mutated gene in Chinese cabbage. J. Am. Soc. Hortic. Sci. 2018, 143, 363–369. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, L.; Safdar, L.B.; Xie, M.; Cheng, X.; Liu, Y.; Xiang, Y.; Tong, C.; Tu, J.; Huang, J.; et al. Characterization and fine mapping of a yellow-virescent gene regulating chlorophyll biosynthesis and early stage chloroplast development in Brassica napus. G3 Genes Genom. Genet. 2020, 10, 3201–3211. [Google Scholar] [CrossRef]
- Chen, X.; Pu, H.; Fang, Y.; Wang, X.; Zhao, S.; Lin, Y.; Zhang, M.; Dai, H.-E.; Gong, W.; Liu, L. Crystal structure of the catalytic subunit of magnesium chelatase. Nat. Plants 2015, 1, 15125. [Google Scholar] [CrossRef]
- Adams, N.B.P.; Marklew, C.J.; Qian, P.; Brindley, A.A.; Davison, P.A.; Bullough, P.A.; Hunter, C.N. Structural and functional consequences of removing the N-terminal domain from the magnesium chelatase ChlH subunit of Thermosynechococcus elongatus. Biochem. J. 2014, 464, 315–322. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Yoo, J.H.; Yoo, S.C.; Cho, S.H.; Koh, H.J.; Seo, H.S.; Paek, N.C. Rice Chlorina-1 and Chlorina-9 Encode ChlD and ChlI Subunits of Mg-Chelatase, a Key Enzyme for Chlorophyll Synthesis and Chloroplast Development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef]
- Nakayama, M.; Masuda, T.; Bando, T.; Yamagata, H.; Ohta, H.; Takamiya, K. Cloning and expression of the soybean ChlH gene encoding a subunit of Mg-chelatase and localization of the Mg2+ concentration-dependent ChlH protein within the chloroplast. Plant Cell Physiol. 1998, 39, 275–284. [Google Scholar] [CrossRef][Green Version]
- Mochizuki, N.; Brusslan, J.A.; Larkin, R.; Nagatani, A.; Chory, J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 2001, 98, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.S.; Li, H. Arabidopsis CHLI2 can substitute for CHLI1. Plant Physiol. 2009, 150, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.Y.; Wang, X.F.; Wu, F.Q.; Du, S.Y.; Cao, Z.; Shang, Y.; Wang, X.L.; Peng, C.C.; Yu, X.C.; Zhu, S.Y.; et al. The Mg-Chelatase H Subunit Is an Abscisic Acid Receptor. Nature 2006, 443, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, N.D.; Froehlich, J.E.; Strand, D.D.; Buck, S.M.; Kramer, D.M.; Larkin, R.M. GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1449–1467. [Google Scholar] [CrossRef] [PubMed]
- Ankele, E.; Kindgren, P.; Pesquet, E.; Strand, Å. In vivo visualization of Mg-protoporphyrinIX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 2007, 19, 1964–1979. [Google Scholar] [CrossRef]
- Strand, Å.; Asami, T.; Alonso, J.; Ecker, J.R.; Chory, J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 2003, 421, 79–83. [Google Scholar] [CrossRef]
- Pontoppidan, B.; Kannangara, C.G. Purification and partial characterisation of barley Glutamyl-tRNAGlu reductase, the enzyme that directs glutamate to chlorophyll biosynthesis. Eur. J. Biochem. 1994, 225, 529–537. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, Z.; Zeng, X.; Gao, J.; Liu, J.; Yi, B.; Ma, C.; Shen, J.; Tu, J.; Fu, T.; et al. Heme oxygenase 1 defects lead to reduced chlorophyll in Brassica napus. Plant Mol. Biol. 2017, 93, 579–592. [Google Scholar] [CrossRef]
- Börner, T.; Aleynikova, A.Y.; Zubo, Y.O.; Kusnetsov, V.V. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta—Bioenerg. 2015, 1847, 761–769. [Google Scholar] [CrossRef]
- Lv, Y.; Shao, G.; Qiu, J.; Jiao, G.; Sheng, Z.; Xie, L.; Wu, Y.; Tang, S.; Wei, X.; Hu, P. White Leaf and Panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice. J. Exp. Bot. 2017, 68, 5147–5160. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Peng, M.; Zhao, X.; Wang, X.; Chen, F.; Zhang, W.; Sun, C.; Zhang, C.; Guan, R.; Zhang, J. Phenotypic identification and genetic analysis of a chlorophyll deficient mutant yl1 in Brassica napus. Chin. J. Oil Crop Sci. 2021, 43, 443, (In Chinese with an English Abstract). [Google Scholar] [CrossRef]
- Wang, X.; Zheng, M.; Liu, H.; Zhang, L.; Chen, F.; Zhang, W.; Fan, S.; Peng, M.; Hu, M.; Wang, H.; et al. Fine-mapping and transcriptome analysis of a candidate gene controlling plant height in Brassica napus L. Biotechnol. Biofuels 2020, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-Seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R.; et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, N.; Liu, Y.; Jia, L.; Fu, Y.; He, X.; Liu, K.; Xu, Z.; Bao, B. Novel molecular markers for high-throughput sex characterization of cynoglossus semilaevis. Aquaculture 2019, 513, 734331. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, R.; Sasaki, T.; Nishio, T. Characterization of DNA methyltransferase genes in Brassica rapa. Genes Genet. Syst. 2006, 81, 235–242. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. BioTechniques 1998, 24, 318–319. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tang, W.; Wang, W.; Chen, D.; Ji, Q.; Jing, Y.; Wang, H.; Lin, R. Transposase-derived proteins FHY3/FAR1 interact with Phytochrome-Interacting Factor1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 2012, 24, 1984–2000. [Google Scholar] [CrossRef]
- Armstrong, G.A.; Runge, S.; Frick, G.; Sperling, U.; Apel, K. Identification of NADPH: Protochlorophyllide oxidoreductases A and B: A branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol. 1995, 108, 1505–1517. [Google Scholar] [CrossRef]
- Fitter, D.W.; Martin, D.J.; Copley, M.J.; Scotland, R.W.; Langdale, J.A. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002, 31, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; He, H.; Li, H.; Tian, H.; Zhang, J.; Zhai, L.; Chen, J.; Wu, H.; Yi, G.; He, Z.-H.; et al. NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and rubisco formation in rice. PLoS ONE 2011, 6, e20015. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Sheng, S.; Wang, J.; Guo, X.; Su, N.; Wang, L.; Jiang, L.; et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef]
- Yuan, G.; Tang, C.; Li, Y.; Chen, B.; He, H.; Peng, H.; Zhang, Y.; Gou, C.; Zou, C.; Pan, G.; et al. Identification and fine mapping of a candidate gene for oil yellow leaf 2 conferring yellow leaf phenotype in maize. Plant Breed. 2021, 140, 100–109. [Google Scholar] [CrossRef]
- Yang, S.; Tian, X.; Wang, Z.; Wei, X.; Zhao, Y.; Su, H.; Zhao, X.; Tian, B.; Yuan, Y.; Zhang, X.W. Fine mapping and candidate gene identification of a white flower gene BrWF3 in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 2021, 12, 646222. [Google Scholar] [CrossRef]
- Ye, J.; Liu, H.; Zhao, Z.; Xu, L.; Li, K.; Du, D. Fine mapping of the QTL cqSPDA2 for chlorophyll content in Brassica napus L. BMC Plant Biol. 2020, 20, 511. [Google Scholar] [CrossRef]
- Zhao, H.; Yu, L.; Huai, Z.; Wang, X.; Ding, G.; Chen, S.; Li, P.; Xu, F. Mapping and candidate gene identification defining BnChd1-1, a locus involved in chlorophyll biosynthesis in Brassica napus. Acta Physiol. Plant 2014, 36, 859–870. [Google Scholar] [CrossRef]
- Gammelvind, L.H.; Schjoerring, J.K.; Mogensen, V.O.; Jensen, C.R.; Bock, J.G.H. Photosynthesis in leaves and siliques of winter oilseed rape (Brassica napus L.). Plant Soil 1996, 186, 227–236. [Google Scholar] [CrossRef]
- Adams, N.B.P.; Bisson, C.; Brindley, A.A.; Farmer, D.A.; Davison, P.A.; Reid, J.D.; Hunter, C.N. The active site of magnesium chelatase. Nat. Plants 2020, 6, 1491–1502. [Google Scholar] [CrossRef]
- Bennett, J. Biosynthesis of the light-harvesting chlorophyll a/b protein. Eur. J. Biochem. 1981, 118, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Bellemare, G.; Bartlett, S.G.; Chua, N.H. Biosynthesis of chlorophyll a/b-binding polypeptides in wild type and the chlorina f2 mutant of barley. J. Biol. Chem. 1982, 257, 7762–7767. [Google Scholar] [CrossRef]
- Kindgren, P.; Kremnev, D.; Blanco, N.E.; de Dios Barajas López, J.; Fernández, A.P.; Tellgren-Roth, C.; Small, I.; Strand, Å. The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 2012, 70, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef] [PubMed]
- Schierholt, A.; Becker, H.C.; Ecke, W. Mapping a high oleic acid mutation in winter oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2000, 101, 897–901. [Google Scholar] [CrossRef]
- Burow, G.; Chopra, R.; Sattler, S.; Burke, J.; Acosta-Martinez, V.; Xin, Z. Deployment of SNP (CAPS and KASP) markers for allelic discrimination and easy access to functional variants for brown midrib genes Bmr6 and Bmr12 in Sorghum bicolor. Mol. Breed. 2019, 39, 115. [Google Scholar] [CrossRef]




| No. | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) | Purpose |
|---|---|---|---|
| 1 | GATCGCGCTTGTGCTTTGGG | TCGGCCTTCCAAGCTCCTCT | qRT-PCR |
| 2 | CAGGAATCGCTGACCGTAT | TCTCCCTTTGAAATCCACAT | |
| 3 | ATGGCTTCACTTATGTATTCACC | CTATCGATCGATCCCTTCAA | Cloning cDNA |
| 4 | GAAGGTGACCAAGTTCATGCTGGAAGATGATCCAGACTTTCTTGG | CCTTCAATCTTGTCTTCAACCTGG | Molecular marker |
| GAAGGTCGGAGTCAACGGATTAGGAAGATGATCCAGACTTTCTTGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, C.; Zhang, W.; Peng, M.; Zhao, X.; Shi, R.; Wu, X.; Chen, F.; Sun, C.; Wang, X.; Zhang, J. Fine Mapping and Characterization of a Major Gene Responsible for Chlorophyll Biosynthesis in Brassica napus L. Biomolecules 2022, 12, 402. https://doi.org/10.3390/biom12030402
Pang C, Zhang W, Peng M, Zhao X, Shi R, Wu X, Chen F, Sun C, Wang X, Zhang J. Fine Mapping and Characterization of a Major Gene Responsible for Chlorophyll Biosynthesis in Brassica napus L. Biomolecules. 2022; 12(3):402. https://doi.org/10.3390/biom12030402
Chicago/Turabian StylePang, Chengke, Wei Zhang, Menlu Peng, Xiaozhen Zhao, Rui Shi, Xu Wu, Feng Chen, Chengming Sun, Xiaodong Wang, and Jiefu Zhang. 2022. "Fine Mapping and Characterization of a Major Gene Responsible for Chlorophyll Biosynthesis in Brassica napus L." Biomolecules 12, no. 3: 402. https://doi.org/10.3390/biom12030402
APA StylePang, C., Zhang, W., Peng, M., Zhao, X., Shi, R., Wu, X., Chen, F., Sun, C., Wang, X., & Zhang, J. (2022). Fine Mapping and Characterization of a Major Gene Responsible for Chlorophyll Biosynthesis in Brassica napus L. Biomolecules, 12(3), 402. https://doi.org/10.3390/biom12030402






