Comprehensive Analysis of Transcriptomics and Genetic Alterations Identifies Potential Mechanisms Underlying Anthracycline Therapy Resistance in Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Download and Analysis
2.2. Differentially Expressed Genes Analysis
2.3. Genetic Alterations and Tumor Mutation Burden (TMB) Estimation
2.4. Immune Infiltration Analysis
2.5. Gene Set Enrichment Analysis (GSEA)
2.6. Statistical Analysis
3. Results
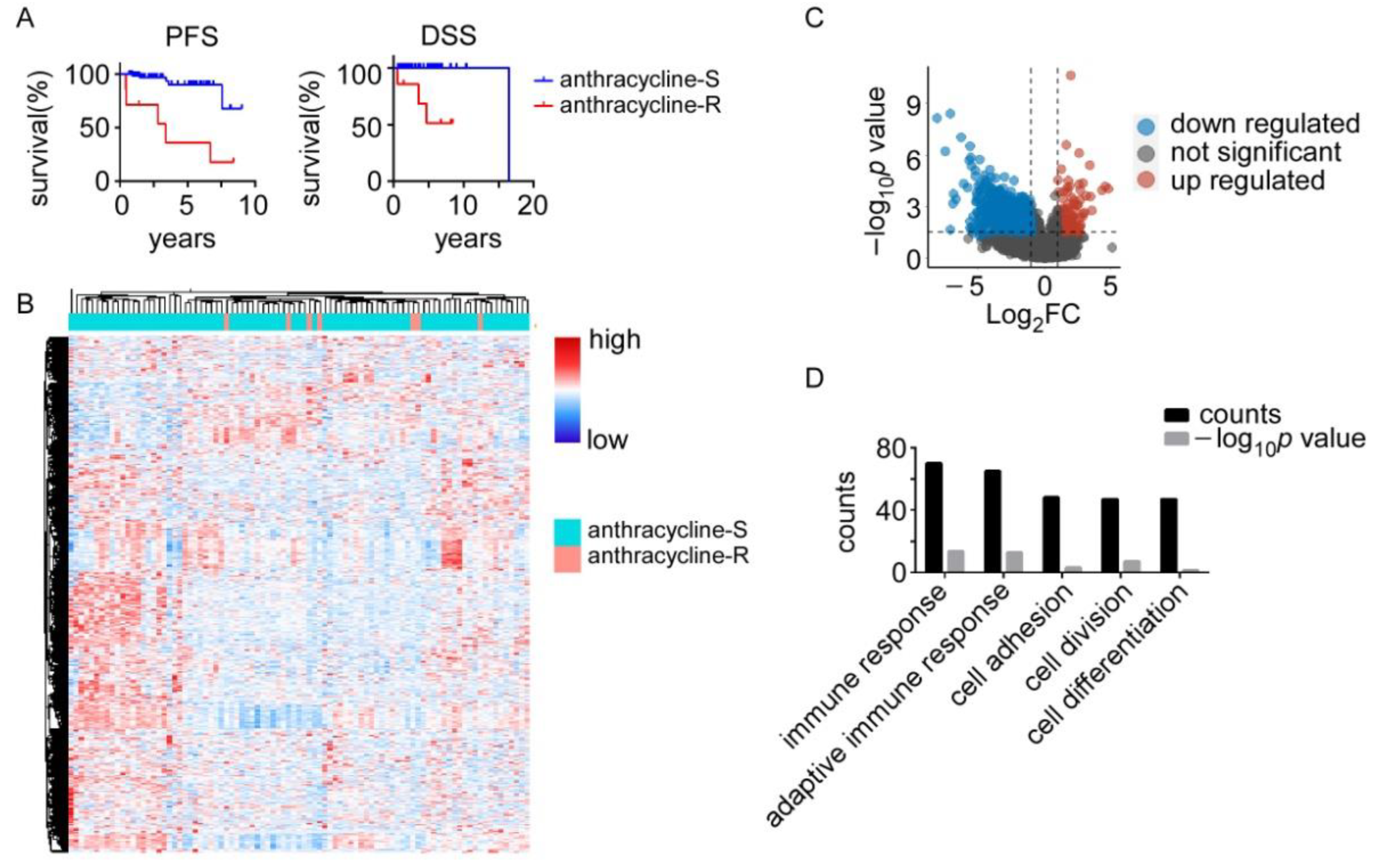
3.1. Anthracycline-Sensitive (Anthracycline-S) and -Resistant (Anthracycline-R) Tumors Display Distinct Transcriptomic Profiles
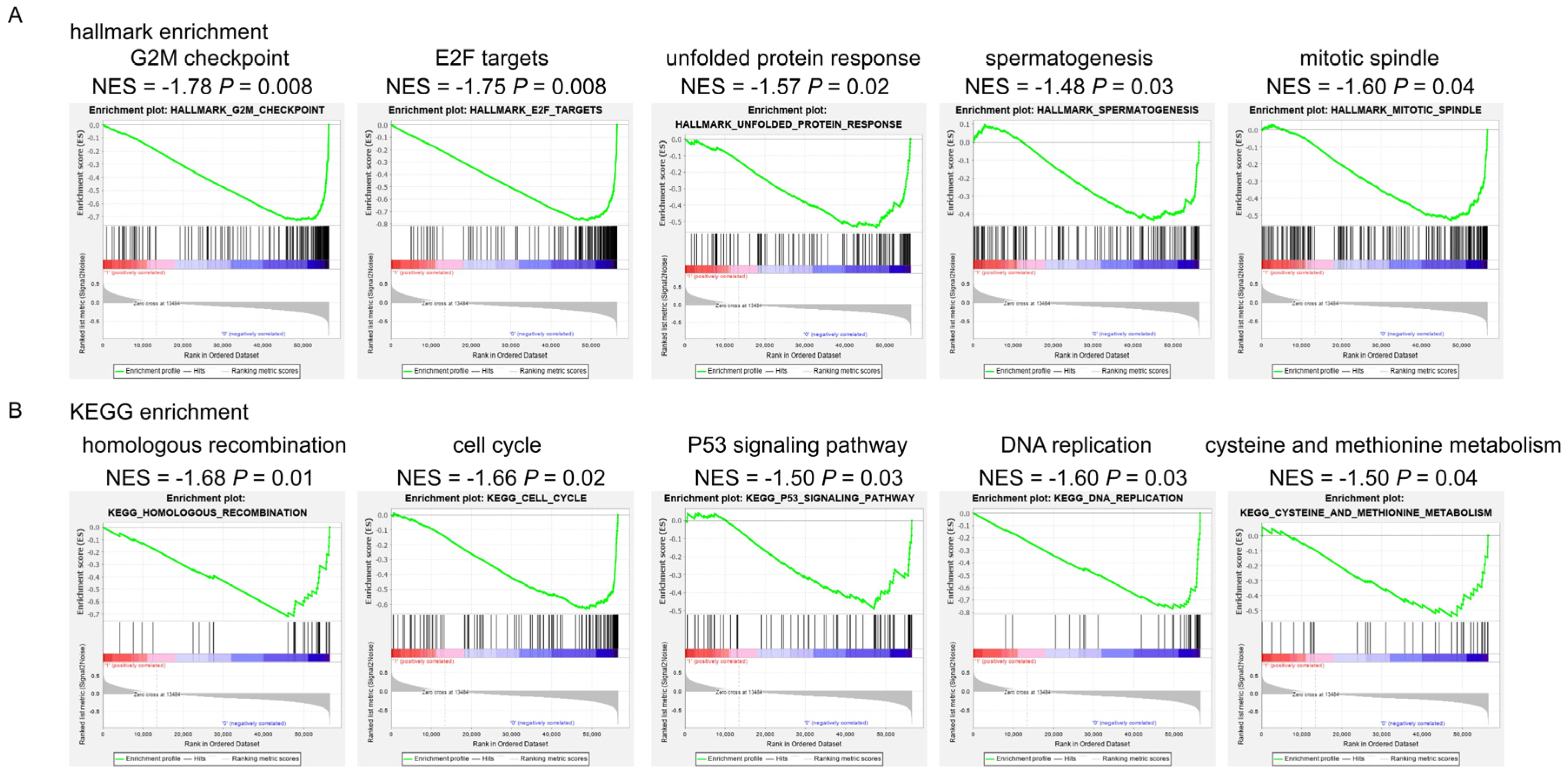
3.2. Signaling Pathways Involved in Anthracycline Sensitivity
3.3. Somatic Genetic Alterations in the Anthracycline-S and Anthracycline-R Tumors
3.4. Immune Infiltration Patterns in the Anthracycline-S and Anthracycline-R Tumors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Longley, D.; Johnston, P. Molecular mechanisms of drug resistance. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Hurvitz, S.A.; McAndrew, N.P.; Bardia, A.; Press, M.F.; Pegram, M.; Crown, J.P.; Fasching, P.A.; Ejlertsen, B.; Yang, E.H.; Glaspy, A.J. A careful reassessment of anthracycline use in curable breast cancer. NPJ Breast Cancer 2021, 7, 134. [Google Scholar] [CrossRef] [PubMed]
- Sritharan, S.; Sivalingam, N. A comprehensive review on time-tested anticancer drug doxorubicin. Life Sci. 2021, 278, 119527. [Google Scholar] [CrossRef] [PubMed]
- Sallustio, B.C.; Boddy, A.V. Is there scope for better individualisation of anthracycline cancer chemotherapy? Br. J. Clin. Pharmacol. 2021, 87, 295–305. [Google Scholar] [CrossRef]
- Martins-Teixeira, M.B.; Carvalho, I. Antitumour anthracyclines: Progress and perspectives. ChemMedChem 2020, 15, 933–948. [Google Scholar] [CrossRef]
- Press, M.F.; Sauter, G.; Buyse, M.; Bernstein, L.; Guzman, R.; Santiago, A.; Villalobos, I.E.; Eiermann, W.; Pienkowski, T.; Martin, M. Alteration of topoisomerase ii–alpha gene in human breast cancer: Association with responsiveness to anthracycline-based chemotherapy. J. Clin. Oncol. 2011, 29, 859. [Google Scholar] [CrossRef]
- Hansen, S.N.; Westergaard, D.; Thomsen, M.B.H.; Vistesen, M.; Do, K.N.; Fogh, L.; Belling, K.C.; Wang, J.; Yang, H.; Gupta, R. Acquisition of docetaxel resistance in breast cancer cells reveals upregulation of abcb1 expression as a key mediator of resistance accompanied by discrete upregulation of other specific genes and pathways. Tumor Biol. 2015, 36, 4327–4338. [Google Scholar] [CrossRef]
- Pljesa-Ercegovac, M.; Savic-Radojevic, A.; Matic, M.; Coric, V.; Djukic, T.; Radic, T.; Simic, T. Glutathione transferases: Potential targets to overcome chemoresistance in solid tumors. Int. J. Mol. Sci. 2018, 19, 3785. [Google Scholar] [CrossRef]
- Guo, B.; Tam, A.; Santi, S.A.; Parissenti, A.M. Role of autophagy and lysosomal drug sequestration in acquired resistance to doxorubicin in mcf-7 cells. BMC Cancer 2016, 16, 762. [Google Scholar] [CrossRef]
- Harbottle, A.; Daly, A.K.; Atherton, K.; Campbell, F.C. Role of glutathione s-transferase p1, p-glycoprotein and multidrug resistance-associated protein 1 in acquired doxorubicin resistance. Int. J. Cancer 2001, 92, 777–783. [Google Scholar] [CrossRef]
- Knappskog, S.; Leirvaag, B.; Gansmo, L.B.; Romundstad, P.; Hveem, K.; Vatten, L.; Lønning, P.E. Prevalence of the chek2 r95* germline mutation. Hered. Cancer Clin. Pract. 2016, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Waks, A.G.; Cohen, O.; Kochupurakkal, B.; Kim, D.; Dunn, C.E.; Buendia, J.B.; Wander, S.; Helvie, K.; Lloyd, M.R.; Marini, L. Reversion and non-reversion mechanisms of resistance to parp inhibitor or platinum chemotherapy in brca1/2-mutant metastatic breast cancer. Ann. Oncol. 2020, 31, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Razavi, P.; Dickler, M.N.; Shah, P.D.; Toy, W.; Brown, D.N.; Won, H.H.; Li, B.T.; Shen, R.; Vasan, N.; Modi, S. Alterations in pten and esr1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat. Cancer 2020, 1, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Bertheau, P.; Plassa, F.; Espie, M.; Turpin, E.; de Roquancourt, A.; Marty, M.; Lerebours, F.; Beuzard, Y.; Janin, A.; de Thé, H. Effect of mutated tp53 on response of advanced breast cancers to high-dose chemotherapy. Lancet 2002, 360, 852–854. [Google Scholar] [CrossRef] [PubMed]
- Bertheau, P.; Turpin, E.; Rickman, D.S.; Espié, M.; de Reynies, A.; Feugeas, J.-P.; Plassa, L.-F.; Soliman, H.; Varna, M.; de Roquancourt, A. Exquisite sensitivity of tp53 mutant and basal breast cancers to a dose-dense epirubicin− cyclophosphamide regimen. PLoS Med. 2007, 4, e90. [Google Scholar] [CrossRef]
- Lehmann-Che, J.; André, F.; Desmedt, C.; Mazouni, C.; Giacchetti, S.; Turpin, E.; Espié, M.; Plassa, L.-F.; Marty, M.; Bertheau, P. Cyclophosphamide dose intensification may circumvent anthracycline resistance of p53 mutant breast cancers. Oncologist 2010, 15, 246–252. [Google Scholar] [CrossRef]
- Varna, M.; Lehmann-Che, J.; Turpin, E.; Marangoni, E.; El-Bouchtaoui, M.; Jeanne, M.; Grigoriu, C.; Ratajczak, P.; Leboeuf, C.; Plassa, F.L. P53 dependent cell-cycle arrest triggered by chemotherapy in xenografted breast tumors. Int. J. Cancer 2009, 124, 991–997. [Google Scholar] [CrossRef]
- King, J.; Mir, H.; Singh, S. Association of cytokines and chemokines in pathogenesis of breast cancer. Prog. Mol. Biol. Transl. Sci. 2017, 151, 113–136. [Google Scholar]
- Cancelliere, R.; di Tinno, A.; di Lellis, A.M.; Contini, G.; Micheli, L.; Signori, E. Cost-effective and disposable label-free voltammetric immunosensor for sensitive detection of interleukin-6. Biosens. Bioelectron. 2022, 213, 114467. [Google Scholar] [CrossRef]
- Micallef, I.; Baron, B. Doxorubicin: An overview of the anti-cancer and chemoresistance mechanisms. Ann. Clin. Toxicol. 2020, 3, 1031. [Google Scholar]
- Aas, T.; Børresen, A.-L.; Geisler, S.; Smith-Sørensen, B.; Johnsen, H.; Varhaug, J.E.; Akslen, L.A.; Lønning, P.E. Specific p53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat. Med. 1996, 2, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Børresen, A.L.; Andersen, T.I.; Eyfjörd, J.E.; Cornelis, R.S.; Thorlacius, S.; Borg, Å.; Johansson, U.; Theillet, C.; Scherneck, S.; Hartman, S. Tp53 mutations and breast cancer prognosis: Particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer 1995, 14, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, D.; Gasco, M.; Hiller, L.; Sullivan, A.; Syed, N.; Trigiante, G.; Yulug, I.; Merlano, M.; Numico, G.; Comino, A. P53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell 2003, 3, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.P.C.; Tsang, W.P.; Chau, P.Y.; Co, N.N.; Tsang, T.Y.; Kwok, T.T. P53-r273h gains new function in induction of drug resistance through down-regulation of procaspase-3. Mol. Cancer Ther. 2007, 6, 1054–1061. [Google Scholar] [CrossRef]
- McVeigh, T.P.; Choi, J.K.; Miller, N.M.; Green, A.J.; Kerin, M.J. Lobular breast cancer in a cdh1 splice site mutation carrier: Case report and review of the literature. Clin. Breast Cancer 2014, 14, e47–e51. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Dai, X.; Wan, L.; Wang, H.; Zhang, J.; Goff, P.S.; Sviderskaya, E.V.; Xuan, Z.; Xu, Z.; Xu, X. The e3 ligase apc/ccdh1 promotes ubiquitylation-mediated proteolysis of pax3 to suppress melanocyte proliferation and melanoma growth. Sci. Signal. 2015, 8, ra87. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Y.; Cui, Z. Identification of adriamycin resistance genes in breast cancer based on microarray data analysis. Transl. Cancer Res. 2020, 9, 7486. [Google Scholar] [CrossRef]
- Parkes, E.E.; Savage, K.I.; Lioe, T.; Boyd, C.; Halliday, S.; Walker, S.M.; Lowry, K.; Knight, L.; Buckley, N.E.; Grogan, A. Activation of a cgas-sting-mediated immune response predicts response to neoadjuvant chemotherapy in early breast cancer. Br. J. Cancer 2022, 126, 247–258. [Google Scholar] [CrossRef]
- Mezni, E.; Behi, K.; Gonçalves, A. Immunotherapy and breast cancer: An overview. Curr. Opin. Oncol. 2022, 34, 587–594. [Google Scholar] [CrossRef]
- Lee, J.S.; Yost, S.E.; Yuan, Y. Neoadjuvant treatment for triple negative breast cancer: Recent progresses and challenges. Cancers 2020, 12, 1404. [Google Scholar] [CrossRef]
- Brown, L.C.; Loi, S. Immune checkpoint inhibition in the treatment of early stage triple negative breast cancer: 2021 update. Breast 2022, 62, S29–S33. [Google Scholar] [CrossRef] [PubMed]
- Ching, T.; Huang, S.; Garmire, L.X. Power analysis and sample size estimation for rna-seq differential expression. Rna 2014, 20, 1684–1696. [Google Scholar] [CrossRef] [PubMed]
- Geistlinger, L.; Csaba, G.; Santarelli, M.; Ramos, M.; Schiffer, L.; Turaga, N.; Law, C.; Davis, S.; Carey, V.; Morgan, M. Toward a gold standard for benchmarking gene set enrichment analysis. Brief. Bioinform. 2021, 22, 545–556. [Google Scholar] [CrossRef] [PubMed]
- AbuHammad, S.; Zihlif, M. Gene expression alterations in doxorubicin resistant mcf7 breast cancer cell line. Genomics 2013, 101, 213–220. [Google Scholar] [CrossRef]
- Ciocan-Cartita, C.A.; Jurj, A.; Zanoaga, O.; Cojocneanu, R.; Pop, L.-A.; Moldovan, A.; Moldovan, C.; Zimta, A.A.; Raduly, L.; Pop-Bica, C. New insights in gene expression alteration as effect of doxorubicin drug resistance in triple negative breast cancer cells. J. Exp. Clin. Cancer Res. 2020, 39, 241. [Google Scholar] [CrossRef]
- He, D.-X.; Gu, F.; Gao, F.; Hao, J.-j.; Gong, D.; Gu, X.-T.; Mao, A.-Q.; Jin, J.; Fu, L.; Ma, X. Genome-wide profiles of methylation, micrornas, and gene expression in chemoresistant breast cancer. Sci. Rep. 2016, 6, 24706. [Google Scholar] [CrossRef]
- McGuirk, S.; Audet-Delage, Y.; Annis, M.G.; Xue, Y.; Vernier, M.; Zhao, K.; St-Louis, C.; Minarrieta, L.; Patten, D.A.; Morin, G. Resistance to different anthracycline chemotherapeutics elicits distinct and actionable primary metabolic dependencies in breast cancer. eLife 2021, 10, e65150. [Google Scholar] [CrossRef]
- Gradishar, W.J.; Moran, M.S.; Abraham, J.; Aft, R.; Agnese, D.; Allison, K.H.; Anderson, B.; Burstein, H.J.; Chew, H.; Dang, C.; et al. Breast cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2022; 20, 691–722. [Google Scholar]
- Gradishar, W.J. NCCN guidelines updates: Management of patients with her2-negative breast cancer. J. Natl. Compr. Cancer Netw. 2022, 20, 561–565. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Gao, J.; Gu, R.; Shi, Y.; Hu, H.; Liu, J.; Huang, J.; Zhong, C.; Zhou, W.; Yang, Y.; et al. Comprehensive Analysis of Transcriptomics and Genetic Alterations Identifies Potential Mechanisms Underlying Anthracycline Therapy Resistance in Breast Cancer. Biomolecules 2022, 12, 1834. https://doi.org/10.3390/biom12121834
Liu Z, Gao J, Gu R, Shi Y, Hu H, Liu J, Huang J, Zhong C, Zhou W, Yang Y, et al. Comprehensive Analysis of Transcriptomics and Genetic Alterations Identifies Potential Mechanisms Underlying Anthracycline Therapy Resistance in Breast Cancer. Biomolecules. 2022; 12(12):1834. https://doi.org/10.3390/biom12121834
Chicago/Turabian StyleLiu, Zihao, Jingbo Gao, Ran Gu, Yu Shi, Hong Hu, Jianlan Liu, Jiefeng Huang, Caineng Zhong, Wenbin Zhou, Yaping Yang, and et al. 2022. "Comprehensive Analysis of Transcriptomics and Genetic Alterations Identifies Potential Mechanisms Underlying Anthracycline Therapy Resistance in Breast Cancer" Biomolecules 12, no. 12: 1834. https://doi.org/10.3390/biom12121834
APA StyleLiu, Z., Gao, J., Gu, R., Shi, Y., Hu, H., Liu, J., Huang, J., Zhong, C., Zhou, W., Yang, Y., & Gong, C. (2022). Comprehensive Analysis of Transcriptomics and Genetic Alterations Identifies Potential Mechanisms Underlying Anthracycline Therapy Resistance in Breast Cancer. Biomolecules, 12(12), 1834. https://doi.org/10.3390/biom12121834




