The Iron Maiden. Cytosolic Aconitase/IRP1 Conformational Transition in the Regulation of Ferritin Translation and Iron Hemostasis
Abstract
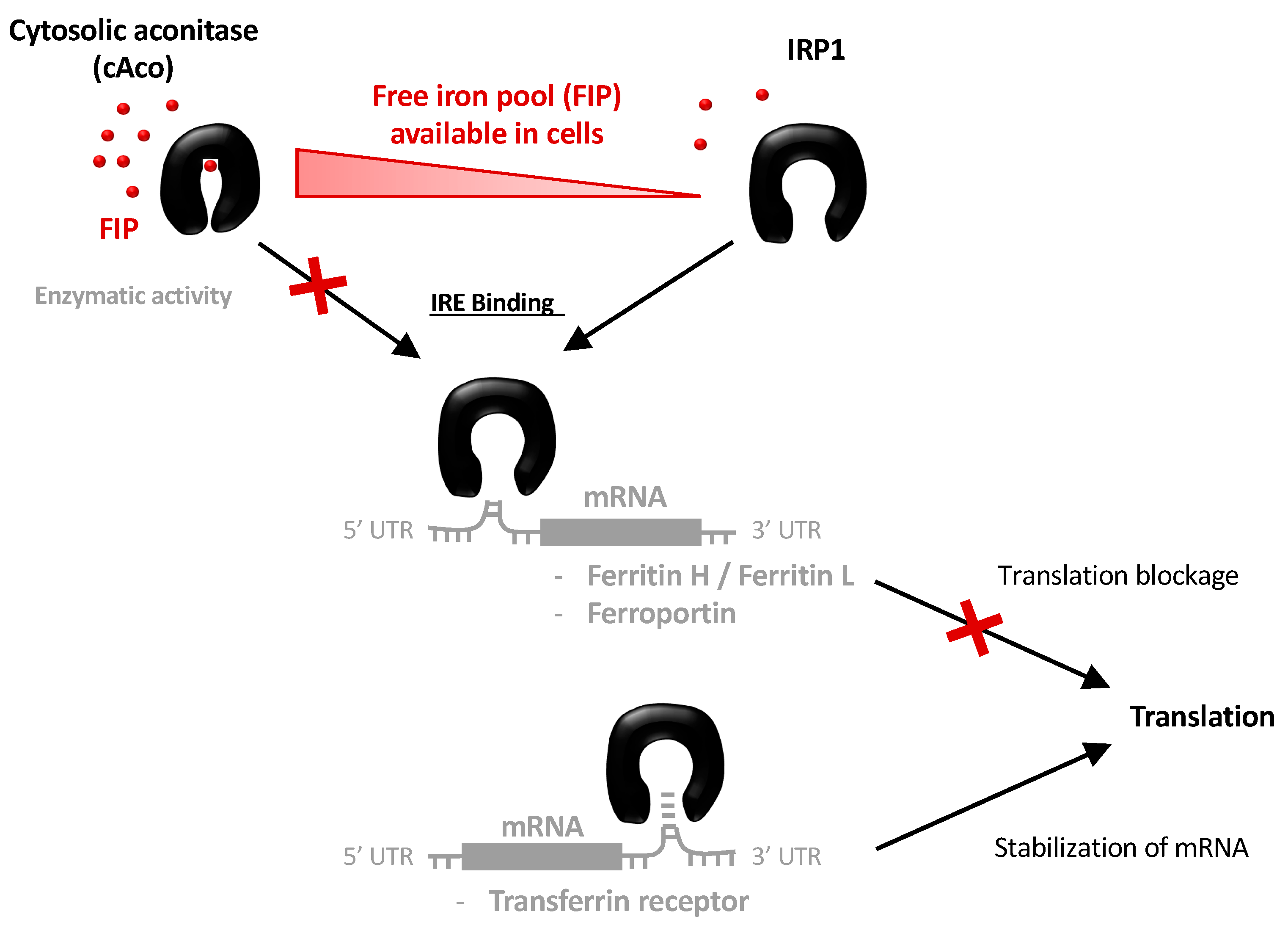
1. Introduction
2. Materials and Methods
3. Results
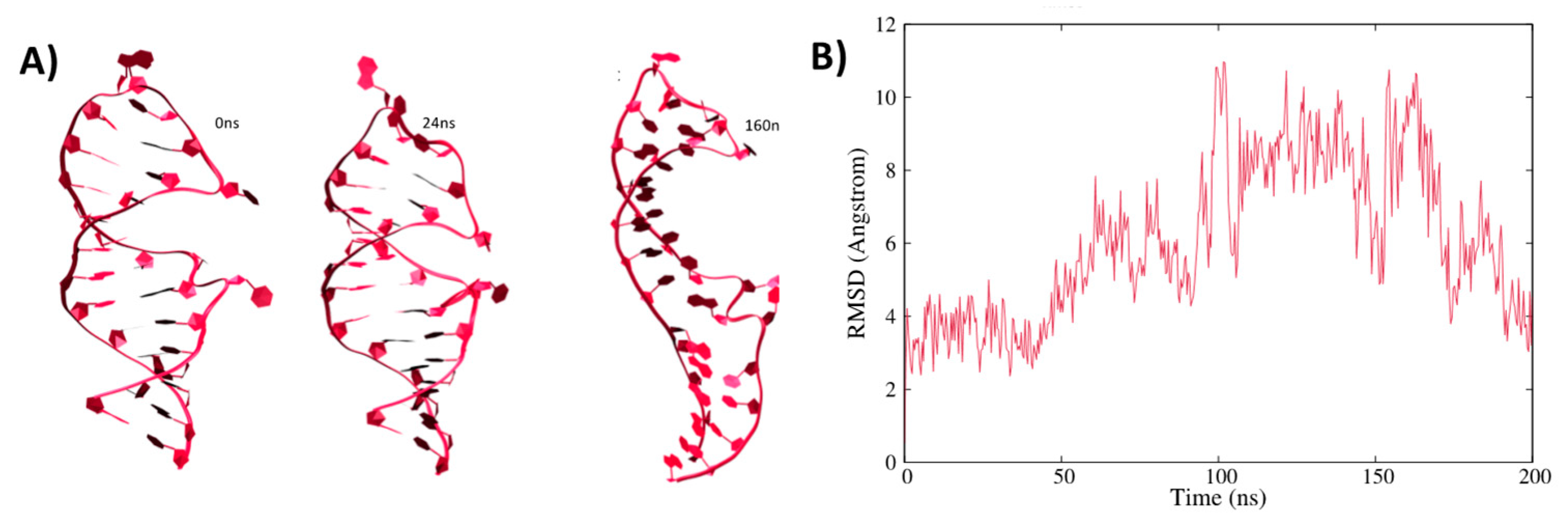
3.1. Structure of IRE mRNA
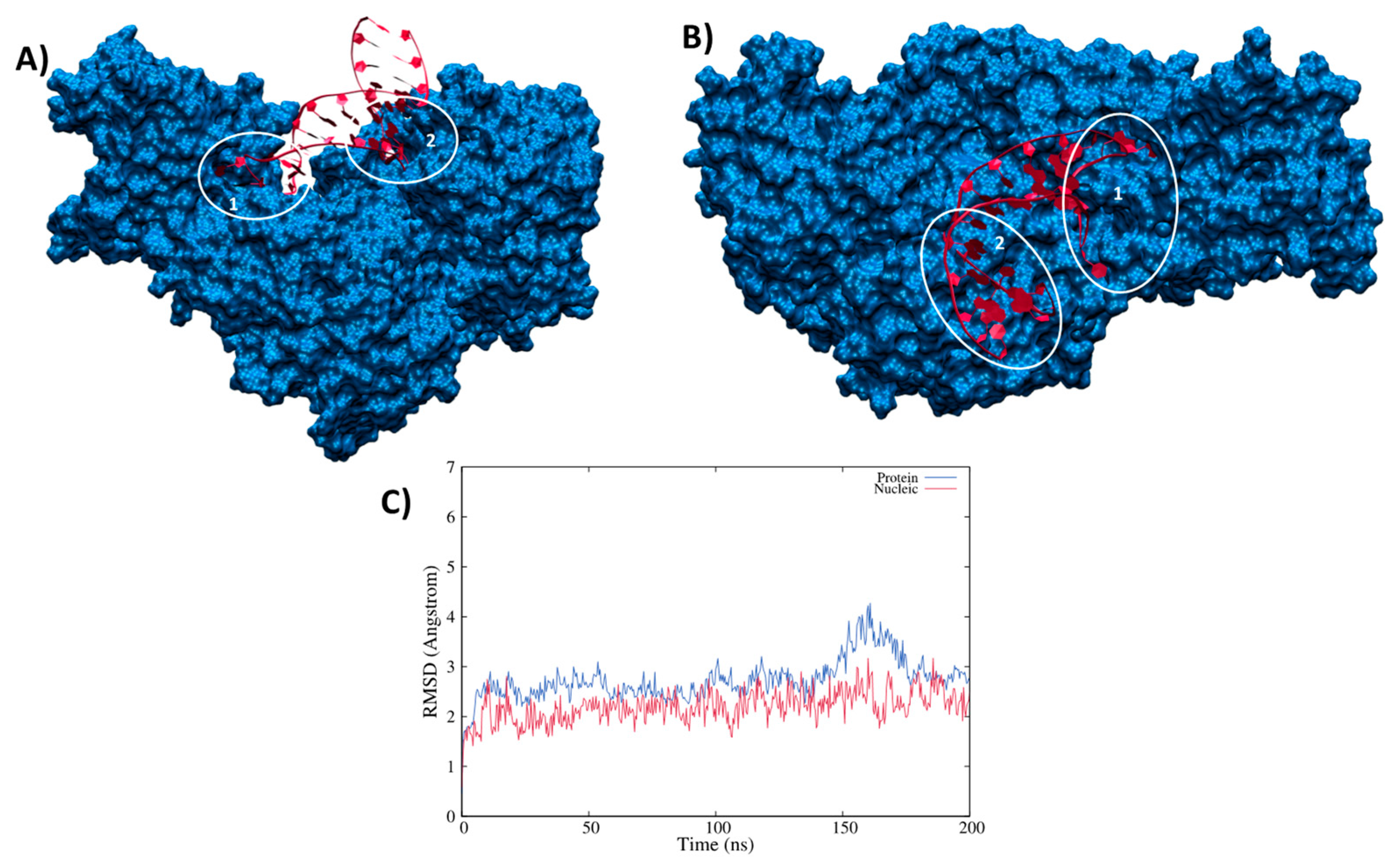
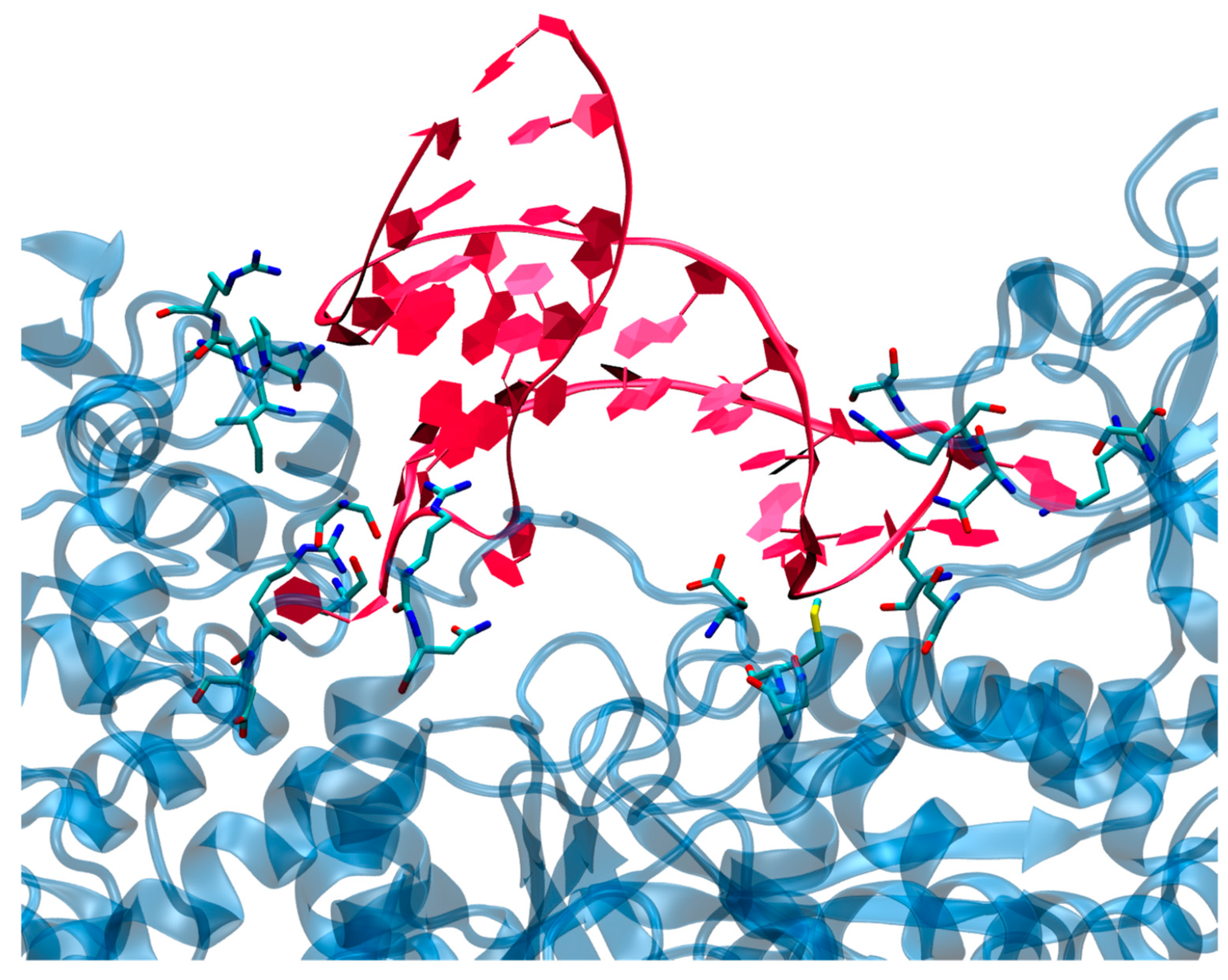
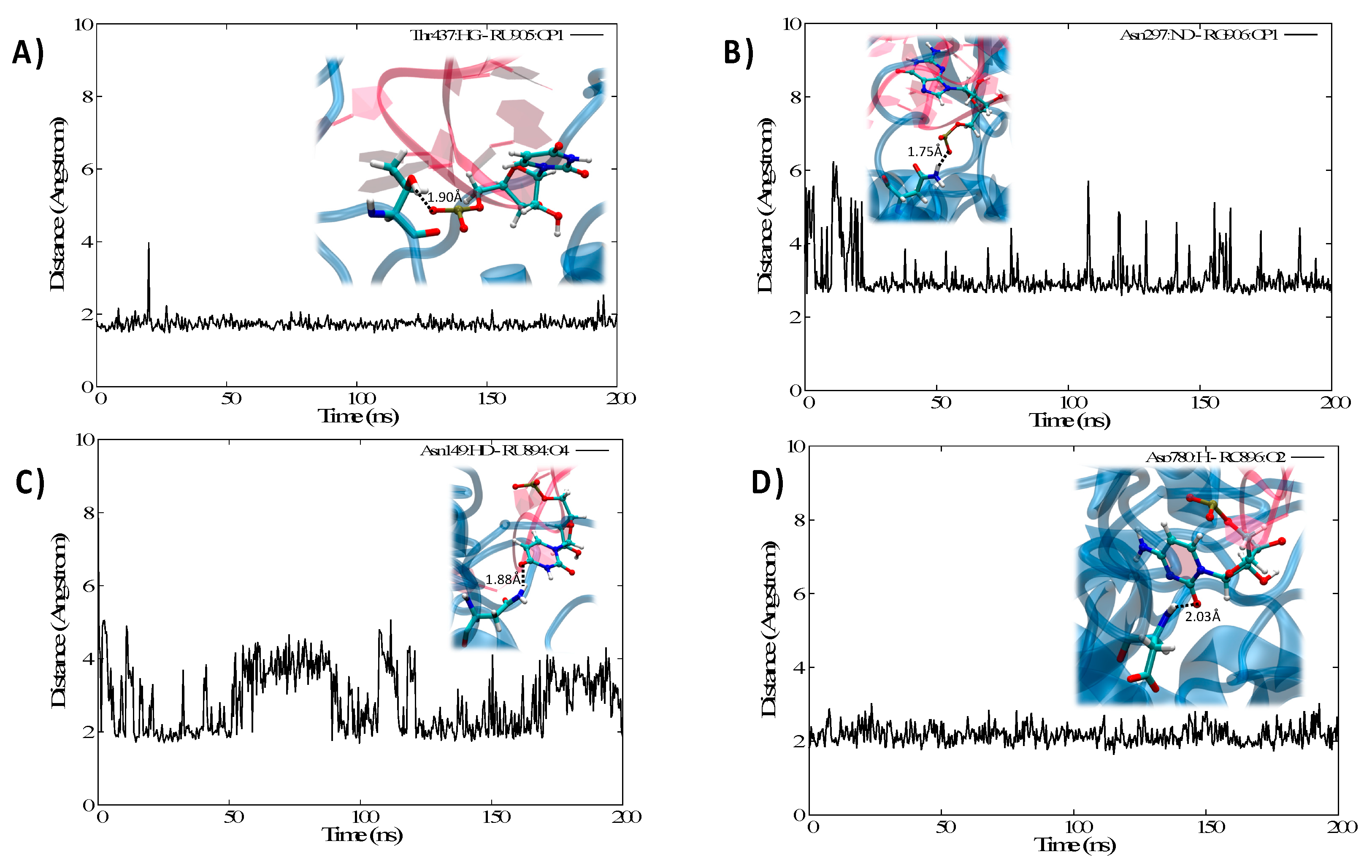
3.2. Structure of the IRP/IRE Complex
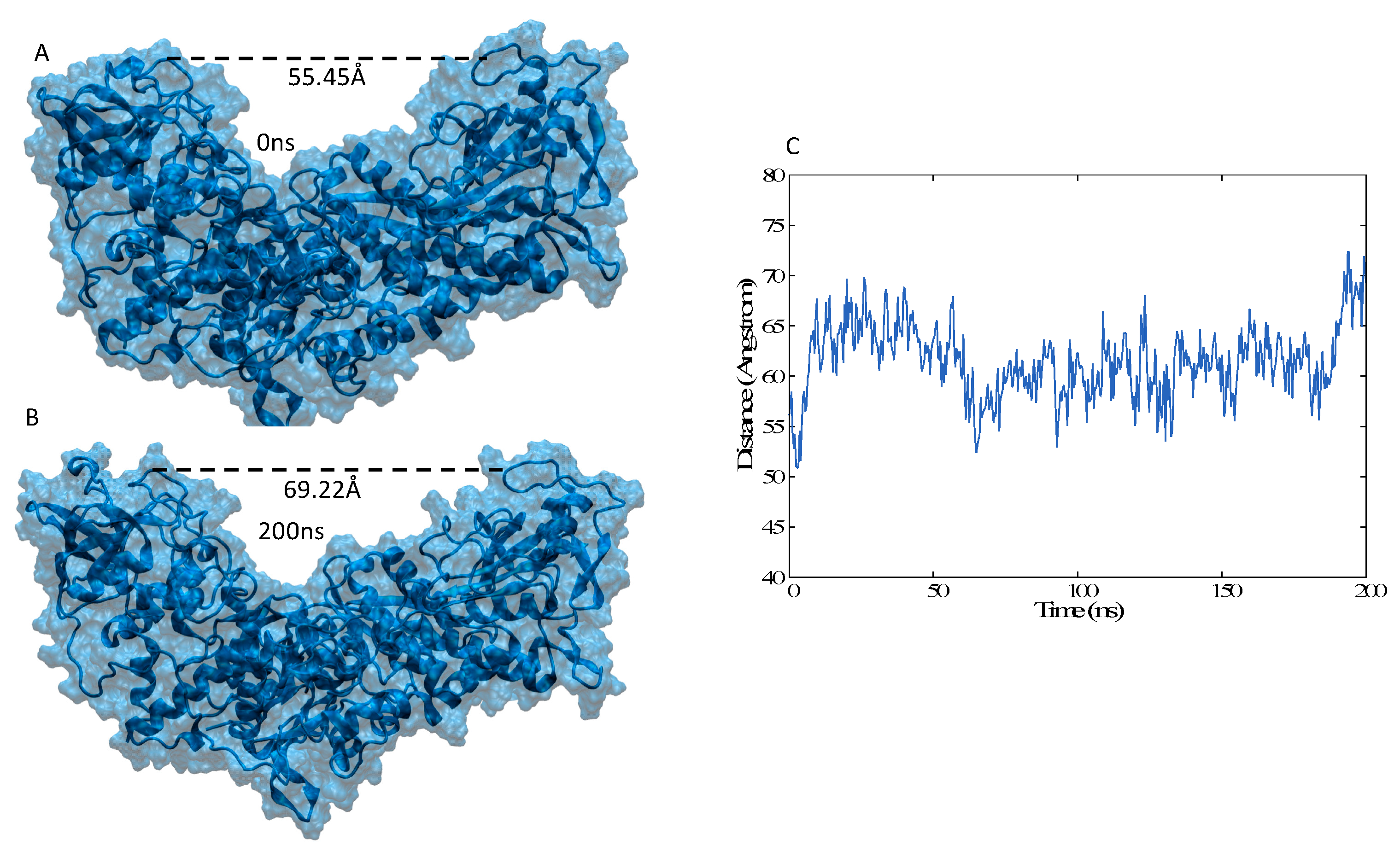
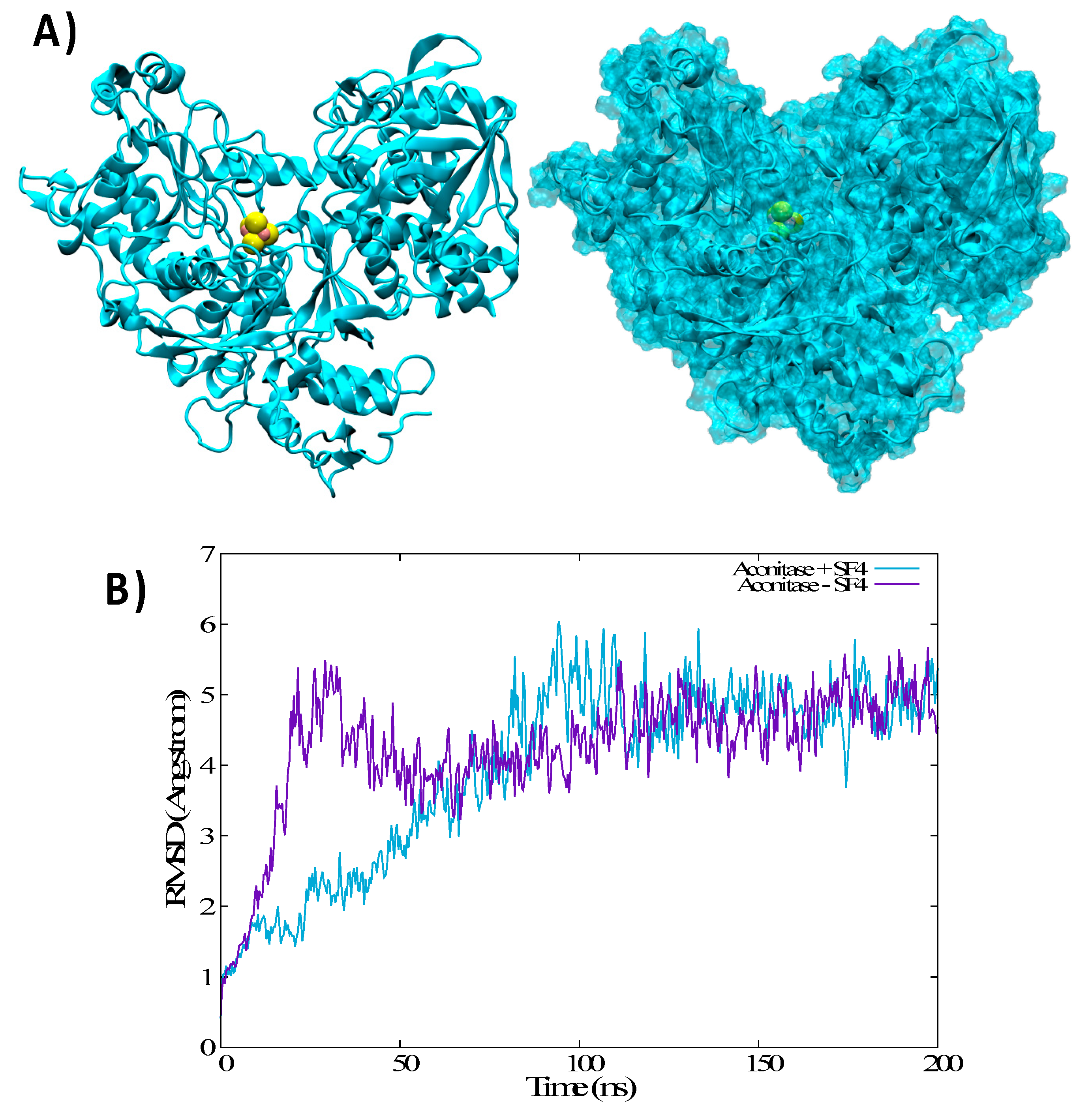
3.3. Structural Features of Unbound IRP1 and cAco
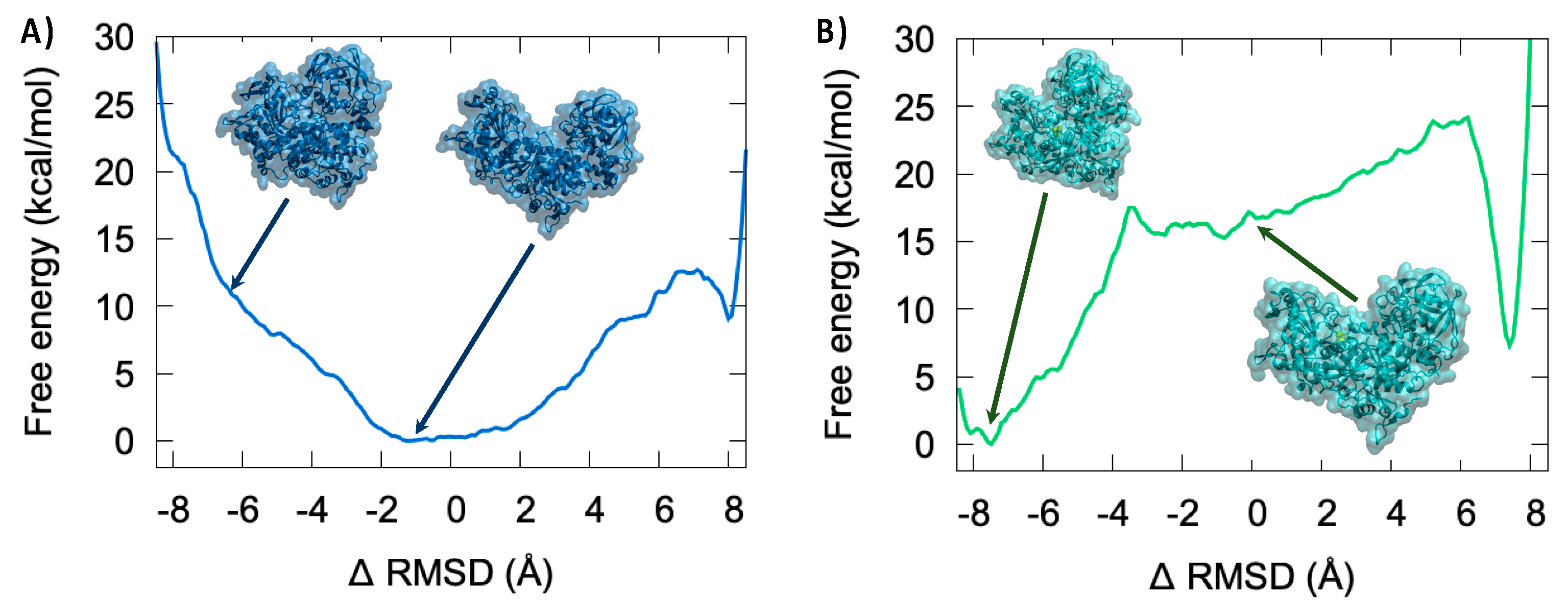
3.4. Thermodynamics of the IRP/Aco Transition
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbate, V.; Hider, R. Iron in biology. Metallomics 2017, 9, 1467–1469. [Google Scholar] [CrossRef]
- Lindley, P.F. Iron in biology: A structural viewpoint. Rep. Prog. Phys. 1996, 59, 867–933. [Google Scholar] [CrossRef]
- Andrews, N.C. Forging a field: The golden age of iron biology. Blood 2008, 112, 219–230. [Google Scholar] [CrossRef]
- Beard, J.L. Iron biology in immune function, muscle metabolism and neuronal functioning. J. Nutr. 2001, 131, 568S–580S. [Google Scholar] [CrossRef]
- Roat-Malone, R.M. Iron-Containing Proteins and Enzymes. In Bioinorganic Chemistry; John Wiley & Sons, Ltd.: New York, NY, USA, 2007; pp. 343–476. [Google Scholar]
- Fuss, J.O.; Tsai, C.L.; Ishida, J.P.; Tainer, J.A. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1253–1271. [Google Scholar] [CrossRef] [PubMed]
- Walden, W.E.; Selezneva, A.; Volz, K. Accommodating variety in iron-responsive elements: Crystal structure of transferrin receptor 1 B IRE bound to iron regulatory protein 1. FEBS Lett. 2012, 586, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Chanet, R.; Baïlle, D.; Golinelli-Cohen, M.-P.; Riquier, S.; Guittet, O.; Lepoivre, M.; Huang, M.-E.; Vernis, L. Fe-S coordination defects in the replicative DNA polymerase delta cause deleterious DNA replication in vivo and subsequent DNA damage in the yeast Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2021, 11, jkab124. [Google Scholar]
- Kruszewski, M. Labile iron pool: The main determinant of cellular response to oxidative stress. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2003, 531, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Chutvanichkul, B.; Vattanaviboon, P.; Mas-oodi, S.; U-pratya, Y.; Wanachiwanawin, W. Labile iron pool as a parameter to monitor iron overload and oxidative stress status in β-thalassemic erythrocytes. Cytom. Part B Clin. Cytom. 2018, 94, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Kakhlon, O.; Cabantchik, Z.I. The labile iron pool: Characterization, measurement, and participation in cellular processes. Free Radic. Biol. Med. 2002, 33, 1037–1046. [Google Scholar] [CrossRef]
- Cabantchik, Z.I. Labile iron in cells and body fluids: Physiology, pathology, and pharmacology. Front. Pharmacol. 2014, 5, 45. [Google Scholar] [CrossRef]
- Bauckman, K.; Haller, E.; Taran, N.; Rockfield, S.; Ruiz-Rivera, A.; Nanjundan, M. Iron alters cell survival in a mitochondria-dependent pathway in ovarian cancer cells. Biochem. J. 2015, 466, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fan, Z.; Yang, Y.; Gu, C. Iron metabolism and its contribution to cancer (Review). Int. J. Oncol. 2019, 54, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.A.M.; Richardson, K.L.; Kabir, T.D.; Trinder, D.; Ganss, R.; Leedman, P.J. Altered Iron Metabolism and Impact in Cancer Biology, Metastasis, and Immunology. Front. Oncol. 2020, 10, 476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, F. Iron homeostasis and tumorigenesis: Molecular mechanisms and therapeutic opportunities. Protein Cell 2015, 6, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.S.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhou, Q.; Wu, D.; Chen, L. Mitochondrial iron metabolism and its role in diseases. Clin. Chim. Acta 2021, 513, 6–12. [Google Scholar] [CrossRef]
- Thomas, C.; Mackey, M.M.; Diaz, A.A.; Cox, D.P. Hydroxyl radical is produced via the Fenton reaction in submitochondrial particles under oxidative stress: Implications for diseases associated with iron accumulation. Redox Rep. 2009, 14, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, R.S.; Blemings, K.P. Iron regulatory proteins, iron responsive elements and iron homeostasis. J. Nutr. 1998, 128, 2295–2298. [Google Scholar] [CrossRef]
- Peyssonnaux, C.; Zinkernagel, A.S.; Schuepbach, R.A.; Rankin, E.; Vaulont, S.; Haase, V.H.; Nizet, V.; Johnson, R.S. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J. Clin. Investig. 2007, 117, 1926–1932. [Google Scholar] [CrossRef]
- Winter, W.E.; Bazydlo, L.A.L.; Harris, N.S. The molecular biology of human iron metabolism. Lab. Med. 2014, 45, 92–102. [Google Scholar] [CrossRef]
- Wallace, D.F. The Regulation of Iron Absorption and Homeostasis. Clin. Biochem. Rev. 2016, 37, 51–62. [Google Scholar]
- Zhou, Z.D.; Tan, E.-K. Iron regulatory protein (IRP)-iron responsive element (IRE) signaling pathway in human neurodegenerative diseases. Mol. Neurodegener. 2017, 12, 1–12. [Google Scholar] [CrossRef]
- Wilkinson, N.; Pantopoulos, K. The IRP/IRE system in vivo: Insights from mouse models. Front. Pharmacol. 2014, 5, 176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Ghosh, M.C.; Rouault, T.A. The physiological functions of iron regulatory proteins in iron homeostasis—An update. Front. Pharmacol. 2014, 5, 124. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, N.; Pantopoulos, K. IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2α mRNA translation. Blood 2013, 122, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Pantopoulos, K. Iron metabolism and the IRE/IRP regulatory system: An update. Ann. N. Y. Acad. Sci. 2004, 1012, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.H.; Rouault, T.A. Metabolic regulation of citrate and iron by aconitases: Role of iron-sulfur cluster biogenesis. BioMetals 2007, 20, 549–564. [Google Scholar] [CrossRef]
- Gourley, B.L.; Parker, S.B.; Jones, B.J.; Zumbrennen, K.B.; Leibold, E.A. Cytosolic aconitase and ferritin are regulated by iron in Caenorhabditis elegans. J. Biol. Chem. 2003, 278, 3227–3234. [Google Scholar] [CrossRef]
- Khan, M.A.; Walden, W.E.; Theil, E.C.; Goss, D.J. Thermodynamic and Kinetic Analyses of Iron Response Element (IRE)-mRNA Binding to Iron Regulatory Protein, IRP1. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Theil, E.C.; McKenzie, R.A.; Sierzputowska-Gracz, H. Structure and function of IREs, the noncoding mRNA sequences regulating synthesis of ferritin, transferrin receptor and (erythroid) 5-aminolevulinate synthase. Adv. Exp. Med. Biol. 1994, 356, 111–118. [Google Scholar] [PubMed]
- Hall, K.B.; Williams, D.J. Dynamics of the IRE RNA hairpin loop probed by 2-aminopurine fluorescence and stochastic dynamics simulations. RNA 2004, 10, 34–47. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cairo, G.; Recalcati, S. Iron-regulatory proteins: Molecular biology and pathophysiological implications. Expert Rev. Mol. Med. 2007, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Olsthoorn, R.C.L. Relevance of the iron-responsive element (IRE) pseudotriloop structure for IRP1/2 binding and validation of IRE-like structures using the yeast three-hybrid system. Gene 2019, 710, 399–405. [Google Scholar] [CrossRef]
- Dupuy, J.; Volbeda, A.; Carpentier, P.; Darnault, C.; Moulis, J.M.; Fontecilla-Camps, J.C. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure 2006, 14, 129–139. [Google Scholar] [CrossRef]
- Autenrieth, F.; Tajkhorshid, E.; Baudry, J.; Luthey-Schulten, Z. Classical force field parameters for the heme prosthetic group of cytochrome c. J. Comput. Chem. 2004, 25, 1613–1622. [Google Scholar] [CrossRef]
- Zgarbová, M.; Šponer, J.; Otyepka, M.; Cheatham, T.E.; Galindo-Murillo, R.; Jurečka, P. Refinement of the Sugar-Phosphate Backbone Torsion Beta for AMBER Force Fields Improves the Description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. [Google Scholar] [CrossRef]
- Dans, P.D.; Ivani, I.; Hospital, A.; Portella, G.; González, C.; Orozco, M. How accurate are accurate force-fields for B-DNA? Nucleic Acids Res. 2017, 45, 4217–4230. [Google Scholar] [CrossRef]
- Carvalho, A.T.P.; Swart, M. Electronic structure investigation and parametrization of biologically relevant iron-sulfur clusters. J. Chem. Inf. Model. 2014, 54, 613–620, Correction in 2015, 55, 1508. [Google Scholar] [CrossRef] [PubMed]
- Sarre, A.; Stelter, M.; Rollo, F.; De Bonis, S.; Seck, A.; Hognon, C.; Ravanat, J.L.J.-L.; Monari, A.; Dehez, F.; Moe, E.; et al. The three Endonuclease III variants of Deinococcus radiodurans possess distinct and complementary DNA repair activities. DNA Repair 2019, 78, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Davidchack, R.L.; Handel, R.; Tretyakov, M.V. Langevin thermostat for rigid body dynamics. J. Chem. Phys. 2009, 130, 234101. [Google Scholar] [CrossRef] [PubMed]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Hopkins, C.W.; Le Grand, S.; Walker, R.C.; Roitberg, A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015, 11, 1864–1874. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N ⋅log( N ) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Zhao, T.; Fu, H.; Lelièvre, T.; Shao, X.; Chipot, C.; Cai, W. The Extended Generalized Adaptive Biasing Force Algorithm for Multidimensional Free-Energy Calculations. J. Chem. Theory Comput. 2017, 13, 1566–1576. [Google Scholar] [CrossRef]
- Barducci, A.; Bonomi, M.; Parrinello, M. Metadynamics. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011, 1, 826–843. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, H.; Chen, H.; Shao, X.; Chipot, C.; Cai, W. Zooming across the Free-Energy Landscape: Shaving Barriers, and Flooding Valleys. J. Phys. Chem. Lett. 2018, 9, 4738–4745. [Google Scholar] [CrossRef]
- Fu, H.; Shao, X.; Cai, W.; Chipot, C. Taming Rugged Free Energy Landscapes Using an Average Force. Acc. Chem. Res. 2019, 52, 3254–3264. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Piccinelli, P.; Samuelsson, T. Evolution of the iron-responsive element. RNA 2007, 13, 952–966. [Google Scholar] [CrossRef]
- Sierzputowska-gracz, H.; Mckenzie, R.A.; Theil, E.C. The importance of a single G in the hairpin loop of the iron responsive element (IRE) in ferritin mRNA for structure: An NMR spectroscopy study. Nucleic Acids Res. 1995, 23, 146–153. [Google Scholar] [CrossRef][Green Version]
- Hognon, C.; Garaude, S.; Timmins, J.; Chipot, C.; Dehez, F.F.; Monari, A. Molecular Bases of DNA Packaging in Bacteria Revealed by All-Atom Molecular Dynamics Simulations: The Case of Histone-Like Proteins in Borrelia burgdorferi. J. Phys. Chem. Lett. 2019, 10, 7200–7207. [Google Scholar] [CrossRef]
- Matoušková, E.; Bignon, E.; Claerbout, V.E.P.; Dršata, T.; Gillet, N.; Monari, A.; Dumont, E.; Lankaš, F. Impact of the Nucleosome Histone Core on the Structure and Dynamics of DNA-Containing Pyrimidine-Pyrimidone (6-4) Photoproduct. J. Chem. Theory Comput. 2020, 16, 5972–5981. [Google Scholar] [CrossRef]
- Hognon, C.; Monari, A. Staring at the Naked Goddess: Unraveling the Structure and Reactivity of Artemis Endonuclease Interacting with a DNA Double Strand. Molecules 2021, 26, 3986. [Google Scholar] [CrossRef]
- Gattuso, H.; Durand, E.; Bignon, E.; Morell, C.; Georgakilas, A.G.; Dumont, E.; Chipot, C.; Dehez, F.; Monari, A. Repair Rate of Clustered Abasic DNA Lesions by Human Endonuclease: Molecular Bases of Sequence Specificity. J. Phys. Chem. Lett. 2016, 7, 3760–3765. [Google Scholar] [CrossRef]
- Bignon, E.; Claerbout, V.E.P.; Jiang, T.; Morell, C.; Gillet, N.; Dumont, E. Nucleosomal embedding reshapes the dynamics of abasic sites. Sci. Rep. 2020, 10, 17314. [Google Scholar] [CrossRef]
- Bignon, E.; Gillet, N.; Jiang, T.; Morell, C.; Dumont, E. A Dynamic View of the Interaction of Histone Tails with Clustered Abasic Sites in a Nucleosome Core Particle. J. Phys. Chem. Lett. 2021, 12, 6014–6019. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hognon, C.; Bignon, E.; Harle, G.; Touche, N.; Grandemange, S.; Monari, A. The Iron Maiden. Cytosolic Aconitase/IRP1 Conformational Transition in the Regulation of Ferritin Translation and Iron Hemostasis. Biomolecules 2021, 11, 1329. https://doi.org/10.3390/biom11091329
Hognon C, Bignon E, Harle G, Touche N, Grandemange S, Monari A. The Iron Maiden. Cytosolic Aconitase/IRP1 Conformational Transition in the Regulation of Ferritin Translation and Iron Hemostasis. Biomolecules. 2021; 11(9):1329. https://doi.org/10.3390/biom11091329
Chicago/Turabian StyleHognon, Cécilia, Emmanuelle Bignon, Guillaume Harle, Nadège Touche, Stéphanie Grandemange, and Antonio Monari. 2021. "The Iron Maiden. Cytosolic Aconitase/IRP1 Conformational Transition in the Regulation of Ferritin Translation and Iron Hemostasis" Biomolecules 11, no. 9: 1329. https://doi.org/10.3390/biom11091329
APA StyleHognon, C., Bignon, E., Harle, G., Touche, N., Grandemange, S., & Monari, A. (2021). The Iron Maiden. Cytosolic Aconitase/IRP1 Conformational Transition in the Regulation of Ferritin Translation and Iron Hemostasis. Biomolecules, 11(9), 1329. https://doi.org/10.3390/biom11091329






