Inter-Site Cooperativity of Calmodulin N-Terminal Domain and Phosphorylation Synergistically Improve the Affinity and Selectivity for Uranyl
Abstract
:1. Introduction
2. Experimental Section
2.1. Engineering, Expression and Purification of CaM Peptides
2.2. In Vitro Phosphorylation of the CaM Peptides
2.3. Chemicals and Stock Solutions
2.4. Mass Spectrometry Analyses
2.5. Spectrofluorimetric Titrations
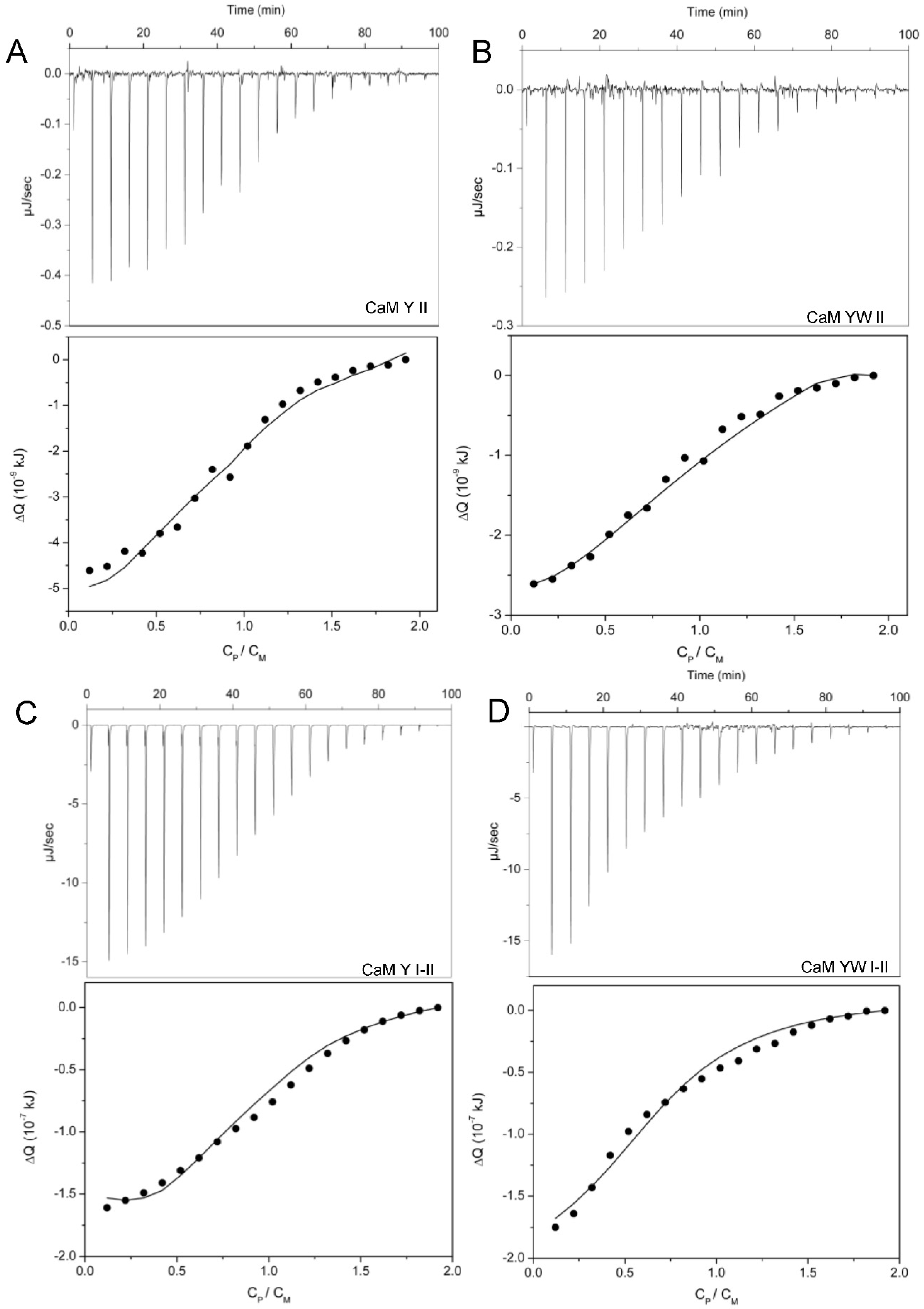
2.6. ITC Titrations
2.7. Calculations
2.7.1. Competition with IDA
2.7.2. Spectrofluorimetric Data Treatment and Evaluation of Macroscopic Constants
M + P ⇄ MP
M + MP ⇄ M2P
2.7.3. Microscopic Constants and Cooperativity Evaluation
2.7.4. Isothermal Titration Calorimetry Data Treatment and Microscopic Thermodynamic Parameters
(ΔHI + ΔHII + ΔHc)(Vj[M]j [MP]j − Vj-1[M]j-1 [MP]j-1)K2
2.8. Molecular Dynamics
3. Results
3.1. Design and Characterization of the Calmodulin Variants
3.2. Uranyl-Binding Properties of Site II Variants of Calmodulin N-Terminal Domain
3.3. Two-Site Peptides and the Cooperative Effect
3.4. Influence of Site I Thr9 Phosphorylation on Uranyl Binding
3.5. Ca2+ Binding to the Peptide Variants
3.6. Structural Modelization of CaM1 Y I-II P Using Molecular Dynamics
4. Discussion
4.1. Site II Affinity for Uranyl
4.2. “Two-Site” N-Ter Domain
4.3. Phosphorylated Site I Considerably Improves Uranyl Binding at Site I and Site II in the N-Ter Domain
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brugge, D.; de Lemos, J.L.; Oldmixon, B. Exposure pathways and health effects associated with chemical and radiological toxicity of natural uranium: A review. Rev. Environ. Health 2005, 20, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Kathren, R.L.; Burklin, R.K. Acute chemical toxicity of uranium. Health Phys. 2008, 94, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.M.; Taylor, S.K. Environmental uranium and human health. Rev. Environ. Health 1997, 12, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Basset, C.; Averseng, O.; Ferron, P.J.; Richaud, N.; Hagege, A.; Pible, O.; Vidaud, C. Revision of the biodistribution of uranyl in serum: Is fetuin-A the major protein target? Chem. Res. Toxicol. 2013, 26, 645–653. [Google Scholar] [CrossRef]
- Brulfert, F.; Safi, S.; Jeanson, A.; Martinez-Baez, E.; Roques, J.; Berthomieu, C.; Solari, P.L.; Sauge-Merle, S.; Simoni, E. Structural Environment and Stability of the Complexes Formed Between Calmodulin and Actinyl Ions. Inorg. Chem. 2016, 55, 2728–2736. [Google Scholar] [CrossRef]
- Huynh, T.S.; Vidaud, C.; Hagege, A. Investigation of uranium interactions with calcium phosphate-binding proteins using ICP/MS and CE-ICP/MS. Met. Integr. Biometal Sci. 2016, 8, 1185–1192. [Google Scholar] [CrossRef]
- Montavon, G.; Apostolidis, C.; Bruchertseifer, F.; Repinc, U.; Morgenstern, A. Spectroscopic study of the interaction of U(VI) with transferrin and albumin for speciation of U(VI) under blood serum conditions. J. Inorg. Biochem. 2009, 103, 1609–1616. [Google Scholar] [CrossRef] [Green Version]
- Pible, O.; Vidaud, C.; Plantevin, S.; Pellequer, J.L.; Quemeneur, E. Predicting the disruption by UO(2)(2+) of a protein-ligand interaction. Protein Sci. 2010, 19, 2219–2230. [Google Scholar] [CrossRef] [Green Version]
- Safi, S.; Creff, G.; Jeanson, A.; Qi, L.; Basset, C.; Roques, J.; Solari, P.L.; Simoni, E.; Vidaud, C.; Den Auwer, C. Osteopontin: A uranium phosphorylated binding-site characterization. Chemistry 2013, 19, 11261–11269. [Google Scholar] [CrossRef]
- Vidaud, C.; Gourion-Arsiquaud, S.; Rollin-Genetet, F.; Torne-Celer, C.; Plantevin, S.; Pible, O.; Berthomieu, C.; Quemeneur, E. Structural consequences of binding of UO2(2+) to apotransferrin: Can this protein account for entry of uranium into human cells? Biochemistry 2007, 46, 2215–2226. [Google Scholar] [CrossRef]
- Michon, J.; Frelon, S.; Garnier, C.; Coppin, F. Determinations of uranium(VI) binding properties with some metalloproteins (transferrin, albumin, metallothionein and ferritin) by fluorescence quenching. J. Fluoresc. 2010, 20, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Scapolan, S. Uranium (VI)-Transferrin System Studied by Time-Resolved Laser-Induced Fluorescence. Radiat. Prot. Dosim. 1998, 79, 505–508. [Google Scholar] [CrossRef]
- Dedieu, A.; Berenguer, F.; Basset, C.; Prat, O.; Quemeneur, E.; Pible, O.; Vidaud, C. Identification of uranyl binding proteins from human kidney-2 cell extracts by immobilized uranyl affinity chromatography and mass spectrometry. J. Chromatogr. 2009, 1216, 5365–5376. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, M.W.; Villalobo, A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. BBA-Mol. Cell Res. 2014, 1843, 398–435. [Google Scholar] [CrossRef]
- Hoeflich, K.P.; Ikura, M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 2002, 108, 739–742. [Google Scholar] [CrossRef] [Green Version]
- Pardoux, R.; Sauge-Merle, S.; Lemaire, D.; Delangle, P.; Guilloreau, L.; Adriano, J.M.; Berthomieu, C. Modulating Uranium Binding Affinity in Engineered Calmodulin EF-Hand Peptides: Effect of Phosphorylation. PLoS ONE 2012, 7, e41922. [Google Scholar] [CrossRef] [Green Version]
- Brulfert, F.; Safi, S.; Jeanson, A.; Foerstendorf, H.; Weiss, S.; Berthomieu, C.; Sauge-Merle, S.; Simoni, E. Enzymatic activity of the CaM-PDE1 system upon addition of actinyl ions. J. Inorg. Biochem. 2017, 172, 46–54. [Google Scholar] [CrossRef]
- Pible, O.; Guilbaud, P.; Pellequer, J.L.; Vidaud, C.; Quemeneur, E. Structural insights into protein-uranyl interaction: Towards an in silico detection method. Biochimie 2006, 88, 1631–1638. [Google Scholar] [CrossRef]
- Van Horn, J.D.; Huang, H. Uranium(VI) bio-coordination chemistry from biochemical, solution and protein structural data. Coord. Chem. Rev. 2006, 250, 765–775. [Google Scholar] [CrossRef]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef]
- Ikura, M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 1996, 21, 14–17. [Google Scholar] [CrossRef]
- Babu, Y.S.; Bugg, C.E.; Cook, W.J. Structure of Calmodulin Refined at 2.2 a Resolution. J. Mol. Biol. 1988, 204, 191–204. [Google Scholar] [CrossRef]
- Babu, Y.S.; Sack, J.S.; Greenhough, T.J.; Bugg, C.E.; Means, A.R.; Cook, W.J. Three-dimensional structure of calmodulin. Nature 1985, 315, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Kretsinger, R.H. Calcium-Binding Proteins. Annu. Rev. Biochem. 1976, 45, 239–266. [Google Scholar] [CrossRef]
- Mcphalen, C.A.; Strynadka, N.C.J.; James, M.N.G. Calcium-Binding Sites in Proteins—A Structural Perspective. Adv. Protein Chem. 1991, 42, 77–144. [Google Scholar]
- Strynadka, N.C.J.; James, M.N.G. Crystal-Structures of the Helix-Loop-Helix Calcium-Binding Proteins. Annu. Rev. Biochem. 1989, 58, 951–998. [Google Scholar] [CrossRef]
- Linse, S.; Helmersson, A.; Forsen, S. Calcium-Binding to Calmodulin and Its Globular Domains. J. Biol. Chem. 1991, 266, 8050–8054. [Google Scholar] [CrossRef]
- Beccia, M.R.; Sauge-Merle, S.; Lemaire, D.; Bremond, N.; Pardoux, R.; Blangy, S.; Guilbaud, P.; Berthomieu, C. Thermodynamics of Calcium binding to the Calmodulin N-terminal domain to evaluate site-specific affinity constants and cooperativity. J. Biol. Inorg. Chem. JBIC A Publ. Soc. Biol. Inorg. Chem. 2015, 20, 905–919. [Google Scholar] [CrossRef]
- Le Clainche, L.; Figuet, M.; Montiardet-Bas, V.; Blanchard, S.; Vita, C. Modulating the affinity and the selectivity of engineered calmodulin EF-hand peptides for lanthanides. Biotechnol. Bioeng. 2006, 95, 29–36. [Google Scholar] [CrossRef]
- Le Clainche, L.; Plancque, G.; Amekraz, B.; Moulin, C.; Pradines-Lecomte, C.; Peltier, G.; Vita, C. Engineering new metal specificity in EF-hand peptides. J. Biol. Inorg. Chem. 2003, 8, 334–340. [Google Scholar] [CrossRef]
- Nitz, M.; Sherawat, M.; Franz, K.J.; Peisach, E.; Allen, K.N.; Imperiali, B. Structural origin of the high affinity of a chemically evolved lanthanide-binding peptide. Angew. Chem. Int. Ed. 2004, 43, 3682–3685. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.M.; Lee, H.W.; Yang, W.; Shealy, S.; Yang, J.J. Probing site-specific calmodulin calcium and lanthanide affinity by grafting. J. Am. Chem. Soc. 2005, 127, 3743–3750. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.H.; Yu, Q.H.; Wen, J.; Li, C.Y.; Guo, Z.H.; Wang, X.L.; Wang, N. Ultrafast and Highly Selective Uranium Extraction from Seawater by Hydrogel-like Spidroin-based Protein Fiber. Angew. Chem. Int. Ed. 2019, 58, 11785–11790. [Google Scholar] [CrossRef] [PubMed]
- Sauge-Merle, S.; Brulfert, F.; Pardoux, R.; Solari, P.L.; Lemaire, D.; Safi, S.; Guilbaud, P.; Simoni, E.; Merroun, M.L.; Berthomieu, C. Structural Analysis of Uranyl Complexation by the EF-Hand Motif of Calmodulin: Effect of Phosphorylation. Chem.-Eur. J. 2017, 23, 15505–15517. [Google Scholar] [CrossRef]
- Lebrun, C.; Starck, M.; Gathu, V.; Chenavier, Y.; Delangle, P. Engineering short peptide sequences for uranyl binding. Chemistry 2014, 20, 16566–16573. [Google Scholar] [CrossRef] [PubMed]
- Starck, M.; Laporte, F.A.; Oros, S.; Sisommay, N.; Gathu, V.; Solari, P.L.; Creff, G.; Roques, J.; Den Auwer, C.; Lebrun, C.; et al. Cyclic Phosphopeptides to Rationalize the Role of Phosphoamino Acids in Uranyl Binding to Biological Targets. Chemistry 2017, 23, 5281–5290. [Google Scholar] [CrossRef]
- Starck, M.; Sisommay, N.; Laporte, F.A.; Oros, S.; Lebrun, C.; Delangle, P. Preorganized Peptide Scaffolds as Mimics of Phosphorylated Proteins Binding Sites with a High Affinity for Uranyl. Inorg. Chem. 2015, 54, 11557–11562. [Google Scholar] [CrossRef]
- Pardoux, R.; Sauge-Merle, S.; Brémond, N.; Beccia, M.R.; Lemaire, D.; Battesti, C.; Delangle, P.; Solari, P.L.; Guilbaud, P.; Berthomieu, C. Optimized co-ordination of uranyl in engineered calmodulin site 1 provides a subnanomolar affinity for uranyl and a strong uranyl versus calcium selec-tivity. Inorg. Chem. 2022; in press. [Google Scholar]
- Kuboniwa, H.; Tjandra, N.; Grzesiek, S.; Ren, H.; Klee, C.B.; Bax, A. Solution Structure of Calcium-Free Calmodulin. Nat. Struct. Biol. 1995, 2, 768–776. [Google Scholar] [CrossRef]
- Zhang, M.; Tanaka, T.; Ikura, M. Calcium-Induced Conformational Transition Revealed by the Solution Structure of Apo Calmodulin. Nat. Struct. Biol. 1995, 2, 758–767. [Google Scholar] [CrossRef]
- Jiang, J.; Renshaw, J.C.; Sarsfield, M.J.; Livens, F.R.; Collison, D.; Charnock, J.M.; Eccles, H. Solution chemistry of uranyl ion with iminodiacetate and oxydiacetate: A combined NMR/EXAFS and potentiometry/calorimetry study. Inorg. Chem. 2003, 42, 1233–1240. [Google Scholar] [CrossRef]
- Velazquez-Campoy, A.; Goni, G.; Peregrina, J.R.; Medina, M. Exact analysis of heterotropic interactions in proteins: Characterization of cooperative ligand binding by isothermal titration calorimetry. Biophys. J. 2006, 91, 1887–1904. [Google Scholar] [CrossRef] [PubMed]
- Brown, A. Analysis of Cooperativity by Isothermal Titration Calorimetry. Int. J. Mol. Sci. 2009, 10, 3457–3477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. Charmm—A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone phi, psi and Side-Chain chi(1) and chi(2) Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [Green Version]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Duvail, M.; Ruas, A.; Venault, L.; Moisy, P.; Guilbaud, P. Molecular Dynamics Studies of Concentrated Binary Aqueous Solutions of Lanthanide Salts: Structures and Exchange Dynamics. Inorg. Chem. 2010, 49, 519–530. [Google Scholar] [CrossRef] [Green Version]
- Xi, C.W.; Schoeters, E.; Vanderleyden, J.; Michiels, J. Symbiosis-specific expression of Rhizobium etli casA encoding a secreted calmodulin-related protein. Proc. Natl. Acad. Sci. USA 2000, 97, 11114–11119. [Google Scholar] [CrossRef] [Green Version]
- Raos, N.; Kasprzak, K.S. Allosteric Binding of Nickel(Ii) to Calmodulin. Fundam. Appl. Toxicol. 1989, 13, 816–822. [Google Scholar] [CrossRef]
- Crouch, T.H.; Klee, C.B. Positive Cooperative Binding of Calcium to Bovine Brain Calmodulin. Biochemistry 1980, 19, 3692–3698. [Google Scholar] [CrossRef]
- Vanscyoc, W.S.; Shea, M.A. Phenylalanine fluorescence studies of calcium binding to N-domain fragments of Paramecium calmodulin mutants show increased calcium affinity correlates with increased disorder. Protein Sci. 2001, 10, 1758–1768. [Google Scholar] [CrossRef] [PubMed]
- Settimo, L.; Donnini, S.; Juffer, A.H.; Woody, R.W.; Marin, O. Conformational changes upon calcium binding and phosphorylation in a synthetic fragment of calmodulin. Biopolymers 2007, 88, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Le Clainche, L.; Vita, C. Selective binding of uranyl cation by a novel calmodulin peptide. Environ. Chem. Lett. 2006, 4, 45–49. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of Protein Association Reactions-Forces Contributing to Stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Grabarek, Z. Insights into modulation of calcium signaling by magnesium in calmodulin, troponin C and related EF-hand proteins. BBA-Mol. Cell Res. 2011, 1813, 913–921. [Google Scholar] [CrossRef] [PubMed]





| Peptide | Site I | Site II | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||||
| CaM Y II | MADQLTDDQISEFKEAFSLF | A | K | A | G | D | G | Y | I | T | T | K | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | T | I | D | F | P | E | FLNLMARK |
| CaM1 Y II | MADQLTDDQISEFKEAFSLF | A | K | A | G | D | G | Y | I | T | A | A | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | T | I | D | F | P | E | FLNLMARK |
| CaM YW II | MADQLTDDQISEFKEAFSLF | A | K | A | G | D | G | Y | I | T | T | K | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | W | I | D | F | P | E | FLNLMARK |
| CaM1 YW II | MADQLTDDQISEFKEAFSLF | A | K | A | G | D | G | Y | I | T | A | A | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | W | I | D | F | P | E | FLNLMARK |
| CaM1 Y II P | MADQLTDDQISEFKEAFSLF | A | K | A | G | D | G | Y | I | TP | A | A | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | T | I | D | F | P | E | FLNLMARK |
| CaM Y I-II | MADQLTDDQISEFKEAFSLF | D | K | D | G | D | G | Y | I | T | T | K | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | T | I | D | F | P | E | FLNLMARK |
| CaM1 Y I-II | MADQLTDDQISEFKEAFSLF | D | K | D | G | D | G | Y | I | T | A | A | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | T | I | D | F | P | E | FLNLMARK |
| CaM YW I-II | MADQLTDDQISEFKEAFSLF | D | K | D | G | D | G | Y | I | T | T | K | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | W | I | D | F | P | E | FLNLMARK |
| CaM1 YW I-II | MADQLTDDQISEFKEAFSLF | D | K | D | G | D | G | Y | I | T | A | A | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | W | I | D | F | P | E | FLNLMARK |
| CaM1 Y I-II P | MADQLTDDQISEFKEAFSLF | D | K | D | G | D | G | Y | I | TP | A | A | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | T | I | D | F | P | E | FLNLMARK |
| CaM1 YW I-II P | MADQLTDDQISEFKEAFSLF | D | K | D | G | D | G | Y | I | TP | A | A | E | LGTVMRSLGQNPTEAELQDMINEV | D | A | D | G | N | G | W | I | D | F | P | E | FLNLMARK |
| Peptide | pH | UO22+ | Ca2+ | ||||
|---|---|---|---|---|---|---|---|
| K1 (M−1) | Kd1 (nM) | K2 (M−1) | Kd2 (nM) | K1 (M−1) | K2 (M−1) | ||
| CaM Y II | 6 | a (3.7 ± 0.5) × 106 b (4.2 ± 1.0) × 106 | 270 ± 42 238 ± 74 | - | c (1.8 ± 0.3) × 104 | - | |
| CaM1 Y II | 6 | a (3.5 ± 0.7) × 106 b (3.9 ± 0.5) × 106 | 286 ± 47 256 ± 39 | - | a (2.2 ± 0.2) × 104 | - | |
| CaM YW II | 6 | a (5.1 ± 0.5) × 106 b (4.9 ± 1.3) × 106 | 196 ± 21 204 ± 74 | - | c (3.1 ± 0.3) × 104 | - | |
| CaM1 YW II | 6 | a (5.9 ± 0.6) × 106 b (9.0 ± 1.2) × 106 | 169 ± 20 111 ± 18 | - | a (4.7 ± 0.2) × 104 | - | |
| CaM Y I | 6 | d (4.0 ± 0.2) × 107 | 25 ± 1 | - | d (2.6 ± 0.1) × 104 | - | |
| CaM1 Y I | 6 | d (3.1 ± 0.1) × 107 | 32 ± 1 | - | d (4.3 ± 0.4) × 104 | - | |
| CaM1 Y II P | 6 7 | a (2.1 ± 0.4) × 107 a (1.8 ± 0.4) × 108 | 48 ± 11 5.5 ± 1 | - | - | - | |
| CaM Y I-II | 6 | a (3.5 ± 1.0) × 107 b (4.4 ± 0.7) × 107 | 29 ± 6 23 ± 3 | a (7.4 ± 0.8) × 106 b (5.7 ± 1.0) × 106 | 135 ± 13 175 ± 26 | c (5.3 ± 0.3) × 104 | c (1.4 ± 0.3) × 105 |
| CaM1 Y I-II | 6 | a (3.0 ± 0.9) × 107 b (4.5 ± 0.6) × 107 | 33 ± 7 22 ± 3 | a (7.0 ± 1.4) × 106 b (6.5 ± 0.7) × 106 | 143 ± 24 154 ± 15 | a (6.0 ± 0.6) × 104 | a (1.2 ± 0.3) × 105 |
| CaM YW I-II | 6 | a (7.4 ± 1.5) × 107 b (6.3 ± 0.5) × 107 | 14 ± 3 16 ± 2 | a (9.7 ± 1.0) × 106 b (7.8 ± 0.3) × 106 | 103 ± 10 128 ± 5 | c (8.1 ± 0.3) × 104 | c (3.1 ± 0.3) × 105 |
| CaM1 YW I-II | 6 | a (7.8 ± 0.3) × 107 b (7.0 ± 0.6) × 107 | 13 ± 1 14 ± 2 | a (9.4 ± 1.5) × 106 b (8.5 ± 0.5) × 106 | 106 ± 15 118 ± 7 | a (7.7 ± 0.4) × 104 | a (2.8 ± 0.2) × 105 |
| CaM1 Y I-II P | 6 7 | a (3.6 ± 1.0) × 108 a (1.0 ± 0.5) × 1011 | 2.8 ± 0.6 0.010 ± 0.008 | a (7.8 ± 0.7) × 107 a (7.1 ± 1.0) × 108 | 13 ± 2 1.4 ± 0.2 | a (7.1 ± 0.2) × 104 a (8.9 ± 0.4) × 104 | a (1.0 ± 0.7) × 105 a (1.1 ± 0.8) × 105 |
| CaM1 YW I-II P | 6 7 | a (3.8 ± 1.0) × 108 a (6.0 ± 0.9) × 1011 | 2.6 ± 0.6 0.0017 ± 0.0003 | a (1.3 ± 0.5) × 108 a (5.1 ± 0.6) × 109 | 7.7 ± 2.2 0.196 ± 0.021 | a (1.0 ± 0.3) × 105 a (9.0 ± 0.5) × 104 | a (2.2 ± 0.1) × 105 a (1.1 ± 0.2) × 105 |
| Peptide | ΔHI (kJ mol−1) | TΔSI (kJ mol−1) | ΔHII (kJ mol−1) | TΔSII (kJ mol−1) | ΔHc(kJ mol−1) | TΔSc (kJ mol−1) |
|---|---|---|---|---|---|---|
| CaM Y II | - | - | −2.0 ± 0.9 | 35.8 ± 0.9 | ||
| CaM1 Y II | - | - | −3.5 ± 1.0 | 34.1 ± 1.0 | ||
| CaM YW II | - | - | −8.9 ± 0.7 | 29.3 ± 0.8 | ||
| CaM1 YW II | - | - | −15.0 ± 2.5 | 24.7 ± 2.5 | ||
| CaM Y I-II | −40.2 ± 0.5 | 3.2 ± 0.5 | −2.0 ± 0.9 | 35.8 ± 0.9 | 25.0 ± 0.1 | 27.0 ± 0.1 |
| CaM1 Y I-II | −26.1 ± 0.3 | 8.6 ± 0.3 | −3.5 ± 1.0 | 34.1 ± 1.0 | 22.0 ± 0.2 | 5.8 ± 0.2 |
| CaM YW I-II | −34.8 ± 0.1 | 18.2 ± 0.1 | −8.9 ± 0.7 | 29.3 ± 0.8 | 3.8 ± 0.1 | 10.7 ± 0.1 |
| CaM1 YW I-II | −26.1 ± 0.3 | 44.5 ± 0.3 | −15.0 ± 5.0 | 24.7 ± 2.5 | 8.9 ± 0.3 | 1.4 ± 0.3 |
| Peptide | pH | ΔΔG (kJ mol−1) | KI (M−1) | KII (M−1) | KI,II (M−1) | KII,I (M−1) |
|---|---|---|---|---|---|---|
| CaM Y I-II | 6 | −2.0 ± 0.4 | (3.1 ± 0.3) × 107 | (3.7 ± 0.5) × 106 | (7.1 ± 0.2) × 107 | (8.3 ± 0.3) × 106 |
| CaM1 Y I-II | 6 | −2.0 ± 1.0 | (2.7 ± 0.3) × 107 | (3.5 ± 0.7) × 106 | (6.0 ± 0.2) × 107 | (7.9 ± 0.9) × 106 |
| CaM YW I-II | 6 | −1.8 ± 0.3 | (6.8 ± 0.2) × 107 | (5.1 ± 0.5) × 106 | (1.4 ± 0.1) × 108 | (1.0 ± 0.2) × 107 |
| CaM1 YW I-II | 6 | −1.3 ± 0.2 | (7.2 ± 0.3) × 107 | (5.9 ± 0.6) × 106 | (1.2 ± 0.1) × 108 | (1.0 ± 0.2) × 107 |
| CaM1 Y I-II P | 6 | −3.4 ± 0.4 | (3.4 ± 0.3) × 108 | (2.1 ± 0.4) × 107 | (1.3 ± 0.2) × 109 | (8.3 ± 0.3) × 107 |
| 7 | −3.4 ± 0.2 | (1.0 ± 0.5) × 1011 | (1.8 ± 0.4) × 108 | (3.9 ± 0.5) × 1011 | (7.1 ± 0.1) × 108 | |
| CaM1 YW I-II P | 6 | −4.7 ± 0.5 | (3.6 ± 0.3) × 108 | (2.1 ± 0.4) × 107 | (2.4 ± 0.2) × 109 | (1.4 ± 0.5) × 108 |
| 7 | −8.3 ± 0.3 | (6.0 ± 0.9) × 1011 | (1.8 ± 0.4) × 108 | (1.7 ± 0.5) × 1013 | (5.1 ± 0.2) × 109 |
| Site I | U1-OAsp3 | U1-OAsp5 | U1-OTyr7 | U1-OGlu12 |
| Mean Distance (Å) | 2.35 | 2.33 | 2.46 | 2.35 2.35 |
| Site II | U2-OAsp3- | U2-O H2O | U2-OAsp9 | U2-OGlu12 |
| Mean Distance (Å) | 2.32 | 2.36 | 2.39 | 2.37 2.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beccia, M.R.; Sauge-Merle, S.; Brémond, N.; Lemaire, D.; Henri, P.; Battesti, C.; Guilbaud, P.; Crouzy, S.; Berthomieu, C. Inter-Site Cooperativity of Calmodulin N-Terminal Domain and Phosphorylation Synergistically Improve the Affinity and Selectivity for Uranyl. Biomolecules 2022, 12, 1703. https://doi.org/10.3390/biom12111703
Beccia MR, Sauge-Merle S, Brémond N, Lemaire D, Henri P, Battesti C, Guilbaud P, Crouzy S, Berthomieu C. Inter-Site Cooperativity of Calmodulin N-Terminal Domain and Phosphorylation Synergistically Improve the Affinity and Selectivity for Uranyl. Biomolecules. 2022; 12(11):1703. https://doi.org/10.3390/biom12111703
Chicago/Turabian StyleBeccia, Maria Rosa, Sandrine Sauge-Merle, Nicolas Brémond, David Lemaire, Pierre Henri, Christine Battesti, Philippe Guilbaud, Serge Crouzy, and Catherine Berthomieu. 2022. "Inter-Site Cooperativity of Calmodulin N-Terminal Domain and Phosphorylation Synergistically Improve the Affinity and Selectivity for Uranyl" Biomolecules 12, no. 11: 1703. https://doi.org/10.3390/biom12111703
APA StyleBeccia, M. R., Sauge-Merle, S., Brémond, N., Lemaire, D., Henri, P., Battesti, C., Guilbaud, P., Crouzy, S., & Berthomieu, C. (2022). Inter-Site Cooperativity of Calmodulin N-Terminal Domain and Phosphorylation Synergistically Improve the Affinity and Selectivity for Uranyl. Biomolecules, 12(11), 1703. https://doi.org/10.3390/biom12111703





