The Halophyte Dehydrin Sequence Landscape
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
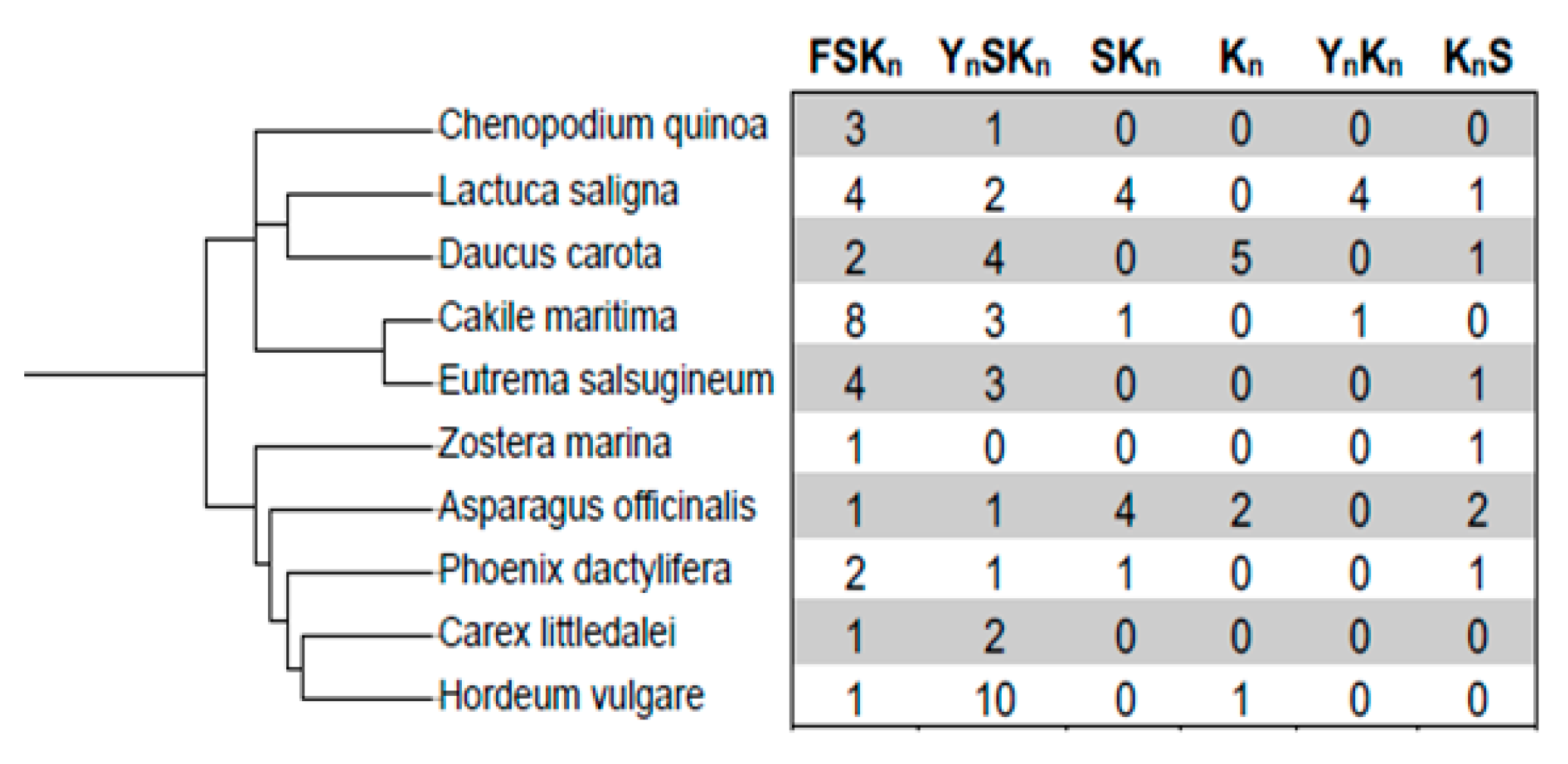
3.1. Dehydrin Architectures
3.2. Comparative Analysis of the Conserved DHN Motifs in Halophytes and Glycophytes
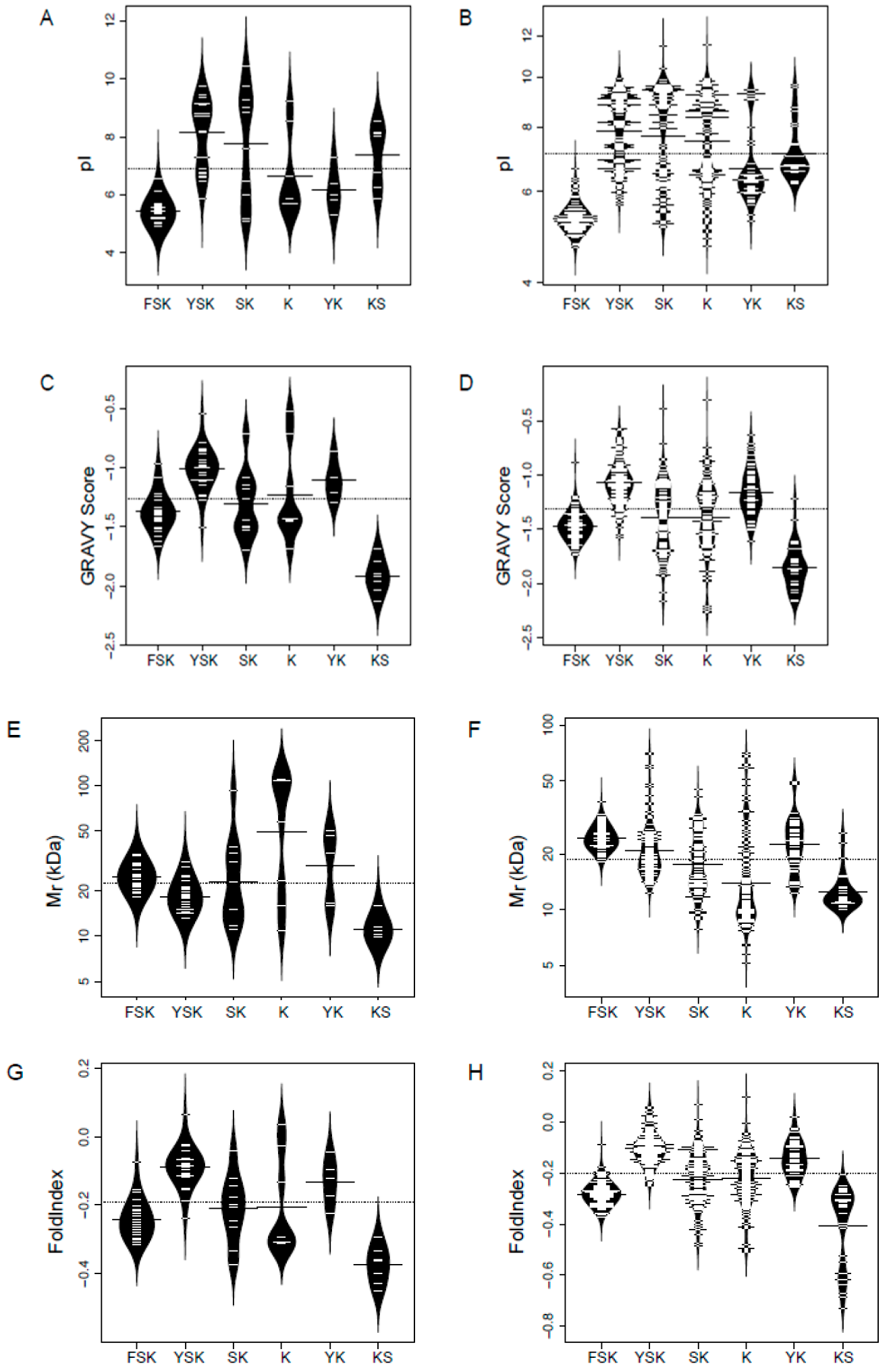
3.3. Physiochemical Characteristics of Dehydrins
3.4. Dehydrin Expression Profiling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, X.; Zhang, M.; Xie, B.; Jiang, X.; Gai, Y. Functional Characteristics Analysis of Dehydrins in Larixkaempferi under Osmotic Stress. Int. J. Mol. Sci. 2021, 22, 1715. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Singh, A. Soil salinization and waterlogging: A threat to environment and agricultural sustainability. Ecol. Indic. 2015, 57, 128–130. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Bendaly, A.; Messedi, D.; Smaoui, A.; Ksouri, R.; Bouchereau, A.; Abdelly, C. Physiological and leaf metabolome changes in the xerohalophyte species Atriplexhalimus induced by salinity. Plant Physiol. Biochem. 2016, 103, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef]
- Ellouzi, H.; Hamed, K.B.; Hernández, I.; Cela, J.; Müller, M.; Magné, C.; Munné-Bosch, S. A comparative study of the early osmotic, ionic, redox and hormonal signaling response in leaves and roots of two halophytes and a glycophyte to salinity. Planta 2014, 240, 1299–1317. [Google Scholar] [CrossRef]
- Taji, T.; Komatsu, K.; Katori, T.; Kawasaki, Y.; Sakata, Y.; Tanaka, S.; Kobayashi, M.; Toyoda, A.; Seki, M.; Shinozaki, K. Comparative genomic analysis of 1047 completely sequenced cDNAs from an Arabidopsis-related model halophyte, Thellungiella halophila. BMC Plant Biol. 2010, 10, 261. [Google Scholar] [CrossRef]
- Galau, G.A.; Hughes, D.W.; Dure, L., III. Abscisic acid induction of cloned cotton late embryogenesis-abundant (Lea) mRNAs. Plant Mol. Biol. 1986, 7, 155–170. [Google Scholar] [CrossRef]
- Close, T.J.; Kortt, A.A.; Chandler, P.M. A cDNA-based comparison of dehydration-induced proteins (dehydrins) in barley and corn. Plant Mol. Biol. 1989, 13, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Halder, T.; Agarwal, T.; Ray, S. Isolation, cloning, and characterization of a novel Sorghum dehydrin (SbDhn2) protein. Protoplasma 2016, 253, 1475–1488. [Google Scholar] [CrossRef] [PubMed]
- Ceccardi, T.L.; Meyer, N.C.; Close, T.J. Purification of a maize dehydrin. Protein Expr. Purif. 1994, 5, 266–269. [Google Scholar] [CrossRef]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.A.; Sabeem, M.; Mullath, S.K.; Brini, F.; Masmoudi, K. Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses. Biomolecules 2021, 11, 1662. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.A.; Veltri, M.; Boddington, K.F.; Singh, K.K.; Graether, S.P. Genome analysis of conserved dehydrin motifs in vascular plants. Front. Plant Sci. 2017, 8, 709. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The Enigmatic LEA Proteins and Other Hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef]
- Shih, M.D.; Hsieh, T.Y.; Jian, W.T.; Wu, M.T.; Yang, S.J.; Hoekstra, F.A.; Hsing, Y.I.C. Functional studies of soybean (Glycine max L.) seed LEA proteins GmPM6, GmPM11, and GmPM30 by CD and FTIR spectroscopy. Plant Sci. 2012, 196, 152–159. [Google Scholar] [CrossRef]
- Findlater, E.E.; Graether, S.P. NMR assignments of the intrinsically disordered K 2 and YSK 2 dehydrins. Biomol. NMR Assign. 2009, 3, 273–275. [Google Scholar] [CrossRef]
- Hughes, S.; Graether, S.P. Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci. 2011, 20, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Allagulova, C.R.; Gimalov, F.R.; Shakirova, F.M.; Vakhitov, V.A. The plant dehydrins: Structure and putative functions. Biochemistry 2003, 68, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Group II late embryogenesis abundant (LEA) proteins: Structural and functional aspects in plant abiotic stress. Plant Growth Regul. 2015, 79, 1–17. [Google Scholar] [CrossRef]
- Hara, M. The multifunctionality of dehydrins: An overview. Plant Signal. Behav. 2010, 5, 503–508. [Google Scholar] [CrossRef]
- Strimbeck, G.R. Hiding in plain sight: The F segment and other conserved features of seed plant SKn dehydrins. Planta 2017, 245, 1061–1066. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Physiologiaplantarum 1996, 97, 795–803. [Google Scholar]
- Boddington, K.F.; Graether, S.P. Binding of a Vitisripariadehydrin to DNA. Plant Sci. 2019, 287, 110172. [Google Scholar] [CrossRef]
- Hernández-Sánchez, I.E.; Martynowicz, D.M.; Rodríguez-Hernández, A.A.; Pérez-Morales, M.B.; Graether, S.P.; Jiménez-Bremont, J.F. A dehydrin-dehydrin interaction: The case of SK3 from Opuntiastreptacantha. Front. Plant Sci. 2014, 5, 520. [Google Scholar] [CrossRef]
- Lv, A.; Su, L.; Liu, X.; Xing, Q.; Huang, B.; An, Y.; Zhou, P. Characterization of Dehydrin protein, CdDHN4-L and CdDHN4-S, and their differential protective roles against abiotic stress in vitro. BMC Plant Biol. 2018, 18, 1–13. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Zhang, T.; Yang, X.; Li, D. Functional characterization of KS-type dehydrin ZmDHN13 and its related conserved domains under oxidative stress. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: A commonalty in the response of plants to dehydration and low temperature. Physiol. Plant. 1997, 100, 291–296. [Google Scholar] [CrossRef]
- Tiwari, P.; Chakrabarty, D. Dehydrin in the past four decades: From chaperones to transcription co-regulators in regulating abiotic stress response. Curr. Res. Biotechnol. 2021, 3, 249–259. [Google Scholar] [CrossRef]
- Peng, Y.; Reyes, J.L.; Wei, H.; Yang, Y.; Karlson, D.; Covarrubias, A.A.; Arora, R. RcDhn5, a cold acclimation-responsive dehydrin from Rhododendron catawbiense rescues enzyme activity from dehydration effects in vitro and enhances freezing tolerance in RcDhn5-overexpressing Arabidopsis plants. Physiol. Plant. 2008, 134, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Brini, F.; Saibi, W.; Amara, I.; Gargouri, A.; Masmoudi, K.; Hanin, M. Wheat dehydrin DHN-5 exerts a heat-protective effect on β-glucosidase and glucose oxidase activities. Biosci. Biotechnol. Biochem. 2010, 74, 1050–1054. [Google Scholar] [CrossRef]
- Drira, M.; Saibi, W.; Brini, F.; Gargouri, A.; Masmoudi, K.; Hanin, M. The K-Segments of the Wheat Dehydrin DHN-5 are Essential for the Protection of Lactate Dehydrogenase and β-Glucosidase Activities In Vitro. Mol. Biotechnol. 2013, 54, 643–650. [Google Scholar] [CrossRef]
- Halder, T.; Upadhyaya, G.; Ray, S. YSK2 Type Dehydrin (SbDhn1) from Sorghum bicolor Showed Improved Protection under High Temperature and Osmotic Stress Condition. Front. Plant Sci. 2017, 8, 918. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Endo, T.; Kamiya, K.; Kameyama, A. The role of hydrophobic amino acids of K-segments in the cryoprotection of lactate dehydrogenase by dehydrins. J. Plant Physiol. 2017, 210, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhang, L.; Lv, H.; Li, H.; Zhang, Y.; Xu, Y.; Yu, J. The K-segments of wheat dehydrin WZY2 are essential for its protective functions under temperature stress. Front. Plant Sci. 2015, 6, 406. [Google Scholar] [CrossRef]
- Drira, M.; Saibi, W.; Amara, I.; Masmoudi, K.; Hanin, M.; Brini, F. Wheat dehydrin K-segments ensure bacterial stress tolerance, antiaggregation and antimicrobial effects. Appl. Biotechnol. 2015, 175, 3310–3321. [Google Scholar] [CrossRef]
- Hara, M.; Uchida, S.; Murata, T.; Wätzig, H. Efficient purification of cryoprotectivedehydrin protein from the radish (Raphanussativus) taproot. Eur. Food Res. Technol. 2014, 239, 339–345. [Google Scholar] [CrossRef][Green Version]
- Yokoyama, T.; Ohkubo, T.; Kamiya, K.; Hara, M. Cryoprotective activity of Arabidopsis KS-type dehydrin depends on the hydrophobic amino acids of two active segments. Arch. Biochem. Biophys. 2020, 691, 108510. [Google Scholar] [CrossRef]
- Koag, M.-C.; Wilkens, S.; Fenton, R.D.; Resnik, J.; Vo, E.; Close, T.J. The K-Segment of Maize DHN1 Mediates Binding to Anionic Phospholipid Vesicles and Concomitant Structural Changes. Plant Physiol. 2009, 150, 1503–1514. [Google Scholar] [CrossRef]
- Graether, S.P.; Boddington, K.F. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 576. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.; Eremina, N.; Barth, A.; Danielsson, J.; Harryson, P. Membrane-induced folding of the plant stress dehydrin Lti30. Plant Physiol. 2016, 171, 932–943. [Google Scholar] [PubMed]
- Clarke, M.W.; Boddington, K.F.; Warnica, J.M.; Atkinson, J.; McKenna, S.; Madge, J.; Barker, C.H.; Graether, S.P. Structural and Functional Insights into the Cryoprotection of Membranes by the Intrinsically Disordered Dehydrins. J. Biol. Chem. 2015, 290, 26900–26913. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.K.; Harryson, P. Dehydrins: Molecular Biology, Structure and Function. In Plant Desiccation Tolerance; Springer: Berlin/Heidelberg, Germany, 2011; pp. 289–305. [Google Scholar]
- Gupta, A.; Marzinek, J.; Jefferies, D.; Bond, P.; Harryson, P.; Wohland, T. The disordered plant dehydrin Lti30 protects the membrane during water-related stress by cross-linking lipids. J. Biol. Chem. 2019, 294, 6468–6482. [Google Scholar] [CrossRef]
- Riley, A.C.; Ashlock, D.A.; Graether, S.P. Evolution of the modular, disordered stress proteins known as dehydrins. PLoS ONE 2019, 14, e0211813. [Google Scholar] [CrossRef]
- Kampstra, P. Beanplot: A Boxplot Alternative for Visual Comparison of Distributions. J. Stat. Softw. 2008, 28, 1–9. [Google Scholar] [CrossRef]
- Guo, R.; Zhao, L.; Zhang, K.; Lu, H.; Bhanbhro, N.; Yang, C. Comparative Genomics and Transcriptomics of the Extreme Halophyte Puccinellia tenuiflora Provides Insights into Salinity Tolerance Differentiation between Halophytes and Glycophytes. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Li, C.; Qi, Y.; Zhao, C.; Wang, X.; Zhang, Q. Transcriptome Profiling of the Salt Stress Response in the Leaves and Roots of Halophytic Eutrema salsugineum. Front. Genet. 2021, 12, 770742. [Google Scholar] [CrossRef]
- Isayenkov, S.; Hilo, A.; Rizzo, P.; Tandron Moya, Y.A.; Rolletschek, H.; Borisjuk, L.; Radchuk, V. Adaptation strategies of halophytic barley Hordeum marinum ssp. marinum to high salinity and osmotic stress. Int. J. Mol. Sci. 2020, 21, 9019. [Google Scholar] [CrossRef]
- Debez, A.; Braun, H.P.; Pich, A.; Taamalli, W.; Koyro, H.W.; Abdelly, C.; Huchzermeyer, B. Proteomic and physiological responses of the halophyte Cakilemaritima to moderate salinity at the germinative and vegetative stages. J. Proteom. 2012, 75, 5667–5694. [Google Scholar] [CrossRef] [PubMed]
- Bueno, M.; Cordovilla, M.P. Ecophysiology and Uses of Halophytes in Diverse Habitats. In Handbook of Halophytes: From Molecules to Ecosystems towards Biosaline Agriculture; Springer: New Delhi, India, 2020; pp. 1–25. [Google Scholar]
- Dagar, J.C. Greening Salty and Waterlogged Lands through Agroforestry Systems for Livelihood Security and Better Environment. In Agroforestry Systems in India: Livelihood Security & Ecosystem Services; Springer: New Delhi, India, 2014; pp. 273–332. [Google Scholar]
- Rorat, T. Plant dehydrins—Tissue location, structure and function. Cell. Mol. Biol. Lett. 2006, 11, 536–556. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, T.; Kameyama, A.; Kamiya, K.; Kondo, M.; Hara, M. F-segments of Arabidopsis dehydrins show cryoprotective activities for lactate dehydrogenase depending on the hydrophobic residues. Phytochemistry 2020, 173, 112300. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sheng, J.; Lv, K.; Ren, L.; Zhang, D. Y2SK2 and SK3 type dehydrins from Agapanthus praecox can improve plant stress tolerance and act as multifunctional protectants. Plant Sci. 2019, 284, 143–160. [Google Scholar] [CrossRef]
- Nguyen, P.N.; Tossounian, M.-A.; Kovacs, D.S.; Thu, T.T.; Stijlemans, B.; Vertommen, D.; Pauwels, J.; Gevaert, K.; Angenon, G.; Messens, J.; et al. Dehydrin ERD14 activates glutathione transferase Phi9 in Arabidopsis thaliana under osmotic stress. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2019, 1864, 129506. [Google Scholar] [CrossRef] [PubMed]
- Fukuchi, S.; Yoshimune, K.; Wakayama, M.; Moriguchi, M.; Nishikawa, K. Unique Amino Acid Composition of Proteins in Halophilic Bacteria. J. Mol. Biol. 2003, 327, 347–357. [Google Scholar] [CrossRef]
- Barua, B.; Lin, J.C.; Williams, V.D.; Kummler, P.; Neidigh, J.W.; Andersen, N.H. The Trp-cage: Optimizing the stability of a globular miniprotein. Protein Eng. Des. Sel. 2008, 21, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Prilusky, J.; Felder, C.E.; Zeev-Ben-Mordehai, T.; Rydberg, E.H.; Man, O.; Beckmann, J.S.; Sussman, J.L. FoldIndex©: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 2005, 21, 3435–3438. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.L.; Schart, V.; Malcolmson, J.; Hogarth, K.A.; Martynowicz, D.M.; Tralman-Baker, E.; Patel, S.N.; Graether, S.P. The Importance of Size and Disorder in the Cryoprotective Effects of Dehydrins. Plant Physiol. 2013, 163, 1376–1386. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghanmi, S.; Graether, S.P.; Hanin, M. The Halophyte Dehydrin Sequence Landscape. Biomolecules 2022, 12, 330. https://doi.org/10.3390/biom12020330
Ghanmi S, Graether SP, Hanin M. The Halophyte Dehydrin Sequence Landscape. Biomolecules. 2022; 12(2):330. https://doi.org/10.3390/biom12020330
Chicago/Turabian StyleGhanmi, Siwar, Steffen P. Graether, and Moez Hanin. 2022. "The Halophyte Dehydrin Sequence Landscape" Biomolecules 12, no. 2: 330. https://doi.org/10.3390/biom12020330
APA StyleGhanmi, S., Graether, S. P., & Hanin, M. (2022). The Halophyte Dehydrin Sequence Landscape. Biomolecules, 12(2), 330. https://doi.org/10.3390/biom12020330






