Muscle-Specific Ablation of Glucose Transporter 1 (GLUT1) Does Not Impair Basal or Overload-Stimulated Skeletal Muscle Glucose Uptake
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. RT-qPCR
2.3. Body Weight & Composition
2.4. Metabolic Cages
2.5. Glucose Tolerance Test
2.6. Unilateral Synergist Muscle Ablation Surgery
2.7. Muscle Glucose Uptake
2.8. Muscle Glycogen
2.9. Muscle Cell Surface GLUT Capture
2.10. Immunoblot Analyses
2.11. GLUT-HEK293 Cell Hexose Uptake
2.12. Statistical Analysis
3. Results
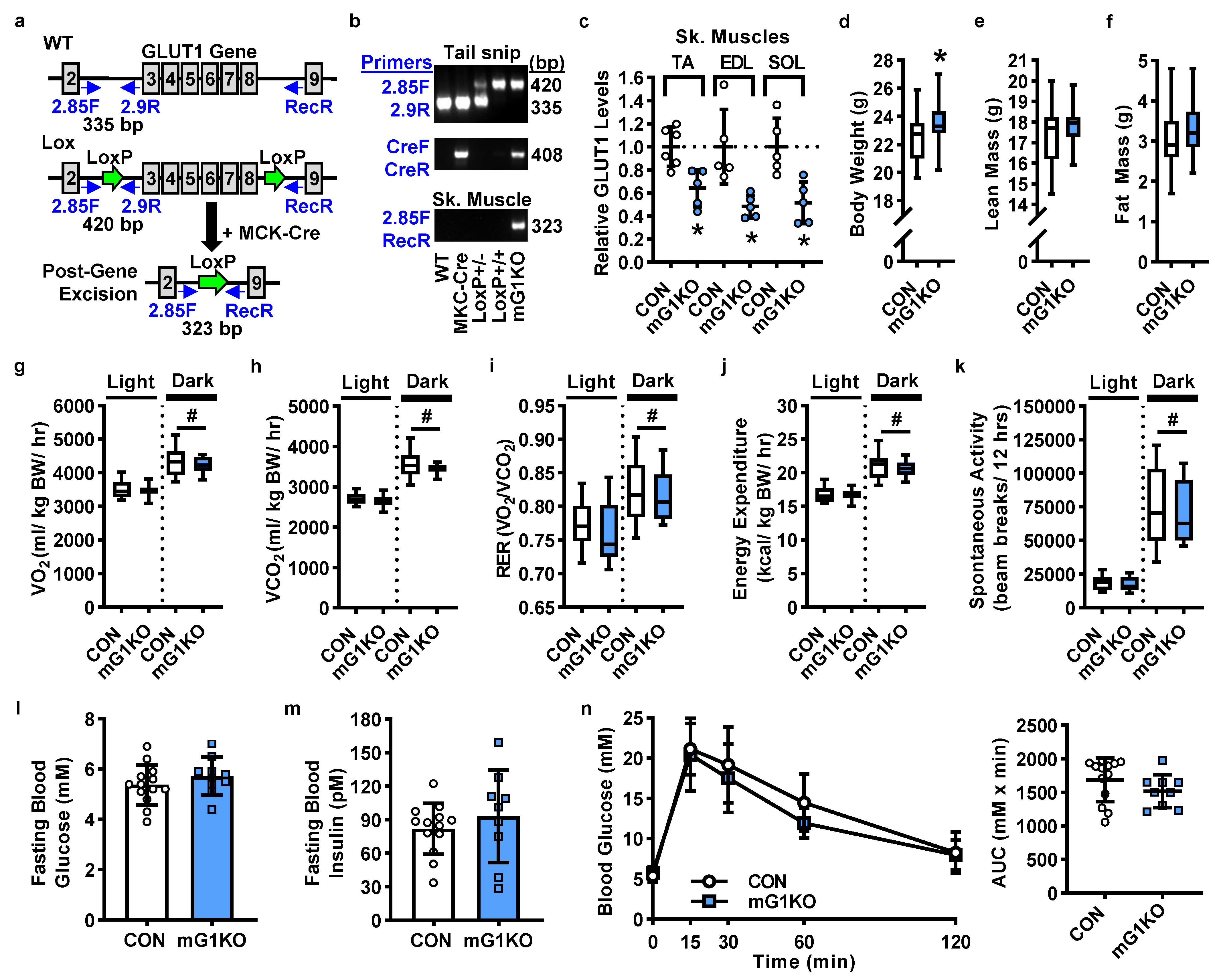
3.1. mGLUT1KO Mice Are Viable with No Alterations in Body Size, Body Composition, Whole Body Metabolism or Systemic Glucose Regulation
3.2. GLUT1 Expression in Muscle Is Not Required for Basal, Insulin-Stimulated or Overload-Stimulated Glucose Uptake
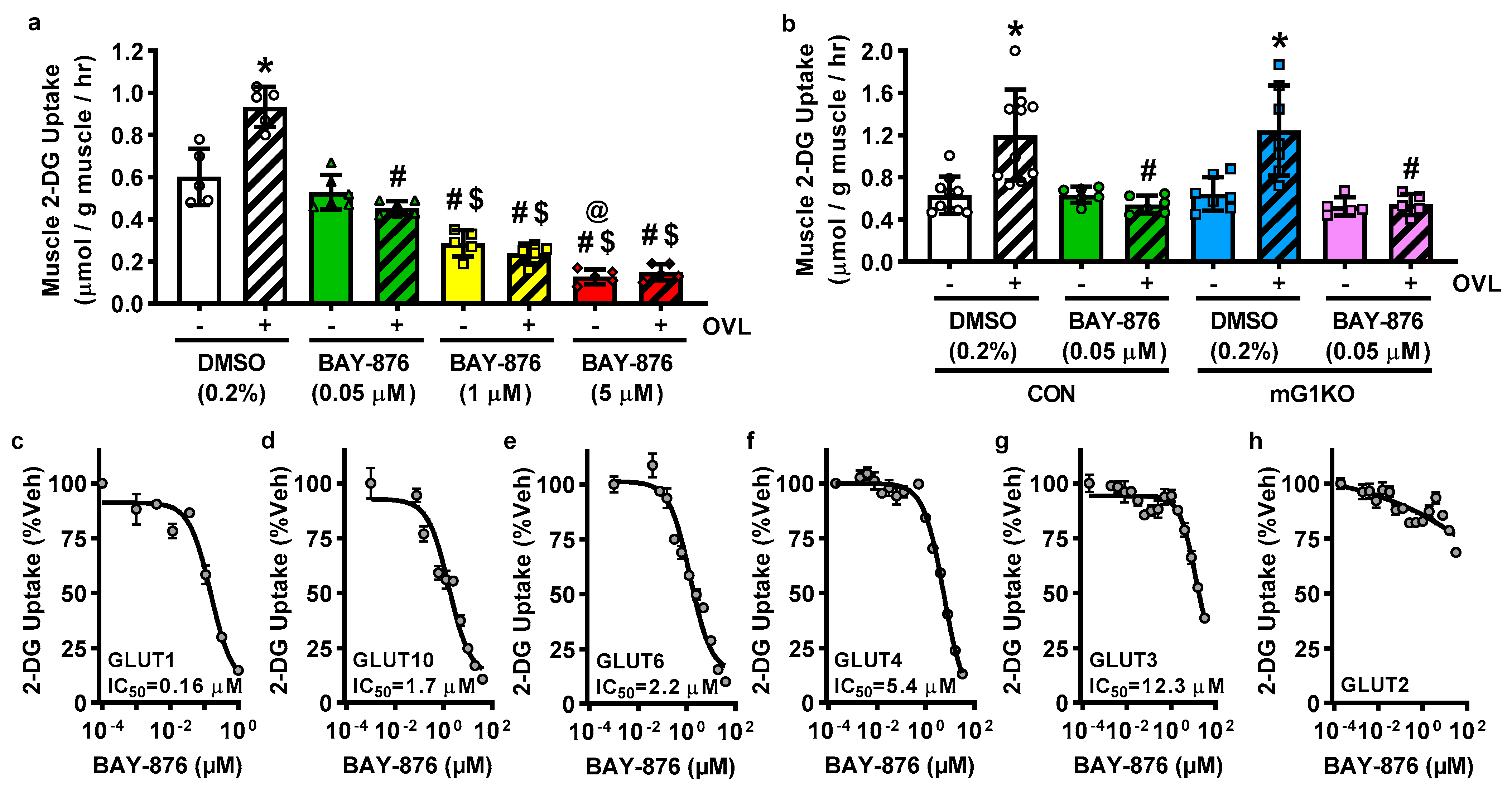
3.3. BAY-876 Inhibits Overload-Stimulated but Not Basal Muscle Glucose Uptake
3.4. BAY-876 Inhibits Glucose Uptake via Multiple Facilitative Glucose Transporters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baron, A.D.; Brechtel, G.; Wallace, P.; Edelman, S.V. Rates and tissue sites of non-insulin- and insulin-mediated glucose uptake in humans. Am. J. Physiol. 1988, 255, E769–E774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron, A.D.; Kolterman, O.G.; Bell, J.; Mandarino, L.J.; Olefsky, J.M. Rates of noninsulin-mediated glucose uptake are elevated in type II diabetic subjects. J. Clin. Investig. 1985, 76, 1782–1788. [Google Scholar] [CrossRef]
- Deshmukh, A.S.; Murgia, M.; Nagaraj, N.; Treebak, J.T.; Cox, J.; Mann, M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell. Proteom. 2015, 14, 841–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klip, A.; McGraw, T.E.; James, D.E. Thirty sweet years of GLUT4. J. Biol. Chem. 2019, 294, 11369–11381. [Google Scholar] [CrossRef] [Green Version]
- Chadt, A.; Al-Hasani, H. Glucose transporters in adipose tissue, liver, and skeletal muscle in metabolic health and disease. Pflug. Arch. Eur. J. Physiol. 2020, 472, 1273–1298. [Google Scholar] [CrossRef] [PubMed]
- Sylow, L.; Tokarz, V.L.; Richter, E.A.; Klip, A. The many actions of insulin in skeletal muscle, the paramount tissue determining glycemia. Cell Metab. 2021, 33, 758–780. [Google Scholar] [CrossRef]
- Holman, G.D. Structure, function and regulation of mammalian glucose transporters of the SLC2 family. Pflug. Arch. Eur. J. Physiol. 2020, 472, 1155–1175. [Google Scholar] [CrossRef]
- Flores-Opazo, M.; McGee, S.L.; Hargreaves, M. Exercise and GLUT4. Exerc. Sport Sci. Rev. 2020, 48, 110–118. [Google Scholar] [CrossRef]
- Marshall, B.A.; Ren, J.M.; Johnson, D.W.; Gibbs, E.M.; Lillquist, J.S.; Soeller, W.C.; Holloszy, J.O.; Mueckler, M. Germline manipulation of glucose homeostasis via alteration of glucose transporter levels in skeletal muscle. J. Biol. Chem. 1993, 268, 18442–18445. [Google Scholar] [CrossRef]
- Etgen, G.J., Jr.; Zavadoski, W.J.; Holman, G.D.; Gibbs, E.M. Insulin-sensitive regulation of glucose transport and GLUT4 translocation in skeletal muscle of GLUT1 transgenic mice. Biochem. J. 1999, 337 Pt 1, 51–57. [Google Scholar] [CrossRef]
- Gulve, E.A.; Ren, J.M.; Marshall, B.A.; Gao, J.; Hansen, P.A.; Holloszy, J.O.; Mueckler, M. Glucose transport activity in skeletal muscles from transgenic mice overexpressing GLUT1. Increased basal transport is associated with a defective response to diverse stimuli that activate GLUT4. J. Biol. Chem. 1994, 269, 18366–18370. [Google Scholar] [CrossRef] [PubMed]
- Zisman, A.; Peroni, O.D.; Abel, E.D.; Michael, M.D.; Mauvais-Jarvis, F.; Lowell, B.B.; Wojtaszewski, J.F.; Hirshman, M.F.; Virkamaki, A.; Goodyear, L.J.; et al. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat. Med. 2000, 6, 924–928. [Google Scholar] [CrossRef]
- Kim, J.K.; Zisman, A.; Fillmore, J.J.; Peroni, O.D.; Kotani, K.; Perret, P.; Zong, H.; Dong, J.; Kahn, C.R.; Kahn, B.B.; et al. Glucose toxicity and the development of diabetes in mice with muscle-specific inactivation of GLUT4. J. Clin. Investig. 2001, 108, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Zierath, J.R.; Tsao, T.S.; Stenbit, A.E.; Ryder, J.W.; Galuska, D.; Charron, M.J. Restoration of hypoxia-stimulated glucose uptake in GLUT4-deficient muscles by muscle-specific GLUT4 transgenic complementation. J. Biol. Chem. 1998, 273, 20910–20915. [Google Scholar] [CrossRef] [Green Version]
- Stenbit, A.E.; Burcelin, R.; Katz, E.B.; Tsao, T.S.; Gautier, N.; Charron, M.J.; Le Marchand-Brustel, Y. Diverse effects of Glut 4 ablation on glucose uptake and glycogen synthesis in red and white skeletal muscle. J. Clin. Investig. 1996, 98, 629–634. [Google Scholar] [CrossRef] [PubMed]
- McMillin, S.L.; Schmidt, D.L.; Kahn, B.B.; Witczak, C.A. GLUT4 Is Not Necessary for Overload-Induced Glucose Uptake or Hypertrophic Growth in Mouse Skeletal Muscle. Diabetes 2017, 66, 1491–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaillou, T.; Lee, J.D.; England, J.H.; Esser, K.A.; McCarthy, J.J. Time course of gene expression during mouse skeletal muscle hypertrophy. J. Appl. Physiol. 2013, 115, 1065–1074. [Google Scholar] [CrossRef]
- Bruning, J.C.; Michael, M.D.; Winnay, J.N.; Hayashi, T.; Horsch, D.; Accili, D.; Goodyear, L.J.; Kahn, C.R. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 1998, 2, 559–569. [Google Scholar] [CrossRef]
- Young, C.D.; Lewis, A.S.; Rudolph, M.C.; Ruehle, M.D.; Jackman, M.R.; Yun, U.J.; Ilkun, O.; Pereira, R.; Abel, E.D.; Anderson, S.M. Modulation of glucose transporter 1 (GLUT1) expression levels alters mouse mammary tumor cell growth in vitro and in vivo. PLoS ONE 2011, 6, e23205. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Ferey, J.L.; Brault, J.J.; Smith, C.A.; Witczak, C.A. Constitutive Activation of CaMKKalpha Signaling is Sufficient But Not Necessary for mTORC1 Activation and Growth in Mouse Skeletal Muscle. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E686–E694. [Google Scholar] [CrossRef] [Green Version]
- Weyrauch, L.A.; McMillin, S.L.; Witczak, C.A. Insulin Resistance Does Not Impair Mechanical Overload-Stimulated Glucose Uptake, but Does Alter the Metabolic Fate of Glucose in Mouse Muscle. Int. J. Mol. Sci. 2020, 21, 4715. [Google Scholar] [CrossRef] [PubMed]
- Hinkley, J.M.; Ferey, J.L.; Brault, J.J.; Smith, C.S.; Gilliam, L.A.A.; Witczak, C.A. Constitutively active CaMKKalpha stimulates skeletal muscle glucose uptake in insulin resistant mice in vivo. Diabetes 2014, 63, 142–151. [Google Scholar] [CrossRef] [Green Version]
- Kramer, H.F.; Witczak, C.A.; Taylor, E.B.; Fujii, N.; Hirshman, M.F.; Goodyear, L.J. AS160 regulates insulin- and contraction-stimulated glucose uptake in mouse skeletal muscle. J. Biol. Chem. 2006, 281, 31478–31485. [Google Scholar] [CrossRef]
- Hashimoto, M.; Hatanaka, Y.; Yang, J.; Dhesi, J.; Holman, G.D. Synthesis of biotinylated bis(D-glucose) derivatives for glucose transporter photoaffinity labelling. Carbohydr. Res. 2001, 331, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Ryder, J.W.; Yang, J.; Galuska, D.; Rincon, J.; Bjornholm, M.; Krook, A.; Lund, S.; Pedersen, O.; Wallberg-Henriksson, H.; Zierath, J.R.; et al. Use of a novel impermeable biotinylated photolabeling reagent to assess insulin- and hypoxia-stimulated cell surface GLUT4 content in skeletal muscle from type 2 diabetic patients. Diabetes 2000, 49, 647–654. [Google Scholar] [CrossRef] [Green Version]
- Kraft, T.E.; Heitmeier, M.R.; Putanko, M.; Edwards, R.L.; Ilagan, M.X.; Payne, M.A.; Autry, J.M.; Thomas, D.D.; Odom, A.R.; Hruz, P.W. A Novel Fluorescence Resonance Energy Transfer-Based Screen in High-Throughput Format To Identify Inhibitors of Malarial and Human Glucose Transporters. Antimicrob. Agents Chemother. 2016, 60, 7407–7414. [Google Scholar] [CrossRef] [Green Version]
- DeBosch, B.J.; Heitmeier, M.R.; Mayer, A.L.; Higgins, C.B.; Crowley, J.R.; Kraft, T.E.; Chi, M.; Newberry, E.P.; Chen, Z.; Finck, B.N.; et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci. Signal. 2016, 9, ra21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, R.O.; Wende, A.R.; Olsen, C.; Soto, J.; Rawlings, T.; Zhu, Y.; Riehle, C.; Abel, E.D. GLUT1 deficiency in cardiomyocytes does not accelerate the transition from compensated hypertrophy to heart failure. J. Mol. Cell. Cardiol. 2014, 72, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zi, Z.; Lee, E.E.; Zhao, J.; Contreras, D.C.; South, A.P.; Abel, E.D.; Chong, B.F.; Vandergriff, T.; Hosler, G.A.; et al. Differential glucose requirement in skin homeostasis and injury identifies a therapeutic target for psoriasis. Nat. Med. 2018, 24, 617–627. [Google Scholar] [CrossRef]
- Winkler, E.A.; Nishida, Y.; Sagare, A.P.; Rege, S.V.; Bell, R.D.; Perlmutter, D.; Sengillo, J.D.; Hillman, S.; Kong, P.; Nelson, A.R.; et al. GLUT1 reductions exacerbate Alzheimer’s disease vasculo-neuronal dysfunction and degeneration. Nat. Neurosci. 2015, 18, 521–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarup, A.; Bell, B.A.; Du, J.; Han, J.Y.S.; Soto, J.; Abel, E.D.; Bravo-Nuevo, A.; FitzGerald, P.G.; Peachey, N.S.; Philp, N.J. Deletion of GLUT1 in mouse lens epithelium leads to cataract formation. Exp. Eye Res. 2018, 172, 45–53. [Google Scholar] [CrossRef]
- Fidler, T.P.; Campbell, R.A.; Funari, T.; Dunne, N.; Balderas Angeles, E.; Middleton, E.A.; Chaudhuri, D.; Weyrich, A.S.; Abel, E.D. Deletion of GLUT1 and GLUT3 Reveals Multiple Roles for Glucose Metabolism in Platelet and Megakaryocyte Function. Cell Rep. 2017, 21, 1705. [Google Scholar] [CrossRef] [PubMed]
- Handberg, A.; Kayser, L.; Høyer, P.E.; Vinten, J. A substantial part of GLUT-1 in crude membranes from muscle originates from perineurial sheaths. Am. J. Physiol. 1992, 262, E721–E727. [Google Scholar] [CrossRef] [PubMed]
- Marette, A.; Richardson, J.M.; Ramlal, T.; Balon, T.W.; Vranic, M.; Pessin, J.E.; Klip, A. Abundance, localization, and insulin-induced translocation of glucose transporters in red and white muscle. Am. J. Physiol. 1992, 263, C443–C452. [Google Scholar] [CrossRef] [PubMed]
- Doria-Medina, C.L.; Lund, D.D.; Pasley, A.; Sandra, A.; Sivitz, W.I. Immunolocalization of GLUT-1 glucose transporter in rat skeletal muscle and in normal and hypoxic cardiac tissue. Am. J. Physiol. 1993, 265, E454–E464. [Google Scholar] [CrossRef]
- dos Santos, J.M.; Benite-Ribeiro, S.A.; Queiroz, G.; Duarte, J.A. The effect of age on glucose uptake and GLUT1 and GLUT4 expression in rat skeletal muscle. Cell Biochem. Funct. 2012, 30, 191–197. [Google Scholar] [CrossRef]
- Kahn, B.B.; Rossetti, L.; Lodish, H.F.; Charron, M.J. Decreased in vivo glucose uptake but normal expression of GLUT1 and GLUT4 in skeletal muscle of diabetic rats. J. Clin. Investig. 1991, 87, 2197–2206. [Google Scholar] [CrossRef] [Green Version]
- Heilig, C.; Brosius, F.; Siu, B.; Concepcion, L.; Mortensen, R.; Heilig, K.; Zhu, M.; Weldon, R.; Wu, G.; Conner, D. Implications of glucose transporter protein type 1 (GLUT1)-haplodeficiency in embryonic stem cells for their survival in response to hypoxic stress. Am. J. Pathol. 2003, 163, 1873–1885. [Google Scholar] [CrossRef] [Green Version]
- Siebeneicher, H.; Cleve, A.; Rehwinkel, H.; Neuhaus, R.; Heisler, I.; Muller, T.; Bauser, M.; Buchmann, B. Identification and Optimization of the First Highly Selective GLUT1 Inhibitor BAY-876. ChemMedChem 2016, 11, 2261–2271. [Google Scholar] [CrossRef]
- Ren, J.M.; Marshall, B.A.; Gulve, E.A.; Gao, J.; Johnson, D.W.; Holloszy, J.O.; Mueckler, M. Evidence from transgenic mice that glucose transport is rate-limiting for glycogen deposition and glycolysis in skeletal muscle. J. Biol. Chem. 1993, 268, 16113–16115. [Google Scholar] [CrossRef] [PubMed]
- Gould, G.W.; Thomas, H.M.; Jess, T.J.; Bell, G.I. Expression of human glucose transporters in Xenopus oocytes: Kinetic characterization and substrate specificities of the erythrocyte, liver, and brain isoforms. Biochemistry 1991, 30, 5139–5145. [Google Scholar] [CrossRef] [PubMed]
- Andrisse, S.; Patel, G.D.; Chen, J.E.; Webber, A.M.; Spears, L.D.; Koehler, R.M.; Robinson-Hill, R.M.; Ching, J.K.; Jeong, I.; Fisher, J.S. ATM and GLUT1-S490 phosphorylation regulate GLUT1 mediated transport in skeletal muscle. PLoS ONE 2013, 8, e66027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uldry, M.; Ibberson, M.; Hosokawa, M.; Thorens, B. GLUT2 is a high affinity glucosamine transporter. FEBS Lett. 2002, 524, 199–203. [Google Scholar] [CrossRef]
- Lambert, M.; Bastide, B.; Cieniewski-Bernard, C. Involvement of O-GlcNAcylation in the Skeletal Muscle Physiology and Physiopathology: Focus on Muscle Metabolism. Front. Endocrinol. 2018, 9, 578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conte, F.; van Buuringen, N.; Voermans, N.C.; Lefeber, D.J. Galactose in human metabolism, glycosylation and congenital metabolic diseases: Time for a closer look. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129898. [Google Scholar] [CrossRef]
- Blazev, R.; Ashwood, C.; Abrahams, J.L.; Chung, L.H.; Francis, D.; Yang, P.; Watt, K.I.; Qian, H.; Quaife-Ryan, G.A.; Hudson, J.E.; et al. Integrated Glycoproteomics Identifies a Role of N-Glycosylation and Galectin-1 on Myogenesis and Muscle Development. Mol. Cell. Proteom. 2021, 20, 100030. [Google Scholar] [CrossRef]
- Doege, H.; Bocianski, A.; Joost, H.G.; Schurmann, A. Activity and genomic organization of human glucose transporter 9 (GLUT9), a novel member of the family of sugar-transport facilitators predominantly expressed in brain and leucocytes. Biochem. J. 2000, 350 Pt 3, 771–776. [Google Scholar] [CrossRef]
- Savini, I.; Catani, M.V.; Duranti, G.; Ceci, R.; Sabatini, S.; Avigliano, L. Vitamin C homeostasis in skeletal muscle cells. Free Radic. Biol. Med. 2005, 38, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Németh, C.E.; Marcolongo, P.; Gamberucci, A.; Fulceri, R.; Benedetti, A.; Zoppi, N.; Ritelli, M.; Chiarelli, N.; Colombi, M.; Willaert, A.; et al. Glucose transporter type 10-lacking in arterial tortuosity syndrome-facilitates dehydroascorbic acid transport. FEBS Lett. 2016, 590, 1630–1640. [Google Scholar] [CrossRef]
- Syu, Y.W.; Lai, H.W.; Jiang, C.L.; Tsai, H.Y.; Lin, C.C.; Lee, Y.C. GLUT10 maintains the integrity of major arteries through regulation of redox homeostasis and mitochondrial function. Hum. Mol. Genet. 2018, 27, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Nagamatsu, S.; Kornhauser, J.M.; Burant, C.F.; Seino, S.; Mayo, K.E.; Bell, G.I. Glucose transporter expression in brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose transporter isoform, and identification of sites of expression by in situ hybridization. J. Biol. Chem. 1992, 267, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Janot, M.; Audfray, A.; Loriol, C.; Germot, A.; Maftah, A.; Dupuy, F. Glycogenome expression dynamics during mouse C2C12 myoblast differentiation suggests a sequential reorganization of membrane glycoconjugates. BMC. Genom. 2009, 10, 483. [Google Scholar] [CrossRef] [Green Version]
- Maedera, S.; Mizuno, T.; Ishiguro, H.; Ito, T.; Soga, T.; Kusuhara, H. GLUT6 is a lysosomal transporter that is regulated by inflammatory stimuli and modulates glycolysis in macrophages. FEBS Lett. 2019, 593, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Tumova, S.; Kerimi, A.; Porter, K.E.; Williamson, G. Transendothelial glucose transport is not restricted by extracellular hyperglycaemia. Vasc. Pharm. 2016, 87, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Feng, N.; Zhou, Y.; Ye, X.; Wang, R.; Zhang, J.; Cui, S.; Ji, S.; Chen, Y.; Zhu, S. Slc2a6 regulates myoblast differentiation by targeting LDHB. Cell. Commun. Signal. 2022, 20, 107. [Google Scholar] [CrossRef]
- Wood, I.S.; Hunter, L.; Trayhurn, P. Expression of Class III facilitative glucose transporter genes (GLUT-10 and GLUT-12) in mouse and human adipose tissues. Biochem. Biophys. Res. Commun. 2003, 308, 43–49. [Google Scholar] [CrossRef]
- Zhou, A.L.; Egginton, S.; Brown, M.D.; Hudlicka, O. Capillary growth in overloaded, hypertrophic adult rat skeletal muscle: An ultrastructural study. Anat. Rec. 1998, 252, 49–63. [Google Scholar] [CrossRef]
- Contat, C.; Ancey, P.B.; Zangger, N.; Sabatino, S.; Pascual, J.; Escrig, S.; Jensen, L.; Goepfert, C.; Lanz, B.; Lepore, M.; et al. Combined deletion of Glut1 and Glut3 impairs lung adenocarcinoma growth. Elife 2020, 9, e53618. [Google Scholar] [CrossRef]
- Ancey, P.B.; Contat, C.; Boivin, G.; Sabatino, S.; Pascual, J.; Zangger, N.; Perentes, J.Y.; Peters, S.; Abel, E.D.; Kirsch, D.G.; et al. GLUT1 Expression in Tumor-Associated Neutrophils Promotes Lung Cancer Growth and Resistance to Radiotherapy. Cancer Res. 2021, 81, 2345–2357. [Google Scholar] [CrossRef]
- Tang, M.; Park, S.H.; Petri, S.; Yu, H.; Rueda, C.B.; Abel, E.D.; Kim, C.Y.; Hillman, E.M.; Li, F.; Lee, Y.; et al. An early endothelial cell-specific requirement for Glut1 is revealed in Glut1 deficiency syndrome model mice. JCI Insight 2021, 6, e145789. [Google Scholar] [CrossRef] [PubMed]

| Gene | GenBank Accession | Primers | Amplicon Size (bp) | Primer Efficiency | Slope | Y-Axis Intercept | R2 Value |
|---|---|---|---|---|---|---|---|
| SLC2A1 | NM_011400 | CACTGGTGTCATCAACGCCC | 94 | 99.1% | −3.34 | 40.5 | 0.99 |
| AGACCAAAGCGTGGTGAGTG | |||||||
| SLC2A3 | NM_011401 | ATGGGGACAACGAAGGTGAC | 107 | 93.5% | −3.49 | 25.6 | 1.00 |
| GTCTCAGGTGCATTGATGACTC | |||||||
| SLC2A6 | NM_172659 | CAGAGTAGCCGAGTGTCGTG | 152 | 92.4% | −3.52 | 26.5 | 1.00 |
| ACCACGGATGTGTTGTCGAA | |||||||
| SLC2A10 | NM_130451 | GCCTGACCTTCGGATATGAGC | 165 | 90.6% | −3.57 | 26.9 | 0.99 |
| TGCCATAGCAGTCAATGAGGA | |||||||
| HAGH | NM_001409599 | CACCACTCACCACCACTGG | 153 | 91.9% | −3.53 | 36.9 | 0.99 |
| ACACTGAGAGACCCCACCTG | |||||||
| RPS17 | NM_009092 | CGGCTATGTCACGCATCTG | 194 | 100.8% | −3.30 | 30.0 | 0.99 |
| TAGAGAGACTGCCAAAGTCCAGG | |||||||
| SRP14 | NM_009273.4 | AGAGCGAGCAGTTCCTGAC | 195 | 96.8% | −3.40 | 33.9 | 0.99 |
| CGGTGCTGATCTTCCTTTTC |
| Transcript Quantified | GLUT-Expressing HEK293 Cell Line | ||||||
|---|---|---|---|---|---|---|---|
| GLUT1 | GLUT2 | GLUT3 | GLUT4 | GLUT5 | GLUT6 | GLUT10 | |
| hGLUT1 | 462,327 ± 4566 | 3725 ± 326 | 4145 ± 501 | 4527 ± 768 | 2139 ± 180 | 3815 ± 29 | 4458 ± 822 |
| hGLUT2 | 5 ± 0 | 84,758 ± 13,107 | ND | 7 ± 4 | 23 ± 4 | 540 ± 9 | 9 ± 4 |
| hGLUT3 | 288 ± 124 | 403 ± 54 | 1,358,605 ± 33,835 | 1223 ± 366 | 205 ± 31 | 12,223 ± 5 | 2269 ± 303 |
| hGLUT4 | 155 ± 76 | 170 ± 49 | 268 ± 83 | 288,864 ± 84,501 | 91 ± 15 | 45 ± 22 | 5 ± 1 |
| hGLUT5 | N/A | N/A | N/A | N/A | 79,938 ± 1625 | ND | ND |
| hGLUT6 | N/A | N/A | N/A | N/A | N/A | 1,532,765 ± 400,123 | N/A |
| hGLUT10 | N/A | N/A | N/A | N/A | 39 ± 9 | 4 ± 1 | 40,431 ± 7235 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McMillin, S.L.; Evans, P.L.; Taylor, W.M.; Weyrauch, L.A.; Sermersheim, T.J.; Welc, S.S.; Heitmeier, M.R.; Hresko, R.C.; Hruz, P.W.; Koumanov, F.; et al. Muscle-Specific Ablation of Glucose Transporter 1 (GLUT1) Does Not Impair Basal or Overload-Stimulated Skeletal Muscle Glucose Uptake. Biomolecules 2022, 12, 1734. https://doi.org/10.3390/biom12121734
McMillin SL, Evans PL, Taylor WM, Weyrauch LA, Sermersheim TJ, Welc SS, Heitmeier MR, Hresko RC, Hruz PW, Koumanov F, et al. Muscle-Specific Ablation of Glucose Transporter 1 (GLUT1) Does Not Impair Basal or Overload-Stimulated Skeletal Muscle Glucose Uptake. Biomolecules. 2022; 12(12):1734. https://doi.org/10.3390/biom12121734
Chicago/Turabian StyleMcMillin, Shawna L., Parker L. Evans, William M. Taylor, Luke A. Weyrauch, Tyler J. Sermersheim, Steven S. Welc, Monique R. Heitmeier, Richard C. Hresko, Paul W. Hruz, Francoise Koumanov, and et al. 2022. "Muscle-Specific Ablation of Glucose Transporter 1 (GLUT1) Does Not Impair Basal or Overload-Stimulated Skeletal Muscle Glucose Uptake" Biomolecules 12, no. 12: 1734. https://doi.org/10.3390/biom12121734
APA StyleMcMillin, S. L., Evans, P. L., Taylor, W. M., Weyrauch, L. A., Sermersheim, T. J., Welc, S. S., Heitmeier, M. R., Hresko, R. C., Hruz, P. W., Koumanov, F., Holman, G. D., Abel, E. D., & Witczak, C. A. (2022). Muscle-Specific Ablation of Glucose Transporter 1 (GLUT1) Does Not Impair Basal or Overload-Stimulated Skeletal Muscle Glucose Uptake. Biomolecules, 12(12), 1734. https://doi.org/10.3390/biom12121734






