Association of Angio-LncRNAs MIAT rs1061540/MALAT1 rs3200401 Molecular Variants with Gensini Score in Coronary Artery Disease Patients Undergoing Angiography
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Cardiovascular Disease (CVD) Risk Assessment
2.3. Echocardiography
2.4. Selective Coronary Angiography
2.5. Sample Collection and Laboratory Investigations
2.6. Allelic Discrimination Analysis
2.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics of the Study Population
3.2. Genotype Analysis
3.3. Association of LncRNA Variants and Disease Outcomes
3.4. Multivariate Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Aparicio, H.J.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics—2021 Update: A Report From the American Heart Association. Circulation 2021, 143, e254–e743. [Google Scholar] [CrossRef]
- Said, M.A.; van de Vegte, Y.; Zafar, M.M.; Van Der Ende, M.Y.; Raja, G.K.; Verweij, N.; Van Der Harst, P. Contributions of Interactions Between Lifestyle and Genetics on Coronary Artery Disease Risk. Curr. Cardiol. Rep. 2019, 21, 1–8. [Google Scholar] [CrossRef]
- Kandaswamy, E.; Zuo, L. Recent Advances in Treatment of Coronary Artery Disease: Role of Science and Technology. Int. J. Mol. Sci. 2018, 19, 424. [Google Scholar] [CrossRef]
- Khera, A.V.; Kathiresan, A.V.K.S. Genetics of coronary artery disease: Discovery, biology and clinical translation. Nat. Rev. Genet. 2017, 18, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Frías-Lasserre, D.; Villagra, C.A. The Importance of ncRNAs as Epigenetic Mechanisms in Phenotypic Variation and Organic Evolution. Front. Microbiol. 2017, 8, 2483. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Wang, Y.; Ding, H.; Xue, D.S.; Qi, H.; Li, P. MicroRNAs or Long Noncoding RNAs in Diagnosis and Prognosis of Coronary Artery Disease. Aging Dis. 2019, 10, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Saygili, H.; Bozgeyik, I.; Yumrutas, O.; Akturk, E.; Bagis, H. Differential Expression of Long Noncoding RNAs in Patients with Coronary Artery Disease. Mol. Syndromol. 2021, 12, 372–378. [Google Scholar] [CrossRef]
- Ismail, N.; Abdullah, N.; Abdul Murad, N.A.; Jamal, R.; Sulaiman, S.A. Long Non-Coding RNAs (lncRNAs) in Cardiovascu-lar Disease Complication of Type 2 Diabetes. Diagnostics 2021, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Garratt, H.; Ashburn, R.; Sopić, M.; Nogara, A.; Caporali, A.; Mitić, T. Long Non-Coding RNA Regulation of Epigenetics in Vascular Cells. Non-Coding RNA 2021, 7, 62. [Google Scholar] [CrossRef]
- Hosen, M.R.; Li, Q.; Liu, Y.; Zietzer, A.; Maus, K.; Goody, P.; Uchida, S.; Latz, E.; Werner, N.; Nickenig, G.; et al. CAD increases the long noncoding RNA PUNISHER in small extracellular vesicles and regulates endothelial cell function via vesicular shuttling. Mol. Ther. Nucleic Acids 2021, 25, 388–405. [Google Scholar] [CrossRef]
- Bell, R.D.; Long, X.; Lin, M.; Bergmann, J.H.; Nanda, V.; Cowan, S.L.; Zhou, Q.; Han, Y.; Spector, D.L.; Zheng, D.; et al. Identification and Initial Functional Characterization of a Human Vascular Cell–Enriched Long Noncoding RNA. Arter. Thromb. Vasc. Biol. 2014, 34, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.; El-Wazir, A.; Alghamdi, S.A.; Alhazmi, A.S.; El-Wazir, M.; Abdel-Daim, M.; Fawzy, M.S. Association of long non-coding RNA MIAT and MALAT1 expression profiles in peripheral blood of coronary artery disease patients with previous cardiac events. Genet. Mol. Biol. 2019, 42, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, Y.; Peng, Y.; Tang, J.; Li, H. Association of polymorphisms in MALAT1 with risk of coronary atherosclerotic heart disease in a Chinese population. Lipids Health Dis. 2018, 17, 1–7. [Google Scholar] [CrossRef]
- Neumann, P.; Jaé, N.; Knau, A.; Glaser, S.F.; Fouani, Y.; Rossbach, O.; Krüger, M.; John, D.; Bindereif, A.; Grote, P.; et al. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shastry, B.S. SNPs: Impact on Gene Function and Phenotype. Single Nucleotide Polymorphisms 2009, 578, 3–22. [Google Scholar] [CrossRef]
- Shahmoradi, N.; Nasiri, M.; Kamfiroozi, H.; Kheiry, M.A. Association of the rs555172 polymorphism in SENCR long non-coding RNA and atherosclerotic coronary artery disease. J. Cardiovasc. Thorac. Res. 2017, 9, 170–174. [Google Scholar] [CrossRef][Green Version]
- Aznaourova, M.; Schmerer, N.; Schmeck, B.; Schulte, L.N. Disease-Causing Mutations and Rearrangements in Long Non-coding RNA Gene Loci. Front. Genet. 2020, 11. [Google Scholar] [CrossRef]
- Mahmood, S.S.; Levy, D.; Vasan, R.S.; Wang, T.J. The Framingham Heart Study and the epidemiology of cardiovascular disease: A historical perspective. Lancet 2013, 383, 999–1008. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Toraih, E.A.; Aly, N.M.; Fakhr-Eldeen, A.; Badran, D.I.; Hussein, M.H. Atherosclerotic and thrombotic genetic and environmental determinants in Egyptian coronary artery disease patients: A pilot study. BMC Cardiovasc. Disord. 2017, 17, 1–19. [Google Scholar] [CrossRef]
- Schussler, J.M. Effectiveness and safety of transradial artery access for cardiac catheterization. Bayl. Univ. Med Cent. Proc. 2011, 24, 205–209. [Google Scholar] [CrossRef]
- Saha, T.; Khalequzzaman; Akanda, A.K.; Saha, S.; Tushar, A.Z.; Ahmed, R.; Saha, G.K.; Ullah, M. Association of GRACE Risk Score with Angiographic Severity of Coronary Artery Disease in patients with ST Elevation Myocardial Infarction. Cardiovasc. J. 2015, 8, 30–34. [Google Scholar] [CrossRef][Green Version]
- Gökdeniz, T.; Kalaycioğlu, E.; Aykan, A.; Boyaci, F.; Turan, T.; Gul, I.; Cavusoglu, G.; Dursun, I. Value of Coronary Artery Calcium Score to Predict Severity or Complexity of Coronary Artery Disease. Arq. Bras. de Cardiol. 2014, 102, 120–127. [Google Scholar] [CrossRef]
- Khandelwal, G. Prediction of Angiographic Extent of Coronary Artery Disease on the Basis of Clinical Risk Scores in Patients of Unstable Angina. J. Clin. Diagn. Res. 2015, 9, OC13–OC16. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, B.; Emül, A.; Özdemir, L.; Sağ, S.; Biçer, M.; Aydınlar, A. Association of Aortic Diameters with Coronary Artery Disease Severity and Albumin Excretion. BioMed Res. Int. 2015, 2015, 1–5. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the Concentration of Low-Density Lipoprotein Cholesterol in Plasma, Without Use of the Preparative Ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, H.M.; A Abdelghany, A.; Al Ageeli, E.; Kattan, S.W.; Hassan, R.; A Toraih, E.; Fawzy, M.S.; Mokhtar, N. Long Non-Coding RNAs Gene Variants as Molecular Markers for Diabetic Retinopathy Risk and Response to Anti-VEGF Therapy. Pharmacogenomics Pers. Med. 2021, 14, 997–1014. [Google Scholar] [CrossRef]
- Kutyavin, I.V. 3’-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000, 28, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, M.S.; Hussein, M.H.; Abdelaziz, E.Z.; Yamany, H.A.; Ismail, H.; Toraih, E.A. Association of MicroRNA-196a2 Variant with Response to Short-Acting β2-Agonist in COPD: An Egyptian Pilot Study. PLoS ONE 2016, 11, e0152834. [Google Scholar] [CrossRef]
- Yu, B.; Wang, S. Angio-LncRs: LncRNAs that regulate angiogenesis and vascular disease. Theranostics 2018, 8, 3654–3675. [Google Scholar] [CrossRef]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef]
- Ma, R.; He, X.; Zhu, X.; Pang, S.; Yan, B. Promoter polymorphisms in the lncRNA-MIAT gene associated with acute myocardial infarction in Chinese Han population: A case–control study. Biosci. Rep. 2020, 40, BSR20191203. [Google Scholar] [CrossRef] [PubMed]
- Arslan, S.; Berkan, O.; Lalem, T.; Özbilüm, N.; Göksel, S.; Korkmaz, Ö.; Çetin, N.; Devaux, Y. Long non-coding RNAs in the atherosclerotic plaque. Atherosclerosis 2017, 266, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Azat, M.; Huojiahemaiti, X.; Gao, R.; Peng, P. Long noncoding RNA MIAT: A potential role in the diagnosis and mediation of acute myocardial infarction. Mol. Med. Rep. 2019, 20, 5216–5222. [Google Scholar] [CrossRef]
- Liao, J.; He, Q.; Li, M.; Chen, Y.; Liu, Y.; Wang, J. LncRNA MIAT: Myocardial infarction associated and more. Gene 2016, 578, 158–161. [Google Scholar] [CrossRef]
- Yan, B.; Yao, J.; Liu, J.-Y.; Li, X.-M.; Wang, X.-Q.; Li, Y.-J.; Tao, Z.-F.; Song, Y.-C.; Yu-Chen, S.; Jiang, Q. lncRNA-MIAT Regulates Microvascular Dysfunction by Functioning as a Competing Endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef]
- Chen, S.; Chen, H.; Yu, C.; Lu, R.; Song, T.; Wang, X.; Tang, W.; Gao, Y. Long noncoding RNA myocardial infarction associated transcript promotes the development of thoracic aortic by targeting microRNA-145 via the PI3K/Akt signaling pathway. J. Cell. Biochem. 2019, 120, 14405–14413. [Google Scholar] [CrossRef]
- Ye, Z.-M.; Yang, S.; Xia, Y.-P.; Hu, R.-T.; Chen, S.; Li, B.-W.; Chen, S.-L.; Luo, X.-Y.; Mao, L.; Li, Y.; et al. LncRNA MIAT sponges miR-149-5p to inhibit efferocytosis in advanced atherosclerosis through CD47 upregulation. Cell Death Dis. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Zhong, X.; Ma, X.; Zhang, L.; Li, Y.; Li, Y.; He, R. MIAT promotes proliferation and hinders apoptosis by modulating miR-181b/STAT3 axis in ox-LDL-induced atherosclerosis cell models. Biomed. Pharmacother. 2018, 97, 1078–1085. [Google Scholar] [CrossRef] [PubMed]
- Esser, C.; Tomiuk, J. Reporting Hardy–Weinberg Tests in Case–Control Studies: Reasons for Caution but not for Panic Reactions. J. Investig. Dermatol. 2005, 124, 1082–1083. [Google Scholar] [CrossRef] [PubMed]
- Graffelman, J.; Jain, D.; Weir, B. A genome-wide study of Hardy–Weinberg equilibrium with next generation sequence data. Qual. Life Res. 2017, 136, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M.; Knight, J. Introduction to Genetic Association Studies. Cold Spring Harb. Protoc. 2012, 2012, 297–306. [Google Scholar] [CrossRef]
- Wang, J.-Z.; Xiang, J.-J.; Wu, L.-G.; Bai, Y.-S.; Chen, Z.-W.; Yin, X.-Q.; Wang, Q.; Guo, W.-H.; Peng, Y.; Guo, H.; et al. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: A survival cohort analysis. BMC Cancer 2017, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, D.; Zhang, Y.; Xu, X.; Bi, L.; Zhang, M.; Yu, B.; Zhang, Y. Association of lncRNA polymorphisms with triglyceride and total cholesterol levels among myocardial infarction patients in Chinese population. Gene 2019, 724, 143684. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Liu, X.; He, Z. Long non-coding RNA H19 and MALAT1 gene variants in patients with ischemic stroke in a northern Chinese Han population. Mol. Brain 2018, 11, 58. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, W.; Zhang, B.; Wu, Y.; Yan, S.; Yuan, Y.; Zhang, H.; Li, J.; Sun, K.; Wang, H.; et al. The MALAT1 gene polymorphism and its relationship with the onset of congenital heart disease in Chinese. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long Noncoding RNAs in Patients With Acute Myocardial Infarction. Circ. Res. 2014, 115, 668–677. [Google Scholar] [CrossRef]
- Sohrabifar, N.; Ghaderian, S.M.H.; Parsa, S.A.; Ghaedi, H.; Jafari, H. Variation in the expression level of MALAT1, MIAT and XIST lncRNAs in coronary artery disease patients with and without type 2 diabetes mellitus. Arch. Physiol. Biochem. 2020, 1–8. [Google Scholar] [CrossRef]
- Qiu, S.; Sun, J. lncRNA-MALAT1 expression in patients with coronary atherosclerosis and its predictive value for in-stent restenosis. Exp. Ther. Med. 2020, 20, 1. [Google Scholar] [CrossRef]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zörnig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long Noncoding RNA MALAT1 Regulates Endothelial Cell Function and Vessel Growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef]
- Huangfu, N.; Xu, Z.; Zheng, W.; Wang, Y.; Cheng, J.; Chen, X. LncRNA MALAT1 regulates oxLDL-induced CD36 expression via activating β-catenin. Biochem. Biophys. Res. Commun. 2018, 495, 2111–2117. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, T.; Duan, J.; Mu, N.; Zhang, T. MALAT1/miR-15b-5p/MAPK1 mediates endothelial progenitor cells autophagy and affects coronary atherosclerotic heart disease via mTOR signaling pathway. Aging 2019, 11, 1089–1109. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell. Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yang, L.; Guo, R.; Lu, N.; Shi, Y.; Wang, X. Long noncoding RNA MALAT1 promotes high glucose-induced human endothelial cells pyroptosis by affecting NLRP3 expression through competitively binding miR-22. Biochem. Biophys. Res. Commun. 2018, 509, 359–366. [Google Scholar] [CrossRef]
- Gordon, A.D.; Biswas, S.; Feng, B.; Chakrabarti, S. MALAT1: A regulator of inflammatory cytokines in diabetic complications. Endocrinol. Diabetes Metab. 2018, 1, e00010. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Yu, X.; Yu, Y.; Zhang, L.; Cen, Y.; Chu, J. Long noncoding RNA MALAT1 promotes high glucose-induced in-flammation and apoptosis of vascular endothelial cells by regulating miR-361-3p/SOCS3 axis. Int. J. Clin. Exp. Pathol. 2020, 13, 1243–1252. [Google Scholar] [PubMed]
- Wang, D.; Xu, H.; Wu, B.; Jiang, S.; Pan, H.; Wang, R.; Chen, J. Long non-coding RNA MALAT1 sponges miR-124-3p.1/KLF5 to promote pulmonary vascular remodeling and cell cycle progression of pulmonary artery hypertension. Int. J. Mol. Med. 2019, 44, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.-R.; Liu, W.; Zhang, Q.; Guo, A.-Y. lncRNASNP2: An updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res. 2017, 46, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Connelly, J.J.; Wang, T.; E Cox, J.; Haynes, C.; Wang, L.; Shah, S.H.; Crosslin, D.R.; Hale, A.B.; Nelson, S.; Crossman, D.C.; et al. GATA2 Is Associated with Familial Early-Onset Coronary Artery Disease. PLoS Genet. 2006, 2, 139. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, T.-N.; Knight, J.K.; Goodwin, J.E. The Glucocorticoid Receptor in Cardiovascular Health and Disease. Cells 2019, 8, 1227. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Dimmeler, S. Long Noncoding RNAs in Cardiovascular Diseases. Circ. Res. 2015, 116, 737–750. [Google Scholar] [CrossRef]
- Thum, T.; Kumarswamy, R. The Smooth Long Noncoding RNA SENCR. Arter. Thromb. Vasc. Biol. 2014, 34, 1124–1125. [Google Scholar] [CrossRef][Green Version]
- Lyu, Q.; Xu, S.; Lyu, Y.; Choi, M.; Christie, C.K.; Slivano, O.J.; Rahman, A.; Jin, Z.-G.; Long, X.; Xu, Y.; et al. SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc. Natl. Acad. Sci. USA 2019, 116, 546–555. [Google Scholar] [CrossRef]
- Liao, Y.; Zhang, B.; Zhang, T.; Zhang, Y.; Wang, F. LncRNA GATA6-AS Promotes Cancer Cell Proliferation and Inhibits Apoptosis in Glioma by Downregulating lncRNA TUG1. Cancer Biotherapy Radiopharm. 2019, 34, 660–665. [Google Scholar] [CrossRef]
- Zhang, L.; Cheng, H.; Yue, Y.; Li, S.; Zhang, D.; He, R. TUG1 knockdown ameliorates atherosclerosis via up-regulating the expression of miR-133a target gene FGF1. Cardiovasc. Pathol. 2018, 33, 6–15. [Google Scholar] [CrossRef]
- Jha, R.; Li, D.; Wu, Q.; Ferguson, K.E.; Forghani, P.; Gibson, G.C.; Xu, C. A long non-coding RNA GATA6-AS1 adjacent to GATA6 is required for cardiomyocyte differentiation from human pluripotent stem cells. FASEB J. 2020, 34, 14336–14352. [Google Scholar] [CrossRef] [PubMed]
- Kurian, L.; Aguirre, A.; Sancho-Martinez, I.; Benner, C.; Hishida, T.; Nguyen, T.B.; Reddy, P.; Nivet, E.; Krause, M.N.; Nelles, D.A.; et al. Identification of Novel Long Noncoding RNAs Underlying Vertebrate Cardiovascular Development. Circulation 2015, 131, 1278–1290. [Google Scholar] [CrossRef]
- Jaé, N.; Dimmeler, S. Noncoding RNAs in Vascular Diseases. Circ. Res. 2020, 126, 1127–1145. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Ding, H.; Ouyang, A.; Zhang, X.; Xu, Q.; Han, Y.; Zhang, X.; Jin, Y. LncRNA MALAT1 gene polymorphisms in coronary artery disease: A case–control study in a Chinese population. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Controls (n = 100) | Patients (n = 100) | p-Value |
|---|---|---|---|
| Demographic data | |||
| Age, years | |||
| Mean ± SD | 56 ± 9.0 | 56.0 ± 9.0 | 0.876 |
| Sex | |||
| Male | 52 (52.0) | 64 (64.0) | 0.023 |
| Weight, kg | 77 ± 6.0 | 84 ± 11 | 0.118 |
| Height, cm | 165 ± 7.0 | 168 ± 6.6 | 0.713 |
| BMI, kg/m2 | 28 ± 2.0 | 30 ± 0.5 | 0.038 |
| Obesity | 2 (2.0) | 44 (44.0) | 0.001 |
| Family history of CVD | 36 (36.0) | 22 (22.0) | 0.123 |
| Smoking | 16 (16.0) | 70 (70.0) | <0.001 |
| Clinical data | |||
| DM | 28 (28.0) | 44 (44.0) | 0.096 |
| HTN | 38 (38.0) | 54 (54.0) | 0.108 |
| Premature CAD | --- | 80 (80.0) | NA |
| Previous events | --- | 80 (80.0) | NA |
| Stroke | --- | 2 (2.0) | NA |
| Aneurysms | --- | 2 (2.0) | NA |
| Echocardiography | |||
| Dias BP, mmHg | --- | 82 ± 14 | NA |
| Pulse, bpm | --- | 87 ± 13 | NA |
| EDD | --- | 52 ± 7.0 | NA |
| ESD | --- | 38 ± 6.0 | NA |
| PW | --- | 9 ± 2.0 | NA |
| SW | --- | 9 ± 2.0 | NA |
| EF | --- | 55 ± 13 | NA |
| Angiography | |||
| Gensini score | --- | 38 ± 43 | NA |
| Vessel score | --- | 2.0 ± 2.0 | NA |
| Laboratory data | |||
| HDL-c | 50 ± 7.0 | 38 ± 13 | <0.001 |
| LDL-c | 77 ± 12 | 145 ± 49 | <0.001 |
| TC | 168 ± 18 | 221 ± 50 | <0.001 |
| TG | 95 ± 35 | 182 ± 72 | <0.001 |
| FBS | --- | 151 ± 67 | NA |
| Gene | Frequency | Variant | All | Controls | Patients | p-Value | |||
|---|---|---|---|---|---|---|---|---|---|
| n | Proportion | n | Proportion | n | Proportion | ||||
| PUNISHER (AGAP2-AS1) rs12318065 | Genotype frequency | A/A | 26 | 0.13 | 10 | 0.1 | 16 | 0.16 | 0.15 |
| C/A | 70 | 0.35 | 44 | 0.44 | 26 | 0.26 | |||
| C/C | 104 | 0.52 | 46 | 0.46 | 58 | 0.58 | |||
| P HWE | 1.00 | ||||||||
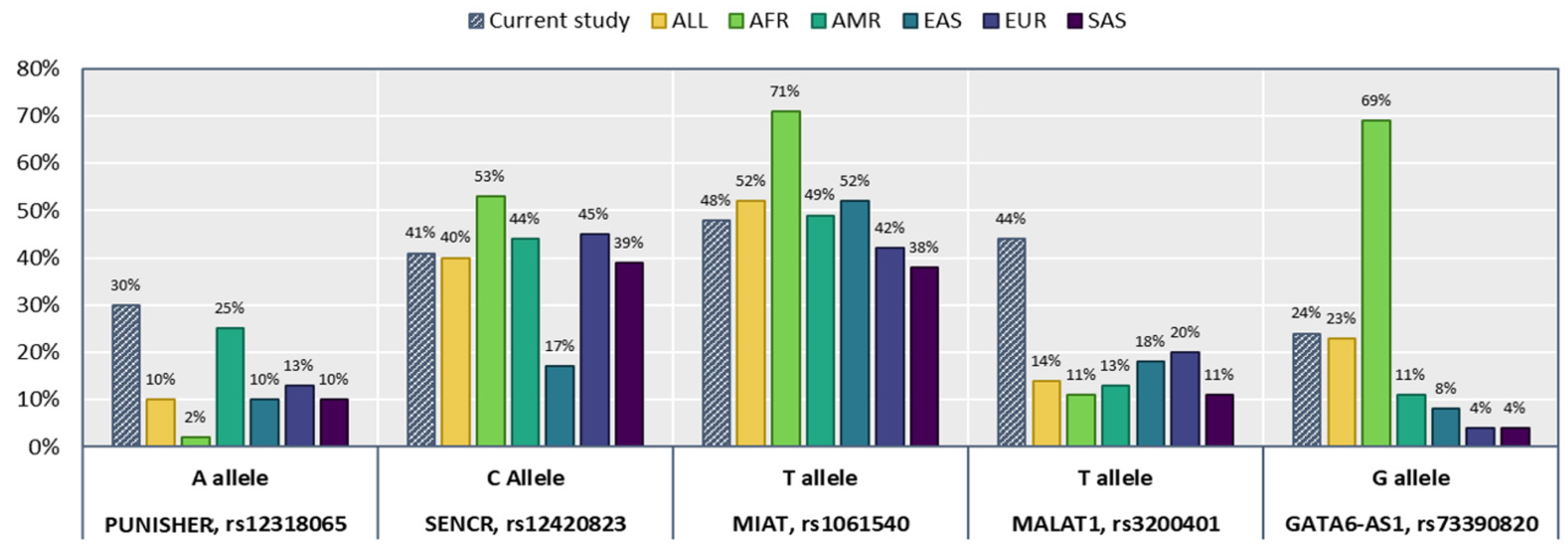
| Allele frequency | C | 278 | 0.7 | 136 | 0.68 | 142 | 0.71 | 0.64 | |
| A | 122 | 0.3 | 64 | 0.32 | 58 | 0.29 | |||
| SENCR (FLI1) rs12420823 | Genotype frequency | C/C | 24 | 0.12 | 12 | 0.12 | 12 | 0.12 | 0.66 |
| T/C | 116 | 0.58 | 62 | 0.62 | 54 | 0.54 | |||
| T/T | 60 | 0.3 | 26 | 0.26 | 34 | 0.34 | |||
| P HWE | 0.08 | ||||||||
| Allele frequency | T | 236 | 0.59 | 114 | 0.57 | 122 | 0.61 | 0.56 | |
| C | 164 | 0.41 | 86 | 0.43 | 78 | 0.39 | |||
| MIAT rs1061540 | Genotype frequency | C/C | 70 | 0.35 | 30 | 0.3 | 40 | 0.4 | 0.47 |
| C/T | 66 | 0.33 | 38 | 0.38 | 28 | 0.28 | |||
| T/T | 64 | 0.32 | 32 | 0.32 | 32 | 0.32 | |||
| P HWE | 0.09 | ||||||||
| Allele frequency | C | 206 | 0.52 | 98 | 0.49 | 108 | 0.54 | 0.47 | |
| T | 194 | 0.48 | 102 | 0.51 | 92 | 0.46 | |||
| MALAT1 rs3200401 | Genotype frequency | C/C | 92 | 0.46 | 38 | 0.38 | 54 | 0.54 | 0.15 |
| C/T | 42 | 0.21 | 28 | 0.28 | 14 | 0.14 | |||
| T/T | 66 | 0.33 | 34 | 0.34 | 32 | 0.32 | |||
| P HWE | 0.001 | ||||||||
| Allele frequency | C | 226 | 0.56 | 104 | 0.52 | 122 | 0.61 | 0.19 | |
| T | 174 | 0.44 | 96 | 0.48 | 78 | 0.39 | |||
| GATA6-AS1 rs73390820 | Genotype frequency | A/A | 114 | 0.57 | 58 | 0.58 | 56 | 0.56 | 0.62 |
| A/G | 74 | 0.37 | 34 | 0.34 | 40 | 0.4 | |||
| G/G | 12 | 0.06 | 8 | 0.08 | 4 | 0.04 | |||
| P HWE | 0.47 | ||||||||
| Allele frequency | A | 302 | 0.76 | 150 | 0.75 | 152 | 0.76 | 0.86 | |
| G | 98 | 0.24 | 50 | 0.25 | 48 | 0.24 | |||
| Gene | Model | Genotype | Controls | Patients | Crude OR (95% CI) | p-Value | Adjusted OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|---|
| PUNISHER | Codominant | C/C | 46 (46%) | 58 (58%) | 1.00 | 0.15 | 1.00 | 0.39 |
| A/C | 44 (44%) | 26 (26%) | 0.47 (0.19–1.13) | 0.47 (0.16–1.41) | ||||
| A/A | 10 (10%) | 16 (16%) | 1.27 (0.37–4.40) | 0.80 (0.17–3.73) | ||||
| Dominant | C/C | 46 (46%) | 58 (58%) | 1.00 | 0.23 | 1.00 | 0.23 | |
| A/C–A/A | 54 (54%) | 42 (42%) | 0.62 (0.28–1.36) | 0.54 (0.20–1.49) | ||||
| Recessive | C/C–A/C | 90 (90%) | 84 (84%) | 1.00 | 0.37 | 1.00 | 0.90 | |
| A/A | 10 (10%) | 16 (16%) | 1.71 (0.52–5.66) | 1.10 (0.26–4.73) | ||||
| Log-additive | --- | --- | --- | 0.89 (0.51–1.55) | 0.67 | 0.76 (0.37–1.53) | 0.44 | |
| SENCR | Codominant | T/T | 26 (26%) | 34 (34%) | 1.00 | 0.67 | 1.00 | 0.77 |
| C/T | 62 (62%) | 54 (54%) | 0.67 (0.27–1.62) | 0.83 (0.26–2.59) | ||||
| C/C | 12 (12%) | 12 (12%) | 0.76 (0.20–2.93) | 1.49 (0.27–8.30) | ||||
| Dominant | T/T | 26 (26%) | 34 (34%) | 1.00 | 0.38 | 1.00 | 0.89 | |
| C/T-C/C | 74 (74%) | 66 (66%) | 0.68 (0.29–1.61) | 0.93 (0.31–2.76) | ||||
| Recessive | T/T-C/T | 88 (88%) | 88 (88%) | 1.00 | 1.00 | 1.00 | 0.52 | |
| C/C | 12 (12%) | 12 (12%) | 1.00 (0.30–3.34) | 1.68 (0.35–8.08) | ||||
| Log-additive | --- | --- | --- | 0.81 (0.43–1.53) | 0.52 | 1.10 (0.49–2.45) | 0.82 | |
| MIAT | Codominant | C/C | 30 (30%) | 40 (40%) | 1.00 | 0.48 | 1.00 | 0.25 |
| C/T | 38 (38%) | 28 (28%) | 0.55 (0.21–1.45) | 0.54 (0.16–1.83) | ||||
| T/T | 32 (32%) | 32 (32%) | 0.75 (0.29–1.97) | 0.35 (0.10–1.28) | ||||
| Dominant | C/C | 30 (30%) | 40 (40%) | 1.00 | 0.29 | 1.00 | 0.12 | |
| C/T-T/T | 70 (70%) | 60 (60%) | 0.64 (0.28–1.47) | 0.44 (0.15–1.27) | ||||
| Recessive | C/C-C/T | 68 (68%) | 68 (68%) | 1.00 | 1.00 | 1.00 | 0.18 | |
| T/T | 32 (32%) | 32 (32%) | 1.00 (0.43–2.32) | 0.46 (0.14–1.48) | ||||
| Log-additive | --- | --- | --- | 0.86 (0.53–1.39) | 0.54 | 0.59 (0.31–1.12) | 0.09 | |
| MALAT1 | Codominant | C/C | 38 (38%) | 54 (54%) | 1.00 | 0.15 | 1.00 | 0.39 |
| T/C | 28 (28%) | 14 (14%) | 0.35 (0.12–1.04) | 0.43 (0.11–1.68) | ||||
| T/T | 34 (34%) | 32 (32%) | 0.66 (0.27–1.63) | 0.53 (0.16–1.73) | ||||
| Dominant | C/C | 38 (38%) | 54 (54%) | 1.00 | 0.11 | 1.00 | 0.18 | |
| T/C-T/T | 62 (62%) | 46 (46%) | 0.52 (0.24–1.16) | 0.49 (0.17–1.40) | ||||
| Recessive | C/C-T/C | 66 (66%) | 68 (68%) | 1.00 | 0.83 | 1.00 | 0.53 | |
| T/T | 34 (34%) | 32 (32%) | 0.91 (0.40–2.10) | 0.71 (0.24–2.08) | ||||
| Log-additive | --- | --- | --- | 0.79 (0.51–1.24) | 0.31 | 0.72 (0.40–1.30) | 0.27 | |
| GATA6-AS1 | Codominant | A/A | 58 (58%) | 56 (56%) | 1.00 | 0.62 | 1.00 | 0.84 |
| A/G | 34 (34%) | 40 (40%) | 1.22 (0.53–2.79) | 1.01 (0.36–2.82) | ||||
| G/G | 8 (8%) | 4 (4%) | 0.52 (0.09–3.06) | 0.45 (0.03–7.02) | ||||
| Dominant | A/A | 58 (58%) | 56 (56%) | 1.00 | 0.84 | 1.00 | 0.90 | |
| A/G-G/G | 42 (42%) | 44 (44%) | 1.09 (0.49–2.40) | 0.94 (0.35–2.56) | ||||
| Recessive | A/A-A/G | 92 (92%) | 96 (96%) | 1.00 | 0.40 | 1.00 | 0.55 | |
| G/G | 8 (8%) | 4 (4%) | 0.48 (0.08–2.74) | 0.45 (0.03–6.78) | ||||
| Log-additive | --- | --- | --- | 0.95 (0.50–1.81) | 0.87 | 0.88 (0.37–2.08) | 0.76 |
| PUNISHER | SENCR | MIAT | MALAT1 | GATA6-AS1 | Total | Controls | Patients | Cumulative Frequency | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C | T | C | T | A | 0.0917 | 0.1292 | 0.0578 | 0.0917 |
| 2 | C | C | T | C | A | 0.0871 | 0.0803 | 0.0923 | 0.1788 |
| 3 | C | T | T | C | G | 0.0817 | 0.0782 | 0.0687 | 0.2605 |
| 4 | C | T | T | T | A | 0.0724 | 0.1059 | 0.0685 | 0.3329 |
| 5 | C | C | C | C | A | 0.072 | 0.0434 | 0.0609 | 0.4049 |
| 6 | C | T | C | C | A | 0.0711 | 0.0508 | 0.1244 | 0.476 |
| 7 | A | C | C | C | A | 0.0645 | 0.1095 | NA | 0.5405 |
| 8 | A | T | T | T | A | 0.0615 | 0.0376 | 0.05 | 0.602 |
| 9 | A | T | C | C | A | 0.0457 | 0.0493 | 0.0626 | 0.6477 |
| 10 | C | C | C | T | A | 0.0377 | 0.0206 | 0.0614 | 0.6854 |
| 11 | C | C | T | T | G | 0.0376 | 0.0341 | 0.0285 | 0.723 |
| 12 | C | C | T | T | A | 0.033 | 0.0409 | 0.013 | 0.756 |
| 13 | A | T | T | C | A | 0.033 | 1e-04 | 0.0777 | 0.789 |
| 14 | A | T | C | C | G | 0.0264 | 0.0224 | 0.0231 | 0.8154 |
| 15 | A | T | C | T | A | 0.0235 | NA |
| Gene | Model | Genotypes | n | Gensini Score Mean (SEM) | Difference (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| PUNISHER | Codominant | C/C | 58 | 36.86 (7.69) | Reference | 0.38 |
| A/C | 26 | 49.92 (13.84) | 14.20 (−13.18, 41.58) | |||
| A/A | 16 | 23.12 (12.31) | −11.08 (−43.92, 21.75) | |||
| Dominant | C/C | 58 | 36.86 (7.69) | Reference | 0.71 | |
| A/C-A/A | 42 | 39.71 (9.99) | 4.61 (−19.13, 28.35) | |||
| Recessive | C/C-A/C | 84 | 40.9 (6.79) | Reference | 0.34 | |
| A/A | 16 | 23.12 (12.31) | −15.51 (−47.23, 16.21) | |||
| Log-additive | --- | --- | --- | −1.70 (−17.34, 13.93) | 0.83 | |
| SENCR | Codominant | T/T | 34 | 40.24 (11.74) | Reference | 0.75 |
| C/T | 54 | 39.56 (7.89) | −0.70 (−26.96, 25.55) | |||
| C/C | 12 | 25.17 (15.95) | −14.39 (−53.90, 25.12) | |||
| Dominant | T/T | 34 | 40.24 (11.74) | Reference | 0.79 | |
| C/T-C/C | 66 | 36.94 (7.04) | −3.39 (−28.44, 21.65) | |||
| Recessive | T/T-C/T | 88 | 39.82 (6.55) | Reference | 0.45 | |
| C/C | 12 | 25.17 (15.95) | −13.97 (−49.91, 21.96) | |||
| Log-additive | --- | --- | --- | −5.39 (−23.64, 12.85) | 0.75 | |
| MIAT | Codominant | C/C | 40 | 28.2 (8.65) | Reference | 0.06 |
| C/T | 28 | 31.86 (9.52) | 5.47 (−22.34, 33.27) | |||
| T/T | 32 | 55.81 (12.51) | 31.88 (5.29, 58.47) | |||
| Dominant | C/C | 40 | 28.2 (8.65) | Reference | 0.11 | |
| C/T-T/T | 60 | 44.63 (8.19) | 19.70 (−3.79, 43.18) | |||
| Recessive | C/C-C/T | 68 | 29.71 (6.33) | Reference | 0.019 | |
| T/T | 32 | 55.81 (12.51) | 29.64 (5.83, 53.46) | |||
| Log-additive | --- | --- | --- | 15.63 (2.40–28.86) | 0.025 | |
| MALAT1 | Codominant | C/C | 54 | 36.67 (8.64) | Reference | 0.044 |
| T/C | 14 | 16.29 (8.33) | −35.10 (−69.90–−0.30) | |||
| T/T | 32 | 49.94 (11) | 13.68 (−11.54–38.90) | |||
| Dominant | C/C | 54 | 36.67 (8.64) | Reference | 0.97 | |
| T/C-T/T | 46 | 39.7 (8.6) | −0.43 (−24.65–23.79) | |||
| Recessive | C/C-T/C | 68 | 32.47 (7.17) | Reference | 0.11 | |
| T/T | 32 | 49.94 (11) | 20.68 (−4.32–45.68) | |||
| Log-additive | --- | --- | --- | 5.42 (−7.80–18.64) | 0.43 | |
| GATA | Codominant | A/A | 56 | 30.25 (7.24) | Reference | 0.25 |
| A/G | 40 | 47.25 (10.37) | 18.19 (−5.68–42.05) | |||
| G/G | 4 | 55.5 (52.5) | 31.75 (−28.87–92.37) | |||
| Dominant | A/A | 56 | 30.25 (7.24) | Reference | 0.1 | |
| A/G-G/G | 44 | 48 (10.04) | 19.44 (−3.54–42.41) | |||
| Recessive | A/A-A/G | 96 | 37.33 (6.1) | Reference | 0.43 | |
| G/G | 4 | 55.5 (52.5) | 24.77 (−35.95–85.49) | |||
| Log-additive | --- | --- | --- | 17.35 (−2.51–37.20) | 0.094 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elwazir, M.Y.; Hussein, M.H.; Toraih, E.A.; Al Ageeli, E.; Esmaeel, S.E.; Fawzy, M.S.; Faisal, S. Association of Angio-LncRNAs MIAT rs1061540/MALAT1 rs3200401 Molecular Variants with Gensini Score in Coronary Artery Disease Patients Undergoing Angiography. Biomolecules 2022, 12, 137. https://doi.org/10.3390/biom12010137
Elwazir MY, Hussein MH, Toraih EA, Al Ageeli E, Esmaeel SE, Fawzy MS, Faisal S. Association of Angio-LncRNAs MIAT rs1061540/MALAT1 rs3200401 Molecular Variants with Gensini Score in Coronary Artery Disease Patients Undergoing Angiography. Biomolecules. 2022; 12(1):137. https://doi.org/10.3390/biom12010137
Chicago/Turabian StyleElwazir, Mohamed Y., Mohammad H. Hussein, Eman A. Toraih, Essam Al Ageeli, Safya E. Esmaeel, Manal S. Fawzy, and Salwa Faisal. 2022. "Association of Angio-LncRNAs MIAT rs1061540/MALAT1 rs3200401 Molecular Variants with Gensini Score in Coronary Artery Disease Patients Undergoing Angiography" Biomolecules 12, no. 1: 137. https://doi.org/10.3390/biom12010137
APA StyleElwazir, M. Y., Hussein, M. H., Toraih, E. A., Al Ageeli, E., Esmaeel, S. E., Fawzy, M. S., & Faisal, S. (2022). Association of Angio-LncRNAs MIAT rs1061540/MALAT1 rs3200401 Molecular Variants with Gensini Score in Coronary Artery Disease Patients Undergoing Angiography. Biomolecules, 12(1), 137. https://doi.org/10.3390/biom12010137








