Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phylogenetic Distributions across the Dus Superfamily
2.2. Dus2 Fusion Analysis
2.3. Dus2 Sequence Logos
2.4. AlphaFold Models
2.5. Cloning of dsRBD of Dus2 from Amphimedon Queenslandica
2.6. Overexpression and Purification of dsRBDAq
2.7. Crystallization, Data Collection, and Structure Determination
3. Results and Discussion
3.1. Phylogenetic Distribution of Dus2 and Domain Analyses
3.2. Structural Analysis of the Dus2 Canonical Domains
3.2.1. Catalytic Domain (TIM-Barrel)
3.2.2. Active Site
3.2.3. Helical Domain
3.3. Structural Extensions of Dus2
3.3.1. Unstructured Extensions
3.3.2. Structured Extensions and the Connecting α-Helix
3.4. Modularity of Dus2
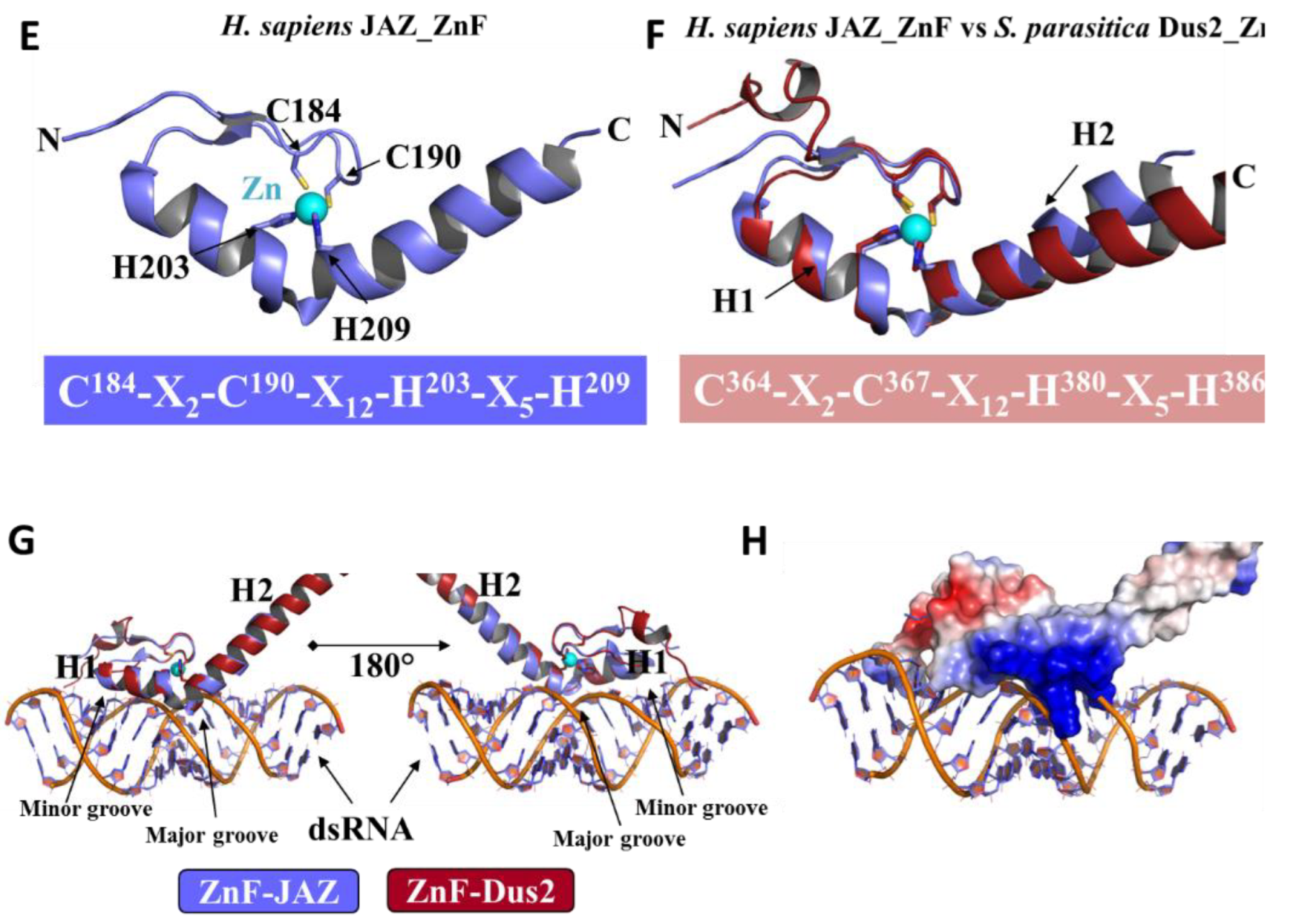
3.4.1. Zinc-Finger Domain
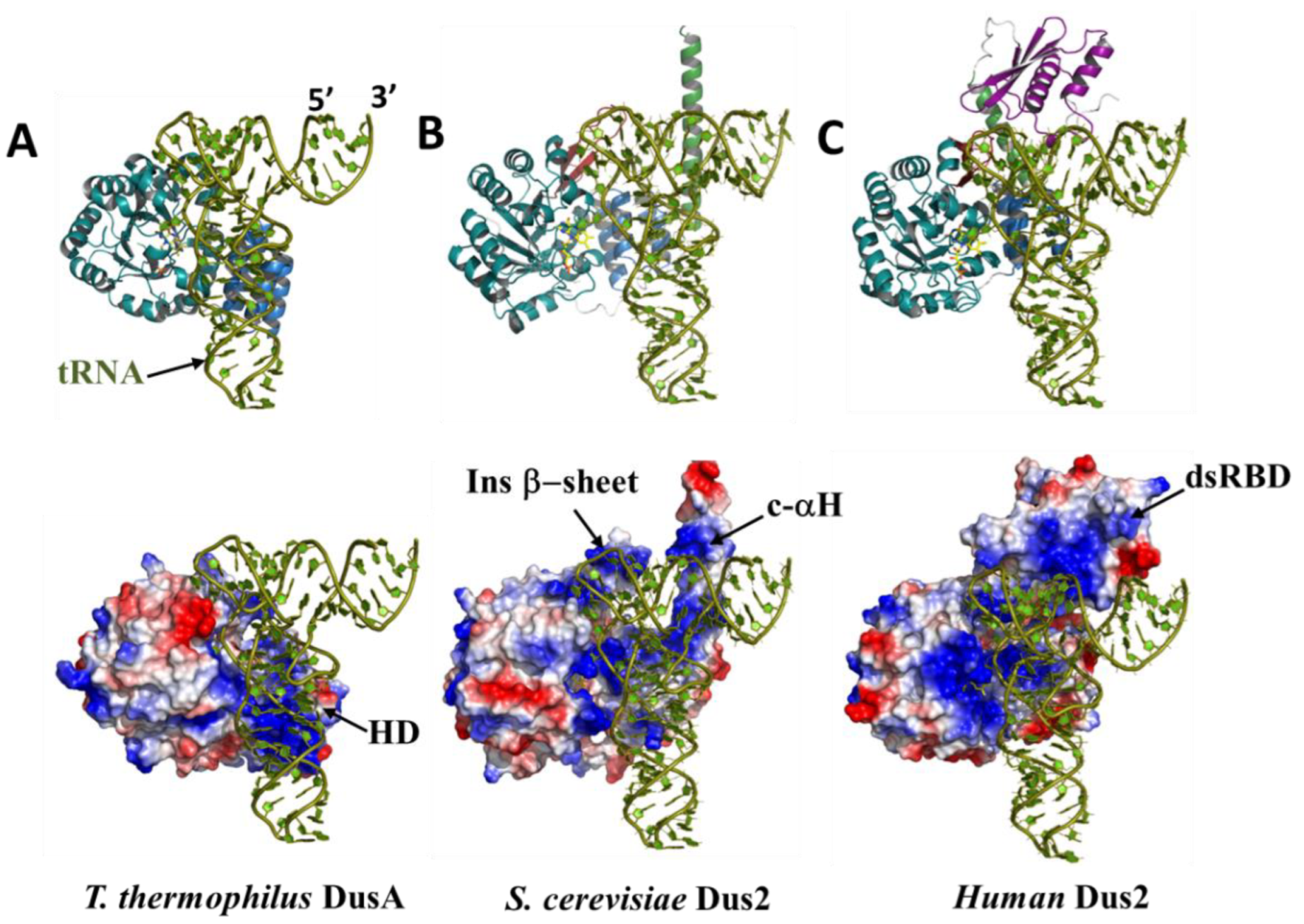
3.4.2. The Double-Stranded Binding Domain
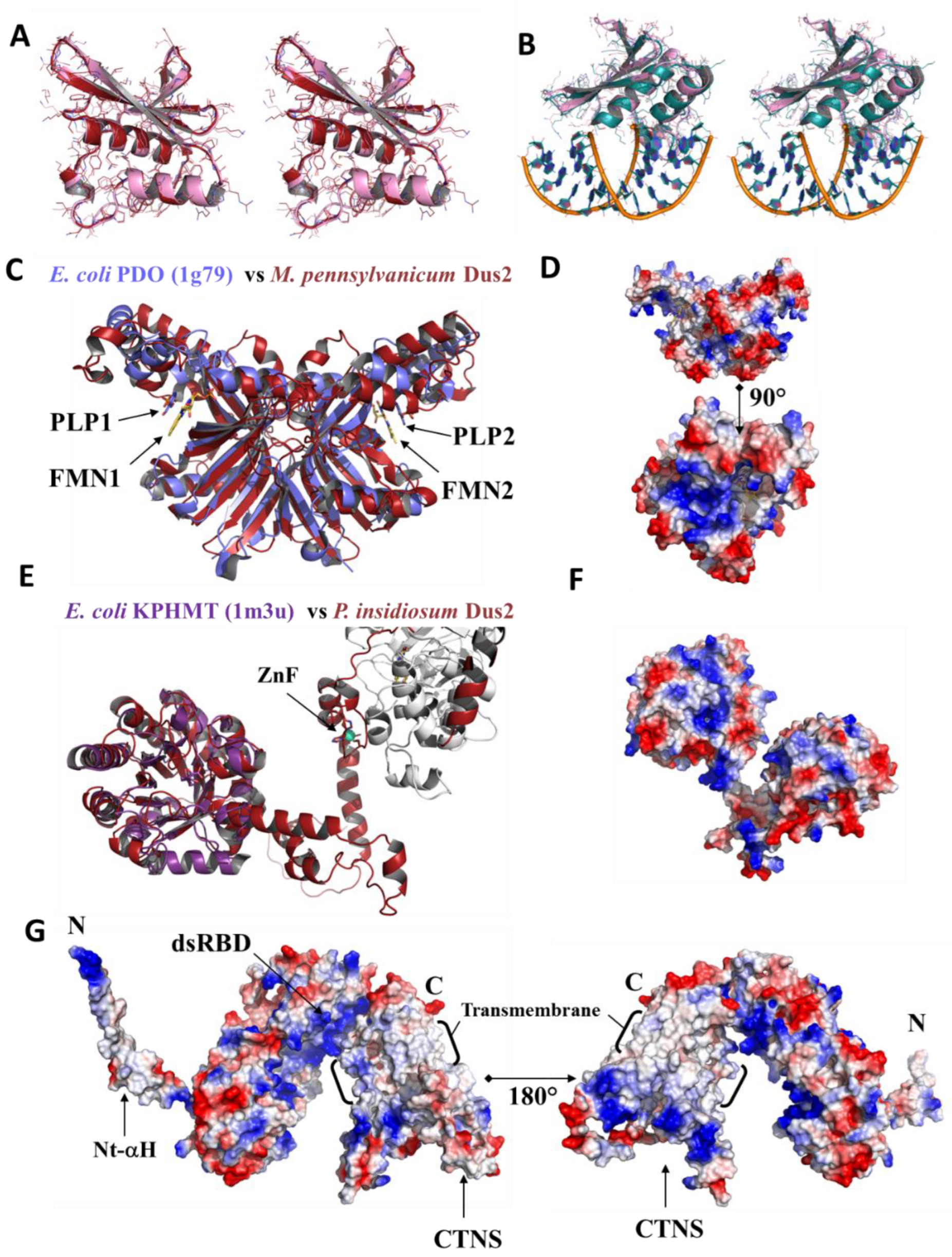
3.4.3. Pyridoxamine 5′-Phosphate Oxidase Domain
3.4.4. A Newly Identified Rossman Fold Domain
3.4.5. ICL_KPHMT and CTNS Domains
3.5. The functional Role of Dus2 Extensions and the Evolution of tRNA Binding
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Finet, O.; Yague-Sanz, C.; Marchand, F.; Hermand, D. The Dihydrouridine landscape from tRNA to mRNA: A perspective on synthesis, structural impact and function. RNA Biol. 2022, 19, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Bregeon, D.; Pecqueur, L.; Toubdji, S.; Sudol, C.; Lombard, M.; Fontecave, M.; de Crecy-Lagard, V.; Motorin, Y.; Helm, M.; Hamdane, D. Dihydrouridine in the Transcriptome: New Life for This Ancient RNA Chemical Modification. ACS Chem. Biol. 2022, 17, 1638–1657. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef] [PubMed]
- Draycott, A.S.; Schaening-Burgos, C.; Rojas-Duran, M.F.; Wilson, L.; Scharfen, L.; Neugebauer, K.M.; Nachtergaele, S.; Gilbert, W.V. Transcriptome-wide mapping reveals a diverse dihydrouridine landscape including mRNA. PLoS Biol. 2022, 20, e3001622. [Google Scholar] [CrossRef]
- Dai, W.; Li, A.; Yu, N.J.; Nguyen, T.; Leach, R.W.; Wuhr, M.; Kleiner, R.E. Activity-based RNA-modifying enzyme probing reveals DUS3L-mediated dihydrouridylation. Nat. Chem. Biol. 2021, 17, 1178–1187. [Google Scholar] [CrossRef]
- Finet, O.; Yague-Sanz, C.; Kruger, L.K.; Tran, P.; Migeot, V.; Louski, M.; Nevers, A.; Rougemaille, M.; Sun, J.; Ernst, F.G.M.; et al. Transcription-wide mapping of dihydrouridine reveals that mRNA dihydrouridylation is required for meiotic chromosome segregation. Mol. Cell 2022, 82, 404–419.e9. [Google Scholar] [CrossRef]
- Kim, S.H.; Suddath, F.L.; Quigley, G.J.; McPherson, A.; Sussman, J.L.; Wang, A.H.; Seeman, N.C.; Rich, A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 1974, 185, 435–440. [Google Scholar] [CrossRef] [Green Version]
- Suddath, F.L.; Quigley, G.J.; McPherson, A.; Sneden, D.; Kim, J.J.; Kim, S.H.; Rich, A. Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0angstroms resolution. Nature 1974, 248, 20–24. [Google Scholar] [CrossRef]
- Dalluge, J.J.; Hashizume, T.; Sopchik, A.E.; McCloskey, J.A.; Davis, D.R. Conformational flexibility in RNA: The role of dihydrouridine. Nucleic Acids Res. 1996, 24, 1073–1079. [Google Scholar] [CrossRef] [Green Version]
- Dyubankova, N.; Sochacka, E.; Kraszewska, K.; Nawrot, B.; Herdewijn, P.; Lescrinier, E. Contribution of dihydrouridine in folding of the D-arm in tRNA. Org. Biomol. Chem. 2015, 13, 4960–4966. [Google Scholar] [CrossRef]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Kuchino, Y.; Borek, E. Tumour-specific phenylalanine tRNA contains two supernumerary methylated bases. Nature 1978, 271, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Daigo, Y.; Hayama, S.; Ishikawa, N.; Yamabuki, T.; Ito, T.; Miyamoto, M.; Kondo, S.; Nakamura, Y. A novel human tRNA-dihydrouridine synthase involved in pulmonary carcinogenesis. Cancer Res. 2005, 65, 5638–5646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalluge, J.J.; Hamamoto, T.; Horikoshi, K.; Morita, R.Y.; Stetter, K.O.; McCloskey, J.A. Posttranscriptional modification of tRNA in psychrophilic bacteria. J. Bacteriol. 1997, 179, 1918–1923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, A.C.; Xu, J.; Johnson, R.C.; Schimmel, P.; de Crecy-Lagard, V. Identification of the tRNA-dihydrouridine synthase family. J. Biol. Chem. 2002, 277, 25090–25095. [Google Scholar] [CrossRef] [Green Version]
- Xing, F.; Hiley, S.L.; Hughes, T.R.; Phizicky, E.M. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004, 279, 17850–17860. [Google Scholar] [CrossRef] [Green Version]
- Lombard, M.; Hamdane, D. Flavin-dependent epitranscriptomic world. Arch. Biochem. Biophys. 2017, 632, 28–40. [Google Scholar] [CrossRef] [Green Version]
- Rider, L.W.; Ottosen, M.B.; Gattis, S.G.; Palfey, B.A. Mechanism of dihydrouridine synthase 2 from yeast and the importance of modifications for efficient tRNA reduction. J. Biol. Chem. 2009, 284, 10324–10333. [Google Scholar] [CrossRef] [Green Version]
- Bou-Nader, C.; Montemont, H.; Guerineau, V.; Jean-Jean, O.; Bregeon, D.; Hamdane, D. Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: Perspectives on the evolution scenario. Nucleic Acids Res. 2018, 46, 1386–1394. [Google Scholar] [CrossRef] [Green Version]
- Kasprzak, J.M.; Czerwoniec, A.; Bujnicki, J.M. Molecular evolution of dihydrouridine synthases. BMC Bioinform. 2012, 13, 153. [Google Scholar] [CrossRef]
- Faivre, B.; Lombard, M.; Fakroun, S.; Vo, C.D.; Goyenvalle, C.; Guerineau, V.; Pecqueur, L.; Fontecave, M.; De Crecy-Lagard, V.; Bregeon, D.; et al. Dihydrouridine synthesis in tRNAs is under reductive evolution in Mollicutes. RNA Biol. 2021, 18, 2278–2289. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Tanaka, Y.; Yamashita, K.; Suzuki, T.; Nakamura, A.; Hirano, N.; Yao, M.; Tanaka, I. Molecular basis of dihydrouridine formation on tRNA. Proc. Natl. Acad. Sci. USA 2011, 108, 19593–19598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusuba, H.; Yoshida, T.; Iwasaki, E.; Awai, T.; Kazayama, A.; Hirata, A.; Tomikawa, C.; Yamagami, R.; Hori, H. In vitro dihydrouridine formation by tRNA dihydrouridine synthase from Thermus thermophilus, an extreme-thermophilic eubacterium. J. Biochem. 2015, 158, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Martzen, M.R.; Phizicky, E.M. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA 2002, 8, 370–381. [Google Scholar] [CrossRef] [Green Version]
- Bou-Nader, C.; Pecqueur, L.; Bregeon, D.; Kamah, A.; Guerineau, V.; Golinelli-Pimpaneau, B.; Guimaraes, B.G.; Fontecave, M.; Hamdane, D. An extended dsRBD is required for post-transcriptional modification in human tRNAs. Nucleic Acids Res. 2015, 43, 9446–9456. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Yu, J.; Tanaka, Y.; Tanaka, M.; Tanaka, I.; Yao, M. Structure of dihydrouridine synthase C (DusC) from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 834–838. [Google Scholar] [CrossRef] [Green Version]
- Byrne, R.T.; Jenkins, H.T.; Peters, D.T.; Whelan, F.; Stowell, J.; Aziz, N.; Kasatsky, P.; Rodnina, M.V.; Koonin, E.V.; Konevega, A.L.; et al. Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases. Proc. Natl. Acad. Sci. USA 2015, 112, 6033–6037. [Google Scholar] [CrossRef] [Green Version]
- Whelan, F.; Jenkins, H.T.; Griffiths, S.C.; Byrne, R.T.; Dodson, E.J.; Antson, A.A. From bacterial to human dihydrouridine synthase: Automated structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1564–1571. [Google Scholar] [CrossRef]
- Bou-Nader, C.; Barraud, P.; Pecqueur, L.; Perez, J.; Velours, C.; Shepard, W.; Fontecave, M.; Tisne, C.; Hamdane, D. Molecular basis for transfer RNA recognition by the double-stranded RNA-binding domain of human dihydrouridine synthase 2. Nucleic Acids Res. 2019, 47, 3117–3126. [Google Scholar] [CrossRef]
- Mittelstadt, M.; Frump, A.; Khuu, T.; Fowlkes, V.; Handy, I.; Patel, C.V.; Patel, R.C. Interaction of human tRNA-dihydrouridine synthase-2 with interferon-induced protein kinase PKR. Nucleic Acids Res. 2008, 36, 998–1008. [Google Scholar] [CrossRef]
- Chen, X.; Ji, B.; Hao, X.; Li, X.; Eisele, F.; Nystrom, T.; Petranovic, D. FMN reduces Amyloid-beta toxicity in yeast by regulating redox status and cellular metabolism. Nat. Commun. 2020, 11, 867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bou-Nader, C.; Bregeon, D.; Pecqueur, L.; Fontecave, M.; Hamdane, D. Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases. Biochemistry 2018, 57, 5407–5414. [Google Scholar] [CrossRef] [PubMed]
- Bou-Nader, C.; Pecqueur, L.; Barraud, P.; Fontecave, M.; Tisne, C.; Sacquin-Mora, S.; Hamdane, D. Conformational Stability Adaptation of a Double-Stranded RNA-Binding Domain to Transfer RNA Ligand. Biochemistry 2019, 58, 2463–2473. [Google Scholar] [CrossRef] [PubMed]
- Nevers, Y.; Kress, A.; Defosset, A.; Ripp, R.; Linard, B.; Thompson, J.D.; Poch, O.; Lecompte, O. OrthoInspector 3.0: Open portal for comparative genomics. Nucleic Acids Res. 2019, 47, D411–D418. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [Green Version]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourao, K.; Ofoegbu, T.C.; Barton, D.; Lui, L.; Menard, A.; Sherstnev, N.; Roldan-Martinez, D.; et al. Alignment of Biological Sequences with Jalview. In Multiple Sequence Alignment; Springer: New York, NY, USA, 2021; Volume 2231, pp. 203–224. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D 2011, 67, 293–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mccoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Cryst. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunkoczi, G.; Read, R.J. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr. D 2011, 67, 303–312. [Google Scholar] [CrossRef] [Green Version]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D 2019, 75, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Dobritzsch, D.; Schneider, G.; Schnackerz, K.D.; Lindqvist, Y. Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J. 2001, 20, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zidek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Lu, D.; Searles, M.A.; Klug, A. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature 2003, 426, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.M. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 2005, 15, 367–373. [Google Scholar] [CrossRef] [Green Version]
- Burge, R.G.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Structural Characterization of Interactions between the Double-Stranded RNA-Binding Zinc Finger Protein JAZ and Nucleic Acids. Biochemistry 2014, 53, 1495–1510. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, R.H. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: Insight into tRNA recognition and reaction mechanism. Proc. Natl. Acad. Sci. USA 2008, 105, 16142–16147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, B.; Bevilacqua, P.C.; Diegelman-Parente, A.; Mathews, M.B. The double-stranded-RNA-binding motif: Interference and much more. Nature reviews. Mol. Cell Biol. 2004, 5, 1013–1023. [Google Scholar] [CrossRef]
- Chang, K.Y.; Ramos, A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005, 272, 2109–2117. [Google Scholar] [CrossRef]
- Masliah, G.; Barraud, P.; Allain, F.H. RNA recognition by double-stranded RNA binding domains: A matter of shape and sequence. Cell. Mol. Life Sci. CMLS 2013, 70, 1875–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefl, R.; Oberstrass, F.C.; Hood, J.L.; Jourdan, M.; Zimmermann, M.; Skrisovska, L.; Maris, C.; Peng, L.; Hofr, C.; Emeson, R.B.; et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell 2010, 143, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, U.; Grey, H.; Cook, A.G. Nuclear factor 90 uses an ADAR2-like binding mode to recognize specific bases in dsRNA. Nucleic Acids Res. 2016, 44, 1924–1936. [Google Scholar] [CrossRef]
- Goto-Ito, S.; Ito, T.; Kuratani, M.; Bessho, Y.; Yokoyama, S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat. Struct. Mol. Biol. 2009, 16, 1109–1115. [Google Scholar] [CrossRef]
- Guo, M.; Yang, X.L.; Schimmel, P. New functions of aminoacyl-tRNA synthetases beyond translation. Nature reviews. Mol. Cell Biol. 2010, 11, 668–674. [Google Scholar] [CrossRef]
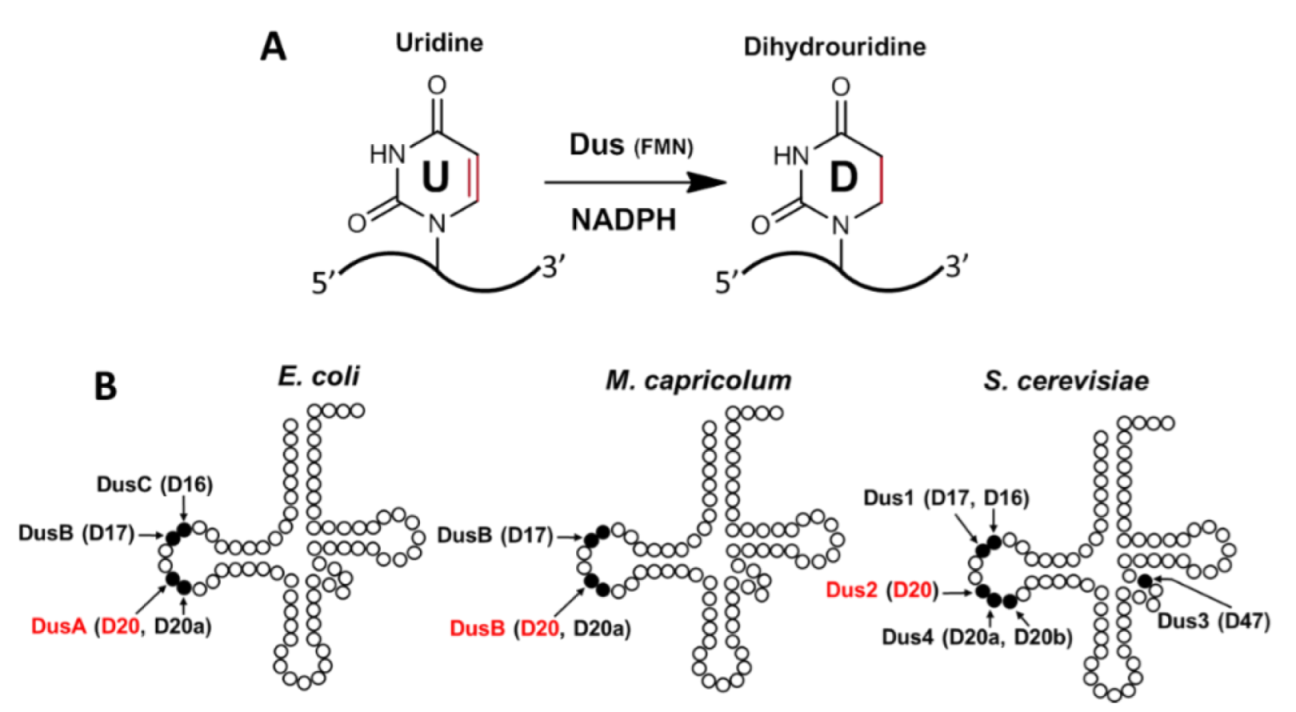
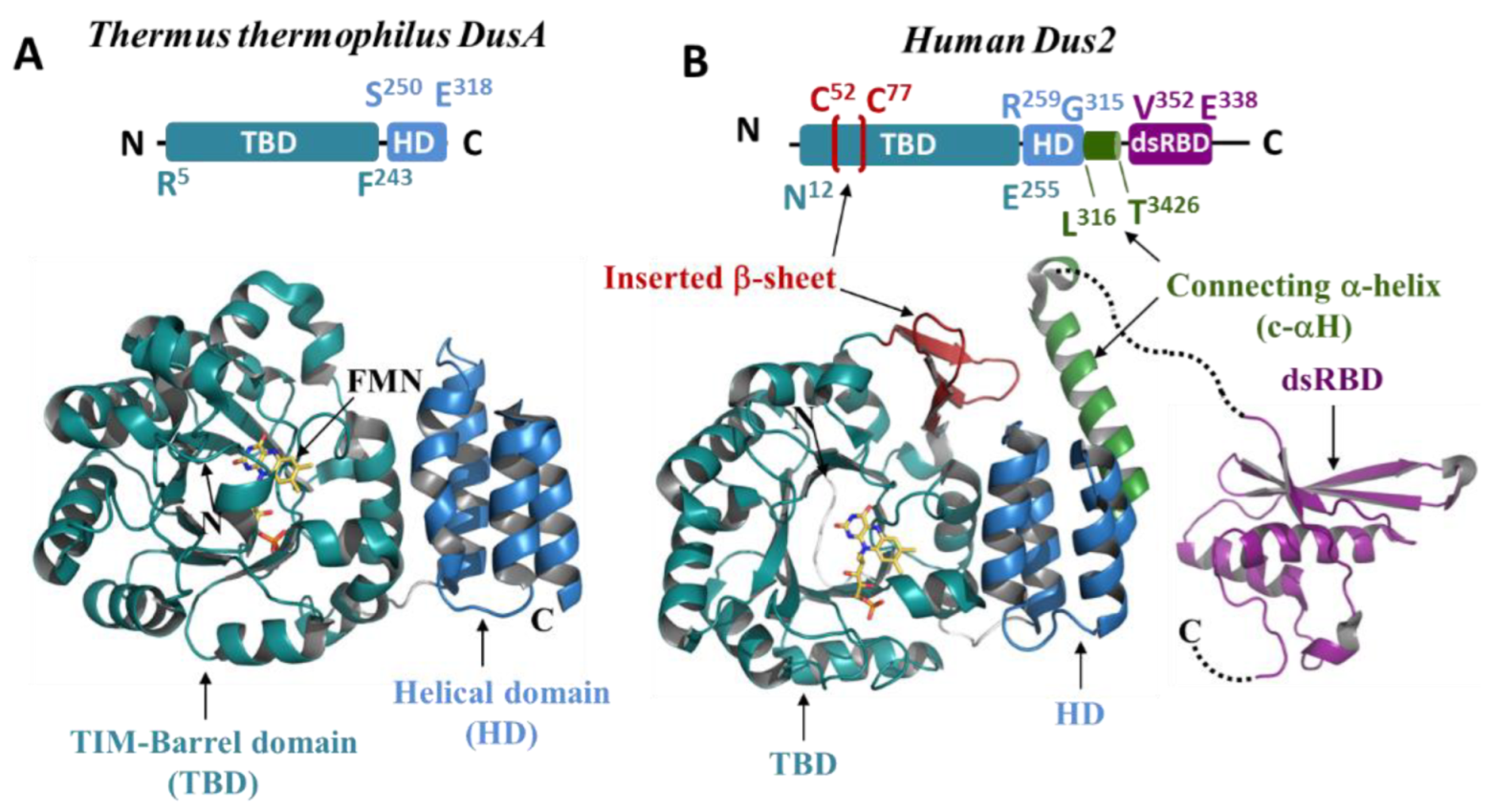

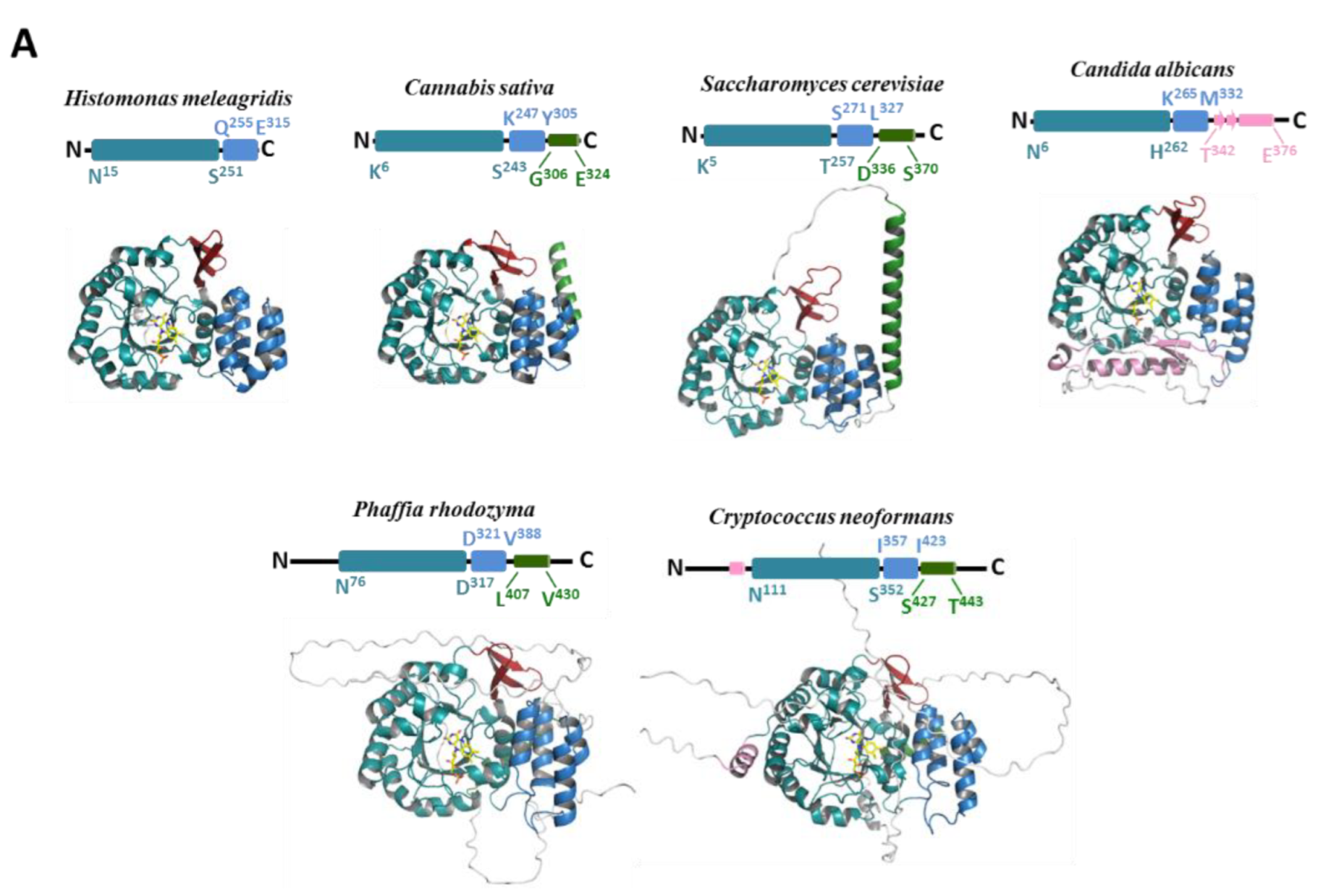
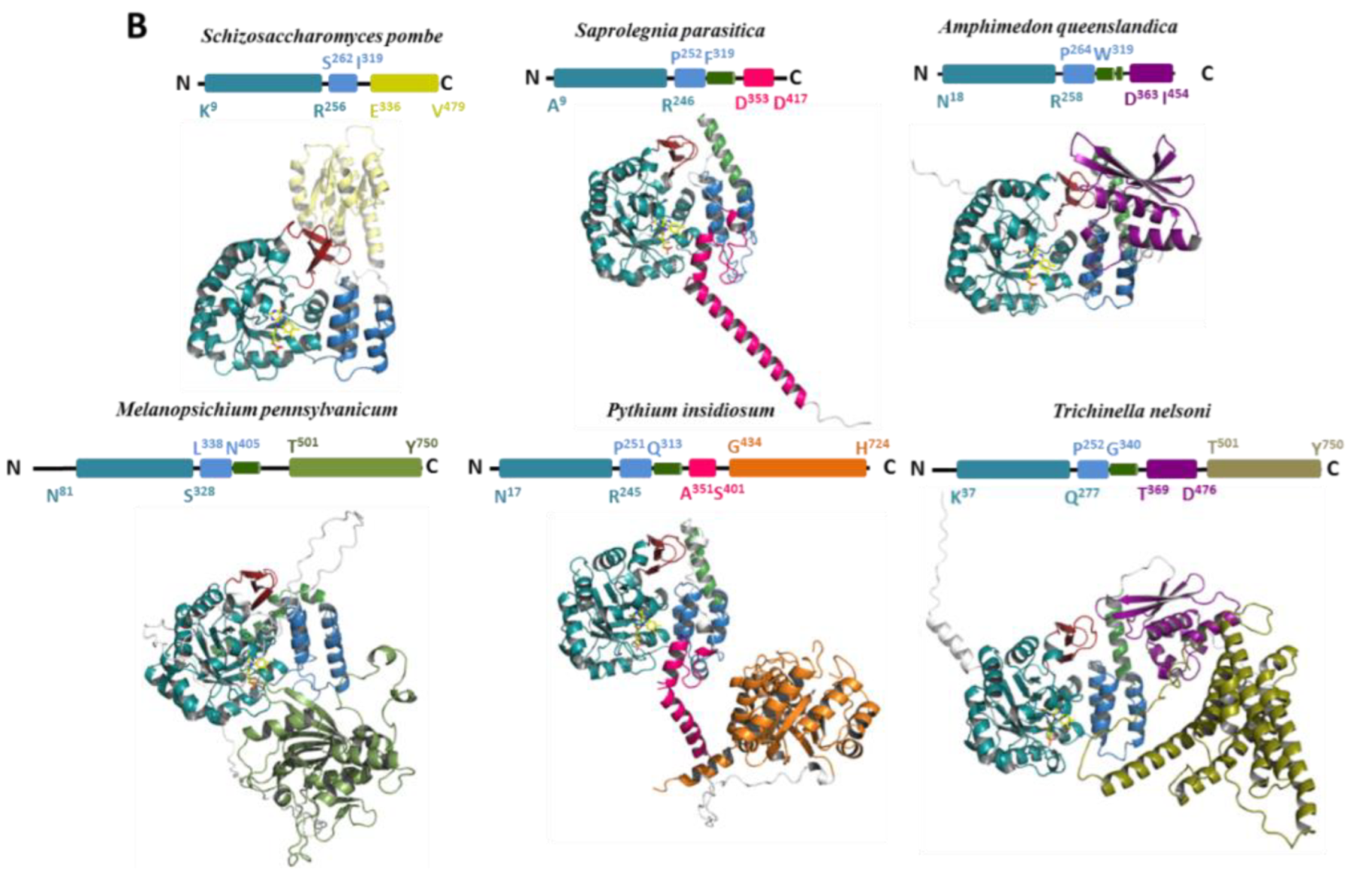



| Proteins | Products | Domain/Complex | PDB Code | Resolution |
|---|---|---|---|---|
| T. thermophilus DusA | D20/D20a | full length | 3B0P | 1.7 |
| T. thermophilus DusA | D20/D20a | full length + RNA fragment | 3B0U | 1.94 |
| T. thermophilus DusA | D20/D20a | full length + tRNAPhe | 3B0V | 3.51 |
| E. coli DusB | D17 | full length | 6EI9 | 2.55 |
| E. coli DusC | D16 | full length | 3W9Z | 2.1 |
| E. coli DusC | D16 | full length | 4BFA | 1.65 |
| E. coli DusC | D16 | full length + tRNATrp | 4YCP | 2.55 |
| E. coli DusC | D16 | full length + tRNAPhe | 4YCO | 2.1 |
| Homo sapiens Dus2 | D20 | TIM Barrel + HD | 4XP7 | 1.9 |
| Homo sapiens Dus2 | D20 | TIM Barrel + HD | 4WFS | 2.68 |
| Homo sapiens Dus2 | D20 | dsRBD | 4WFT | 1.7 |
| Homo sapiens Dus2 | D20 | dsRBD + dsRNA | 5OC6 | 3.2 |
| dsRBDaq | |
|---|---|
| PDB code | 8B02 |
| Data collection | |
| Wavelength (Å) | 0.9801 |
| Resolution range (Å) | 42.37–1.68 (1.70–1.68) |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 29.077, 56.895, 63.587 |
| α, β, γ (°) | 90.00, 93.05, 90.00 |
| Multiplicity | 2.9 (2.0) |
| Completeness (%) | 97.9 (79.7) |
| Mean I/sigma(I) | 7.8 (0.8) |
| Wilson B-factor (Å2) | 24.07 |
| R-meas | 0.090 (1.245) |
| R-pim | 0.051 (0.762) |
| CC1/2 | 0.997 (0.351) |
| Refinement | |
| Reflections used in refinement | 23356 (1170) |
| R-work / R-free (%) | 20.35/22.78 (32.80/36.10) |
| Number of non-hydrogen atoms | |
| macromolecules | 1548 |
| ligands | 28 |
| solvent | 139 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.02 |
| Ramachandran plot (%) | |
| favored | 98.96 |
| allowed | 1.04 |
| outliers | 0.00 |
| Average B-factor (Å2) | |
| Overall | 28.45 |
| macromolecules | 26.91 |
| ligands | 42.77 |
| solvent | 42.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lombard, M.; Reed, C.J.; Pecqueur, L.; Faivre, B.; Toubdji, S.; Sudol, C.; Brégeon, D.; de Crécy-Lagard, V.; Hamdane, D. Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family. Biomolecules 2022, 12, 1760. https://doi.org/10.3390/biom12121760
Lombard M, Reed CJ, Pecqueur L, Faivre B, Toubdji S, Sudol C, Brégeon D, de Crécy-Lagard V, Hamdane D. Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family. Biomolecules. 2022; 12(12):1760. https://doi.org/10.3390/biom12121760
Chicago/Turabian StyleLombard, Murielle, Colbie J. Reed, Ludovic Pecqueur, Bruno Faivre, Sabrine Toubdji, Claudia Sudol, Damien Brégeon, Valérie de Crécy-Lagard, and Djemel Hamdane. 2022. "Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family" Biomolecules 12, no. 12: 1760. https://doi.org/10.3390/biom12121760
APA StyleLombard, M., Reed, C. J., Pecqueur, L., Faivre, B., Toubdji, S., Sudol, C., Brégeon, D., de Crécy-Lagard, V., & Hamdane, D. (2022). Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family. Biomolecules, 12(12), 1760. https://doi.org/10.3390/biom12121760






