Single-Cell RNA Sequencing Reveals Heterogeneity in the Tumor Microenvironment between Young-Onset and Old-Onset Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Sample Collection
2.2. Single-Cell Isolation
2.3. scRNA-Seq
2.4. Single-Cell Gene Expression Matrix Acquisition and Cell Clustering
2.5. Identifying Marker Genes and Differentially Expressed Genes
2.6. Cell Developmental Trajectory
2.7. Pathway Enrichment Analysis
2.8. The Cancer Genome Atlas Data Analysis
2.9. InferCNV Analysis to Identify Malignant Cells
2.10. Cell-Cell Communication Analysis
2.11. Statistical Analysis
3. Results
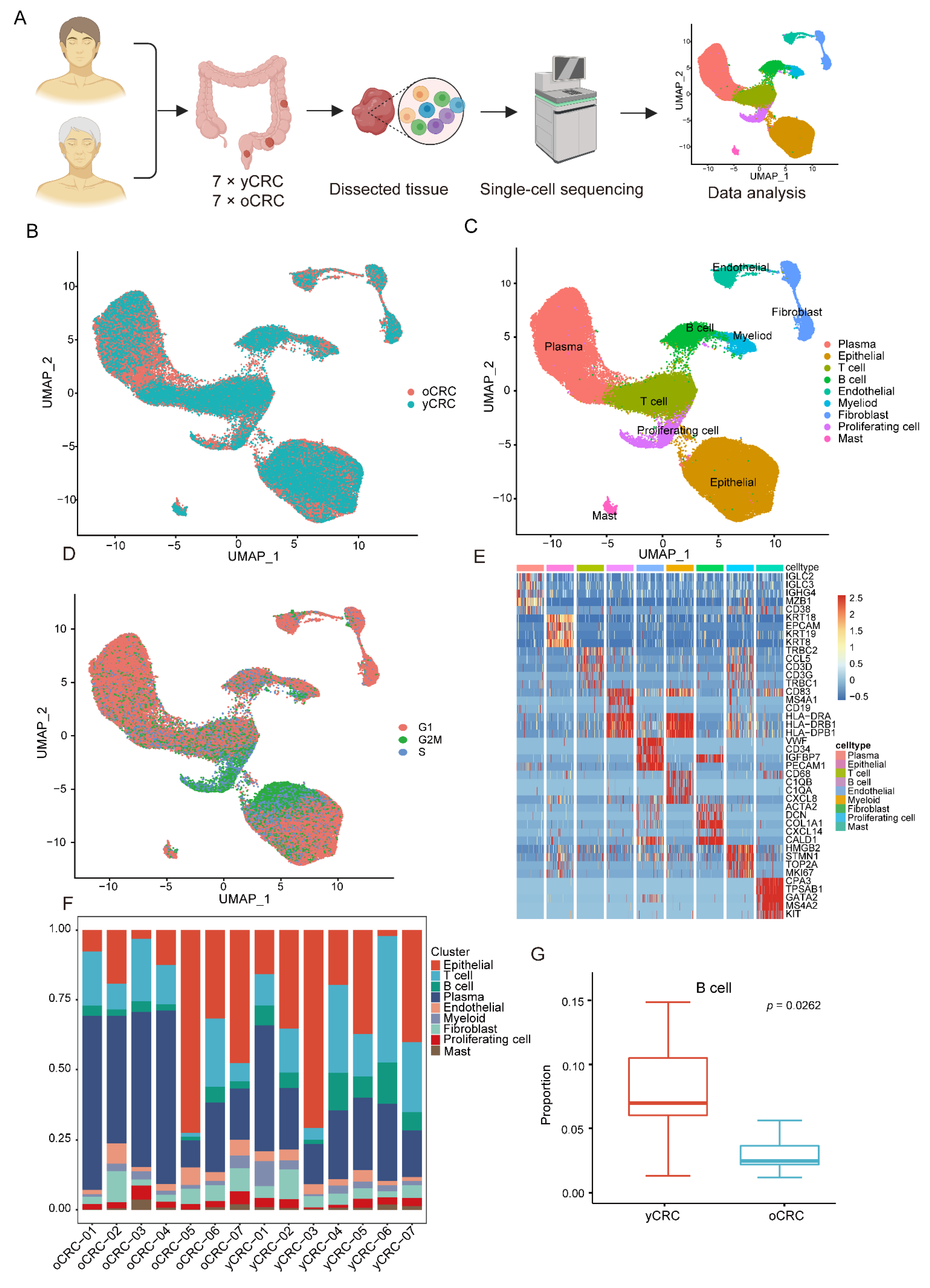
3.1. Single-Cell Transcriptomic Profiling of yCRC and oCRC
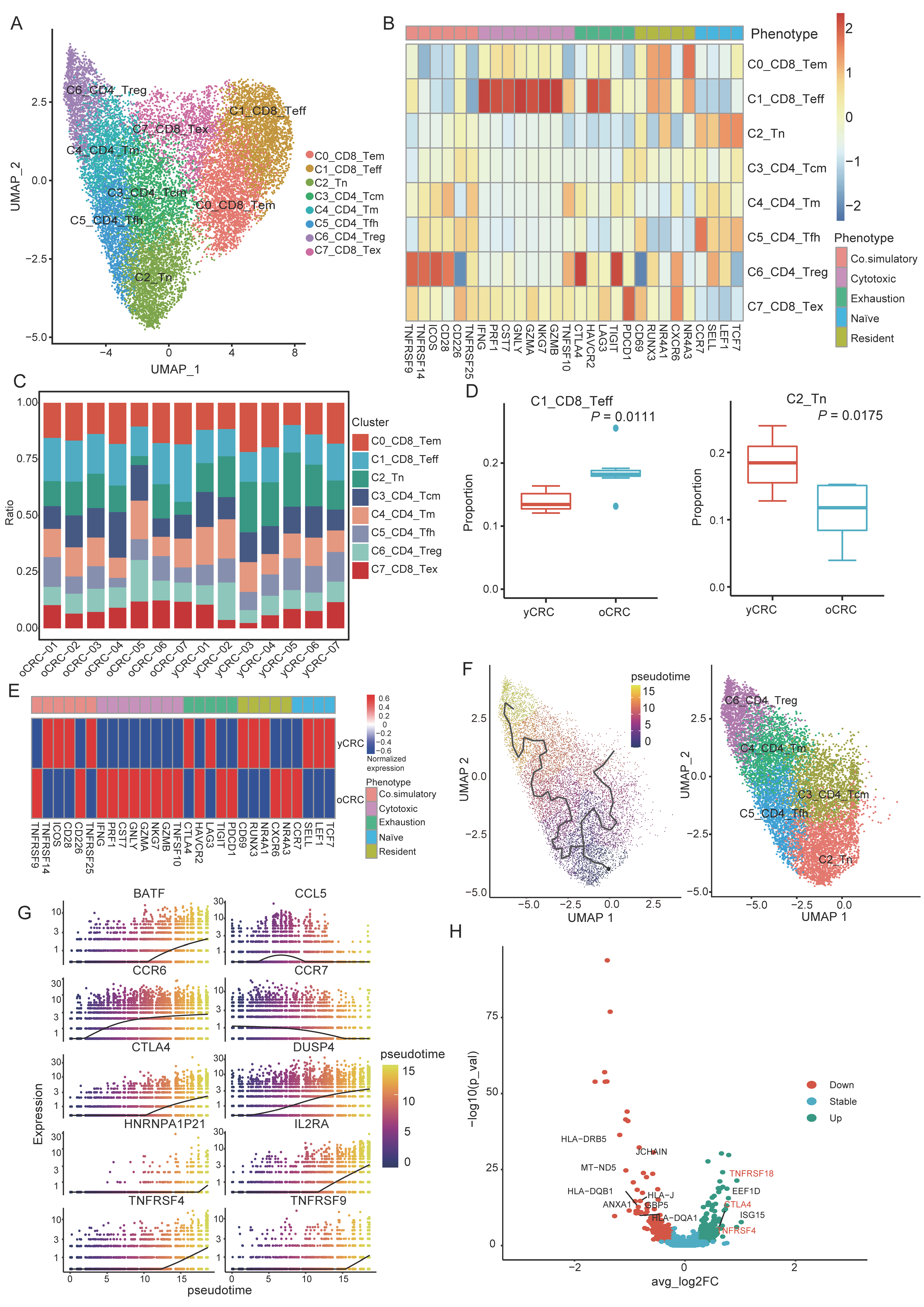
3.2. Features and Heterogeneity of T-Cell Subtypes in yCRC and oCRC
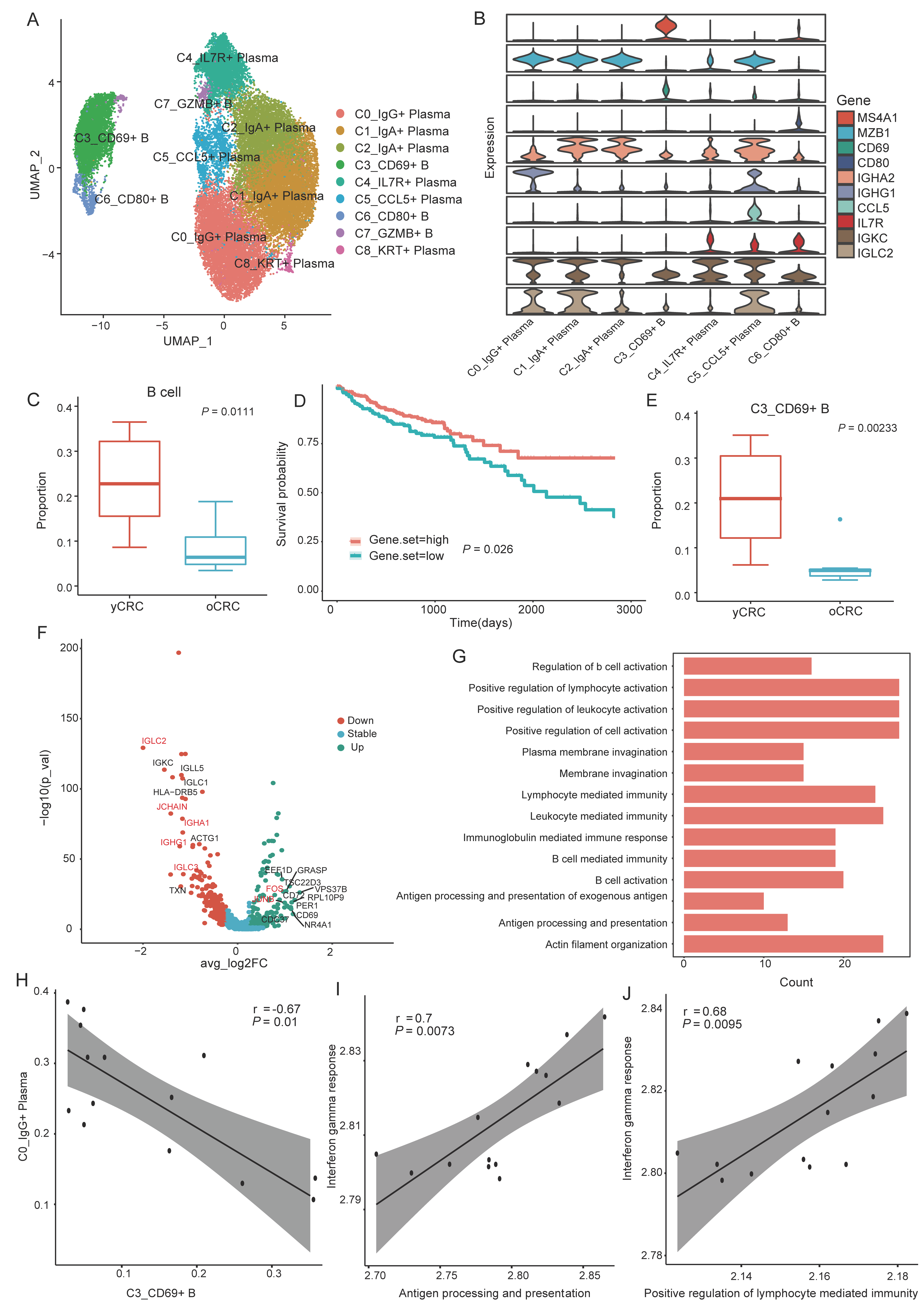
3.3. Heterogeneity of B Cells and Plasma Cells in the TME of yCRC and oCRC
3.4. Antitumor Immunity of Myeloid Cells Declines in yCRC
3.5. Heterogeneity of Malignant Cells and Interactions with Immune Cells in the TME
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Levine, O.; Zbuk, K. Colorectal cancer in adolescents and young adults: Defining a growing threat. Pediatr. Blood Cancer 2019, 66, e27941. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.D.; You, Y.N. Young-onset colorectal cancer: Diagnosis and management. Semin. Colon Rectal. Surg. 2018, 29, 98–101. [Google Scholar] [CrossRef]
- Kirzin, S.; Marisa, L.; Guimbaud, R.; De Reynies, A.; Legrain, M.; Laurent-Puig, P.; Cordelier, P.; Pradère, B.; Bonnet, D.; Meggetto, F.; et al. Sporadic Early-Onset Colorectal Cancer Is a Specific Sub-Type of Cancer: A Morphological, Molecular and Genetics Study. PLoS ONE 2014, 9, e103159. [Google Scholar] [CrossRef] [PubMed]
- Silla, I.O.; Rueda, D.; Rodríguez, Y.; García, J.L.; Vigo, F.D.L.C.; Perea, J. Early-onset colorectal cancer: A separate subset of colorectal cancer. World J. Gastroenterol. 2014, 20, 17288–17296. [Google Scholar] [CrossRef]
- Ballester, V.; Rashtak, S.; Boardman, L. Clinical and molecular features of young-onset colorectal cancer. World J. Gastroenterol. 2016, 22, 1736–1744. [Google Scholar] [CrossRef]
- Yang, Y.; Du, L.; Shi, D.; Kong, C.; Liu, J.; Liu, G.; Li, X.; Ma, Y. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat. Commun. 2021, 12, 6757. [Google Scholar] [CrossRef]
- Ugai, T.; Väyrynen, J.P.; Lau, M.C.; Borowsky, J.; Akimoto, N.; Väyrynen, S.A.; Zhao, M.; Zhong, R.; Haruki, K.; Costa, A.D.; et al. Immune cell profiles in the tumor microenvironment of early-onset, intermediate-onset, and later-onset colorectal cancer. Cancer Immunol. Immunother. 2022, 71, 933–942. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Wu, T.; Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef]
- Fane, M.; Weeraratna, A.T. How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 2020, 20, 89–106. [Google Scholar] [CrossRef]
- Kugel, C.H., III; Douglass, S.M.; Webster, M.R.; Kaur, A.; Liu, Q.; Yin, X.; Weiss, S.A.; Darvishian, F.; Al-Rohil, R.N.; Ndoye, A.; et al. Age Correlates with Response to Anti-PD1, Reflecting Age-Related Differences in Intratumoral Effector and Regulatory T-Cell Populations. Clin. Cancer Res. 2018, 24, 5347–5356. [Google Scholar] [CrossRef]
- Machiraju, D.; Schäfer, S.; Hassel, J.C. Potential Reasons for Unresponsiveness to Anti-PD1 Immunotherapy in Young Patients with Advanced Melanoma. Life 2021, 11, 1318. [Google Scholar] [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Chen, H.; Ye, F.; Guo, G. Revolutionizing immunology with single-cell RNA sequencing. Cell. Mol. Immunol. 2019, 16, 242–249. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018, 37, 38–44. [Google Scholar] [CrossRef]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., II; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.-H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Wang, W.; Zhong, Y.; Zhuang, Z.; Xie, J.; Lu, Y.; Huang, C.; Sun, Y.; Wu, L.; Yin, J.; Yu, H.; et al. Multiregion single-cell sequencing reveals the transcriptional landscape of the immune microenvironment of colorectal cancer. Clin. Transl. Med. 2021, 11, e253. [Google Scholar] [CrossRef]
- Qian, J.; Olbrecht, S.; Boeckx, B.; Vos, H.; Laoui, D.; Etlioglu, E.; Wauters, E.; Pomella, V.; Verbandt, S.; Busschaert, P.; et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020, 30, 745–762. [Google Scholar] [CrossRef]
- Cheng, S.; Li, Z.; Gao, R.; Xing, B.; Gao, Y.; Yang, Y.; Qin, S.; Zhang, L.; Ouyang, H.; Du, P.; et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 2021, 184, 792–809.e23. [Google Scholar] [CrossRef]
- Pusztaszeri, M.P.; Seelentag, W.; Bosman, F.T. Immunohistochemical Expression of Endothelial Markers CD31, CD34, von Willebrand Factor, and Fli-1 in Normal Human Tissues. J. Histochem. Cytochem. 2006, 54, 385–395. [Google Scholar] [CrossRef]
- Angelin, A.; Gil-De-Gómez, L.; Dahiya, S.; Jiao, J.; Guo, L.; Levine, M.H.; Wang, Z.; Quinn, W.J., III; Kopinski, P.K.; Wang, L.; et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 2017, 25, 1282–1293.e7. [Google Scholar] [CrossRef] [PubMed]
- Polanczyk, M.J.; Walker, E.; Haley, D.; Guerrouahen, B.S.; Akporiaye, E.T. Blockade of TGF-beta signaling to enhance the antitumor response is accompanied by dysregulation of the functional activity of CD4(+)CD25(+)Foxp3(+) and CD4(+)CD25(-)Foxp3(+) T cells. J. Transl. Med. 2019, 17, 219. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Tokumaru, Y.; Angarita, F.A.; Lee, L.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Adipogenesis in triple-negative breast cancer is associated with unfavorable tumor immune microenvironment and with worse survival. Sci. Rep. 2021, 11, 12541. [Google Scholar] [CrossRef] [PubMed]
- Battaglin, F.; Puccini, A.; Intini, R.; Schirripa, M.; Ferro, A.; Bergamo, F.; Lonardi, S.; Zagonel, V.; Lenz, H.-J.; Loupakis, F. The role of tumor angiogenesis as a therapeutic target in colorectal cancer. Expert Rev. Anticancer. Ther. 2018, 18, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Maraskovsky, E.; Chen, W.F.; Shortman, K. IL-2 and IFN-gamma are two necessary lymphokines in the development of cytolytic T cells. J. Immunol. 1989, 143, 1210–1214. [Google Scholar]
- Curtsinger, J.M.; Agarwal, P.; Lins, D.C.; Mescher, M.F. Autocrine IFN-gamma promotes naive CD8 T cell differentiation and synergizes with IFN-alpha to stimulate strong function. J. Immunol. 2012, 189, 659–668. [Google Scholar] [CrossRef]
- Allie, S.R.; Bradley, J.E.; Mudunuru, U.; Schultz, M.D.; Graf, B.A.; Lund, F.E.; Randall, T.D. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 2019, 20, 97–108. [Google Scholar] [CrossRef]
- Freeman, G.J.; Freedman, A.S.; Segil, J.M.; Lee, G.; Whitman, J.F.; Nadler, L.M. B7, a new member of the Ig superfamily with unique expression on activated and neoplastic B cells. J. Immunol. 1989, 143, 2714–2722. [Google Scholar]
- Sahoo, N.C.; Rao, K.V.S.; Natarajan, K. CD80 Expression is Induced on Activated B Cells Following Stimulation by CD86. Scand. J. Immunol. 2002, 55, 577–584. [Google Scholar] [CrossRef]
- Lenschow, D.J.; Su, G.H.; Zuckerman, L.A.; Nabavi, N.; Jellis, C.L.; Gray, G.S.; Miller, J.; Bluestone, J.A. Expression and functional significance of an additional ligand for CTLA-4. Proc. Natl. Acad. Sci. USA 1993, 90, 11054–11058. [Google Scholar] [CrossRef]
- Schmidt, M.; Hellwig, B.; Hammad, S.; Othman, A.; Lohr, M.; Chen, Z.; Boehm, D.; Gebhard, S.; Petry, I.; Lebrecht, A.; et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin. Cancer Res. 2012, 18, 2695–2703. [Google Scholar] [CrossRef]
- Lohr, M.; Edlund, K.; Botling, J.; Hammad, S.; Hellwig, B.; Othman, A.; Berglund, A.; Lambe, M.; Holmberg, L.; Ekman, S.; et al. The prognostic relevance of tumour-infiltrating plasma cells and immunoglobulin kappa C indicates an important role of the humoral immune response in non-small cell lung cancer. Cancer Lett. 2013, 333, 222–228. [Google Scholar] [CrossRef]
- Kroeger, D.R.; Milne, K.; Nelson, B.H. Tumor-Infiltrating Plasma Cells Are Associated with Tertiary Lymphoid Structures, Cytolytic T-Cell Responses, and Superior Prognosis in Ovarian Cancer. Clin. Cancer Res. 2016, 22, 3005–3015. [Google Scholar] [CrossRef]
- Inada, K.; Okada, S.; Phuchareon, J.; Hatano, M.; Sugimoto, T.; Moriya, H.; Tokuhisa, T. c-Fos induces apoptosis in germinal center B cells. J. Immunol. 1998, 161, 3853–3861. [Google Scholar]
- Zhang, Q.; He, Y.; Luo, N.; Patel, S.J.; Han, Y.; Gao, R.; Modak, M.; Carotta, S.; Haslinger, C.; Kind, D.; et al. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 2019, 179, 829–845.e20. [Google Scholar] [CrossRef]
- Baitsch, D.; Bock, H.H.; Engel, T.; Telgmann, R.; Müller-Tidow, C.; Varga, G.; Bot, M.; Herz, J.; Robenek, H.; von Eckardstein, A.; et al. Apolipoprotein E Induces Antiinflammatory Phenotype in Macrophages. Arter. Thromb. Vasc. Biol. 2011, 31, 1160–1168. [Google Scholar] [CrossRef]
- Rao, G.; Wang, H.; Li, B.; Huang, L.; Xue, D.; Wang, X.; Jin, H.; Wang, J.; Zhu, Y.; Lu, Y.; et al. Reciprocal Interactions between Tumor-Associated Macrophages and CD44-Positive Cancer Cells via Osteopontin/CD44 Promote Tumorigenicity in Colorectal Cancer. Clin. Cancer Res. 2013, 19, 785–797. [Google Scholar] [CrossRef]
- Qin, X.; Yan, M.; Wang, X.; Xu, Q.; Wang, X.; Zhu, X.; Shi, J.; Li, Z.; Zhang, J.; Chen, W. Cancer-associated Fibroblast-derived IL-6 Promotes Head and Neck Cancer Progression via the Osteopontin-NF-kappa B Signaling Pathway. Theranostics 2018, 8, 921–940. [Google Scholar] [CrossRef]
- Chen, R.-H.; Xiao, Z.-W.; Yan, X.-Q.; Han, P.; Liang, F.-Y.; Wang, J.-Y.; Yu, S.-T.; Zhang, T.-Z.; Chen, S.-Q.; Zhong, Q.; et al. Tumor Cell-Secreted ISG15 Promotes Tumor Cell Migration and Immune Suppression by Inducing the Macrophage M2-Like Phenotype. Front. Immunol. 2020, 11, 594775. [Google Scholar] [CrossRef]
- Marciscano, A.E.; Anandasabapathy, N. The role of dendritic cells in cancer and anti-tumor immunity. Semin. Immunol. 2021, 52, 101481. [Google Scholar] [CrossRef]
- Verneau, J.; Sautés-Fridman, C.; Sun, C.-M. Dendritic cells in the tumor microenvironment: Prognostic and theranostic impact. Semin. Immunol. 2020, 48, 101410. [Google Scholar] [CrossRef]
- Klement, J.D.; Paschall, A.V.; Redd, P.S.; Ibrahim, M.L.; Lu, C.; Yang, D.; Celis, E.; Abrams, S.I.; Ozato, K.; Liu, K. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J. Clin. Investig. 2018, 128, 5549–5560. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Sun, Y.M. The IL-6/JAK/STAT3 pathway: Potential therapeutic strategies in treating colorectal cancer (Review). Int. J. Oncol. 2014, 44, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Ng, A.S.; Cai, S.; Li, Q.; Yang, L.; Kerr, D. Novel therapeutic strategies: Targeting epithelial–mesenchymal transition in colorectal cancer. Lancet Oncol. 2021, 22, e358–e368. [Google Scholar] [CrossRef] [PubMed]
- Poon, E.; Harris, A.; Ashcroft, M. Targeting the hypoxia-inducible factor (HIF) pathway in cancer. Expert Rev. Mol. Med. 2009, 11, e26. [Google Scholar] [CrossRef]
- Gong, Y.; Liu, Y.; Wang, T.; Li, Z.; Gao, L.; Chen, H.; Shu, Y.; Li, Y.; Xu, H.; Zhou, Z.; et al. Age-Associated Proteomic Signatures and Potential Clinically Actionable Targets of Colorectal Cancer. Mol. Cell. Proteom. 2021, 20, 100115. [Google Scholar] [CrossRef]
- Kulkarni, N.; Meitei, H.T.; Sonar, S.A.; Sharma, P.K.; Mujeeb, V.R.; Srivastava, S.; Boppana, R.; Lal, G. CCR6 signaling inhibits suppressor function of induced-Treg during gut inflammation. J. Autoimmun. 2018, 88, 121–130. [Google Scholar] [CrossRef]
- Nobre, C.C.G.; De Araújo, J.M.G.; Fernandes, T.A.A.D.M.; Cobucci, R.N.O.; Lanza, D.C.F.; Andrade, V.S.; Fernandes, J.V. Macrophage Migration Inhibitory Factor (MIF): Biological Activities and Relation with Cancer. Pathol. Oncol. Res. 2017, 23, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Noe, J.T.; Mitchell, R.A. MIF-Dependent Control of Tumor Immunity. Front. Immunol. 2020, 11, 609948. [Google Scholar] [CrossRef]
- Oh, J.; Magnuson, A.; Benoist, C.; Pittet, M.J.; Weissleder, R. Age-related tumor growth in mice is related to integrin α 4 in CD8+ T cells. J. Clin. Investig. 2018, 3, e122961. [Google Scholar] [CrossRef]
- Saule, P.; Trauet, J.; Dutriez, V.; Lekeux, V.; Dessaint, J.-P.; Labalette, M. Accumulation of memory T cells from childhood to old age: Central and effector memory cells in CD4+ versus effector memory and terminally differentiated memory cells in CD8+ compartment. Mech. Ageing Dev. 2006, 127, 274–281. [Google Scholar] [CrossRef]
- Kumar, P.; Bhattacharya, P.; Prabhakar, B.S. A comprehensive review on the role of co-signaling receptors and Treg homeostasis in autoimmunity and tumor immunity. J. Autoimmun. 2018, 95, 77–99. [Google Scholar] [CrossRef]
- Zappasodi, R.; Serganova, I.; Cohen, I.J.; Maeda, M.; Shindo, M.; Senbabaoglu, Y.; Watson, M.J.; Leftin, A.; Maniyar, R.; Verma, S.; et al. CTLA-4 blockade drives loss of Treg stability in glycolysis-low tumours. Nature 2021, 591, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Oberle, N.; Krammer, P.H. Molecular Mechanisms of Treg-Mediated T Cell Suppression. Front. Immunol. 2012, 3, 51. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, V.; Di Rella, F.; Di Giacomo, A.; Matarese, G. Regulatory T cells as suppressors of anti-tumor immunity: Role of metabolism. Cytokine Growth Factor Rev. 2017, 35, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-A.; Li, X.-L.; Mo, Y.-Z.; Fan, C.-M.; Tang, L.; Xiong, F.; Guo, C.; Xiang, B.; Zhou, M.; Ma, J.; et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol. Cancer 2018, 17, 168. [Google Scholar] [CrossRef] [PubMed]
- Tsaknaridis, L.; Spencer, L.; Culbertson, N.; Hicks, K.; LaTocha, D.; Chou, Y.K.; Whitham, R.H.; Bakke, A.; Jones, R.E.; Offner, H.; et al. Functional assay for human CD4+CD25+ Treg cells reveals an age-dependent loss of suppressive activity. J. Neurosci. Res. 2003, 74, 296–308. [Google Scholar] [CrossRef]
- Scholz, J.L.; Diaz, A.; Riley, R.L.; Cancro, M.P.; Frasca, D. A comparative review of aging and B cell function in mice and humans. Curr. Opin. Immunol. 2013, 25, 504–510. [Google Scholar] [CrossRef]
- Tesar, B.M.; Walker, W.E.; Unternaehrer, J.; Joshi, N.S.; Chandele, A.; Haynes, L.; Kaech, S.; Goldstein, D.R. Murine [corrected] myeloid dendritic cell-dependent toll-like receptor immunity is preserved with aging. Aging Cell 2006, 5, 473–486. [Google Scholar] [CrossRef]
- Asquith, M.; Haberthur, K.; Brown, M.; Engelmann, F.; Murphy, A.; Al-Mahdi, Z.; Messaoudi, I. Age-dependent changes in innate immune phenotype and function in rhesus macaques (Macaca mulatta). Pathobiol. Aging Age-Relat. Dis. 2012, 2, 18052. [Google Scholar] [CrossRef]
- Liu, X.; Yu, H.; Yan, C.; Mei, Y.; Lin, C.; Hong, Y.; Lin, X.; Zhang, Q.; Yu, J. Plasmacytoid Dendritic Cells and ICOS+ Regulatory T Cells Predict Poor Prognosis in Gastric Cancer: A Pilot Study. J. Cancer 2019, 10, 6711–6715. [Google Scholar] [CrossRef]
- Treilleux, I.; Blay, J.-Y.; Bendriss-Vermare, N.; Ray-Coquard, I.; Bachelot, T.; Guastalla, J.-P.; Bremond, A.; Goddard, S.; Pin, J.-J.; Barthelemy-Dubois, C.; et al. Dendritic Cell Infiltration and Prognosis of Early Stage Breast Cancer. Clin. Cancer Res. 2004, 10, 7466–7474. [Google Scholar] [CrossRef]
- Yan, X.; Orentas, R.J.; Johnson, B.D. Tumor-derived macrophage migration inhibitory factor (MIF) inhibits T lymphocyte activation. Cytokine 2006, 33, 188–198. [Google Scholar] [CrossRef]
- Klasen, C.; Ohl, K.; Sternkopf, M.; Shachar, I.; Schmitz, C.; Heussen, N.; Hobeika, E.; Levit-Zerdoun, E.; Tenbrock, K.; Reth, M.; et al. MIF Promotes B Cell Chemotaxis through the Receptors CXCR4 and CD74 and ZAP-70 Signaling. J. Immunol. 2014, 192, 5273–5284. [Google Scholar] [CrossRef]
- Wang, X.; Chen, T.; Leng, L.; Fan, J.; Cao, K.; Duan, Z.; Zhang, X.; Shao, C.; Wu, M.; Tadmori, I.; et al. MIF Produced by Bone Marrow–Derived Macrophages Contributes to Teratoma Progression after Embryonic Stem Cell Transplantation. Cancer Res. 2012, 72, 2867–2878. [Google Scholar] [CrossRef]
- White, E.; Strom, S.R.B.; Wys, N.L.; Arenberg, D. Non-Small Cell Lung Cancer Cells Induce Monocytes to Increase Expression of Angiogenic Activity. J. Immunol. 2001, 166, 7549–7555. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.-M.; Xiao, G.-Z.; Qin, P.-F.; Wan, X.-Y.; Fu, Y.-J.; Zheng, Y.-H.; Luo, M.-Y.; Ren, D.-L.; Liu, S.-P.; Chen, H.-X.; et al. Single-Cell RNA Sequencing Reveals Heterogeneity in the Tumor Microenvironment between Young-Onset and Old-Onset Colorectal Cancer. Biomolecules 2022, 12, 1860. https://doi.org/10.3390/biom12121860
Li G-M, Xiao G-Z, Qin P-F, Wan X-Y, Fu Y-J, Zheng Y-H, Luo M-Y, Ren D-L, Liu S-P, Chen H-X, et al. Single-Cell RNA Sequencing Reveals Heterogeneity in the Tumor Microenvironment between Young-Onset and Old-Onset Colorectal Cancer. Biomolecules. 2022; 12(12):1860. https://doi.org/10.3390/biom12121860
Chicago/Turabian StyleLi, Gui-Ming, Guo-Zhong Xiao, Peng-Fei Qin, Xing-Yang Wan, Yuan-Ji Fu, Yi-Hui Zheng, Min-Yi Luo, Dong-Lin Ren, Shi-Ping Liu, Hua-Xian Chen, and et al. 2022. "Single-Cell RNA Sequencing Reveals Heterogeneity in the Tumor Microenvironment between Young-Onset and Old-Onset Colorectal Cancer" Biomolecules 12, no. 12: 1860. https://doi.org/10.3390/biom12121860
APA StyleLi, G.-M., Xiao, G.-Z., Qin, P.-F., Wan, X.-Y., Fu, Y.-J., Zheng, Y.-H., Luo, M.-Y., Ren, D.-L., Liu, S.-P., Chen, H.-X., & Lin, H.-C. (2022). Single-Cell RNA Sequencing Reveals Heterogeneity in the Tumor Microenvironment between Young-Onset and Old-Onset Colorectal Cancer. Biomolecules, 12(12), 1860. https://doi.org/10.3390/biom12121860




