Post-Transcriptional Control of Mating-Type Gene Expression during Gametogenesis in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Sequencing and Data Analysis
2.2. Quantitative Reverse Transcription PCR
2.3. Microscopy
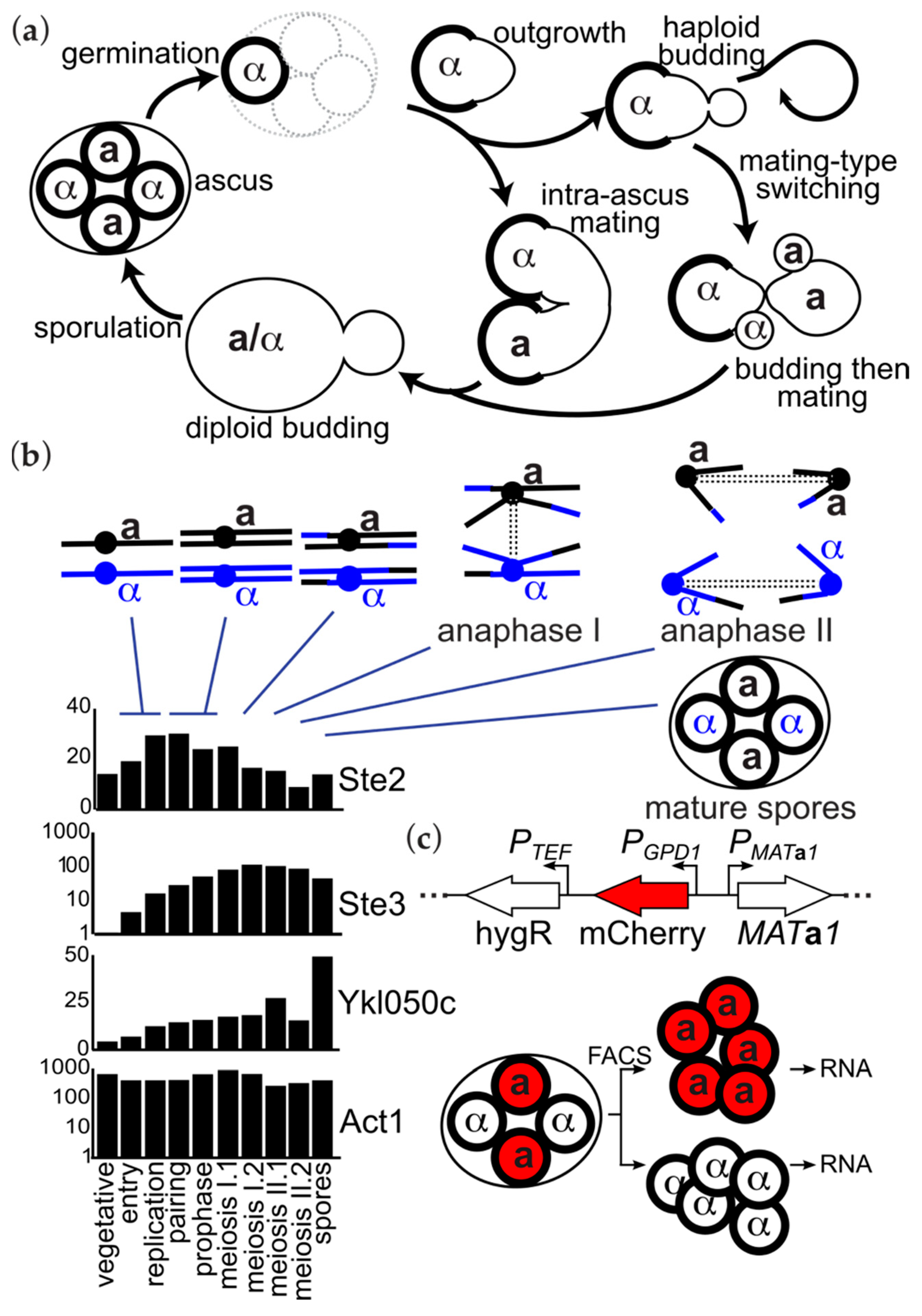
3. Results
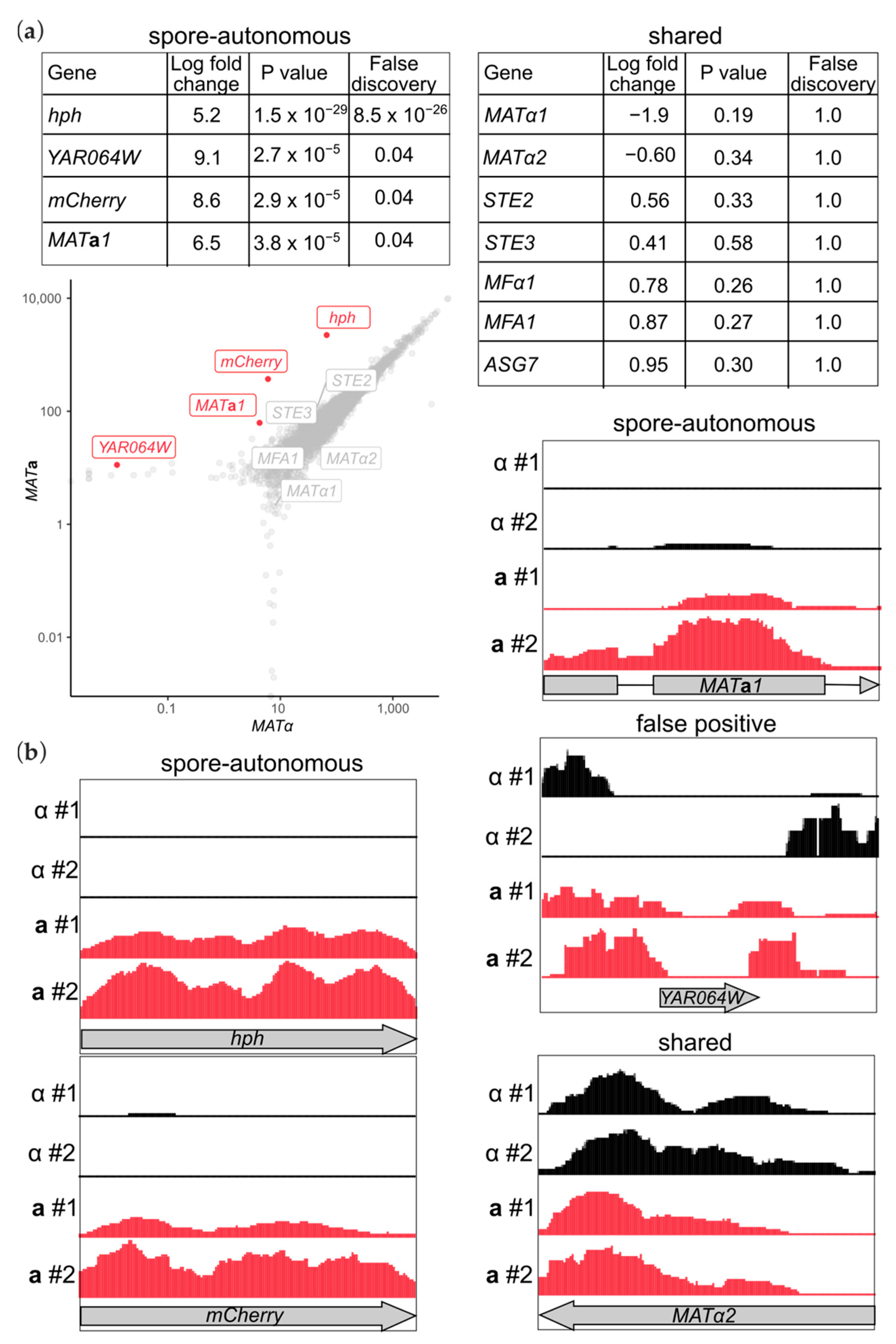
3.1. Only One Transcript Distinguishes the Transcriptomes of Spores of Distinct Mating Types
3.2. Accumulation of Unspliced Mata1 Transcripts in MATa Spores
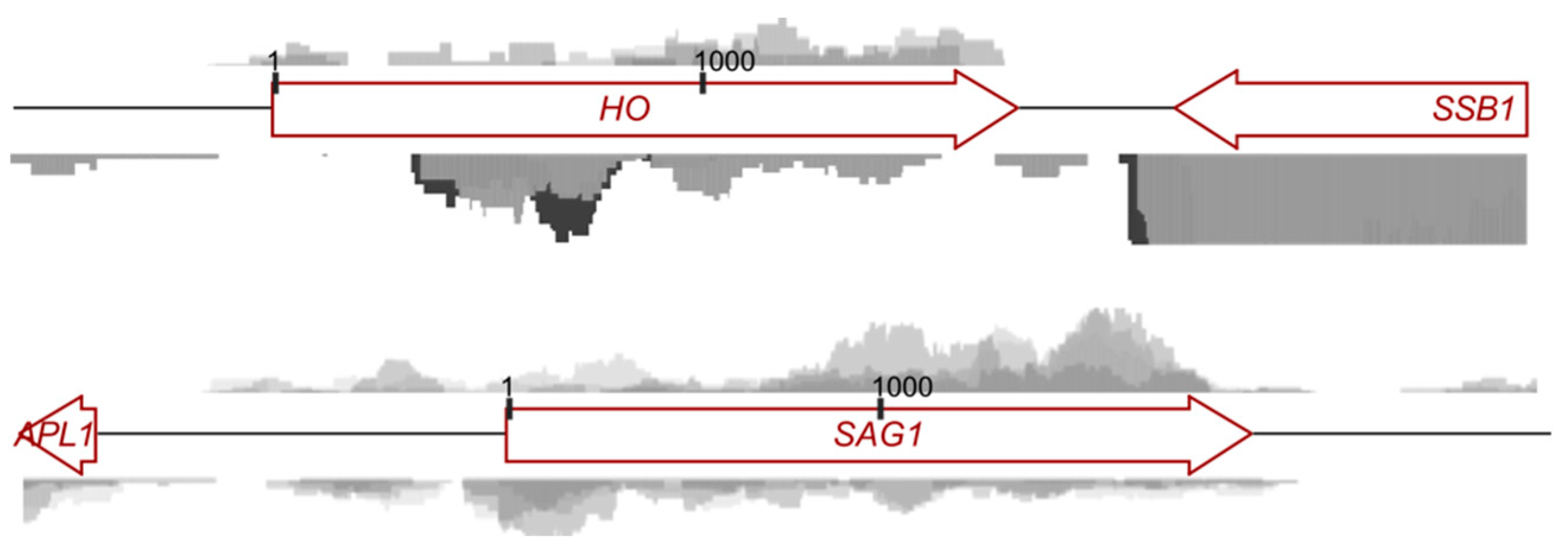
3.3. Antisense Transcripts Corresponding to “Mating-Type-Specific” Genes Are Pervasive in Spores
3.4. Translational Repression in Spores Mediated by the Promoters of Mating-Type-Specific Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sprague, G.F.; Blair, L.C.; Thorner, J. Cell interactions and regulation of cell type in the yeast Saccharomyces cerevisiae. Annu. Rev. Microbiol. 1983, 37, 623–660. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.; Shore, D. Transcriptional regulation in the yeast life cycle. Science 1987, 237, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Herskowitz, I. A regulatory hierarchy for cell specialization in yeast. Nature 1989, 342, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Haber, J.E. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 2012, 191, 33–64. [Google Scholar] [CrossRef]
- Galgoczy, D.J.; Cassidy-Stone, A.; Llinás, M.; O’Rourke, S.M.; Herskowitz, I.; DeRisi, J.L.; Johnson, A.D. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2004, 101, 18069–18074. [Google Scholar] [CrossRef]
- Brar, G.A.; Yassour, M.; Friedman, N.; Regev, A.; Ingolia, N.T.; Weissman, J.S. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 2012, 335, 552–557. [Google Scholar] [CrossRef]
- Chin, B.L.; Frizzell, M.A.; Timberlake, W.E.; Fink, G.R. FASTER MT: Isolation of Pure Populations of a and α Ascospores from Saccharomycescerevisiae. G3 Genes Genomes Genet. 2012, 2, 449–452. [Google Scholar] [CrossRef][Green Version]
- Herskowitz, I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 1988, 52, 536–553. [Google Scholar] [CrossRef]
- Hicks, J.B.; Herskowitz, I. Interconversion of Yeast Mating Types I. Direct Observations of the Action of the Homothallism (HO) Gene. Genetics 1976, 83, 245–258. [Google Scholar] [CrossRef]
- Laney, J.D.; Hochstrasser, M. Ubiquitin-dependent degradation of the yeast Mat(alpha)2 repressor enables a switch in developmental state. Genes Dev. 2003, 17, 2259–2270. [Google Scholar] [CrossRef]
- Hirsch, J.P.; Cross, F.R. The pheromone receptors inhibit the pheromone response pathway in Saccharomyces cerevisiae by a process that is independent of their associated G alpha protein. Genetics 1993, 135, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Chia, M.; van Werven, F.J. Temporal Expression of a Master Regulator Drives Synchronous Sporulation in Budding Yeast. G3 Genes Genomes Genet. 2016, 6, 3553–3560. [Google Scholar] [CrossRef] [PubMed]
- Deutschbauer, A.M.; Davis, R.W. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 2005, 37, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Treusch, S.; Albert, F.W.; Bloom, J.S.; Kotenko, I.E.; Kruglyak, L. Genetic mapping of MAPK-mediated complex traits Across S. cerevisiae. PLoS Genet. 2015, 11, e1004913. [Google Scholar] [CrossRef]
- Bushkin, G.G.; Pincus, D.; Morgan, J.T.; Richardson, K.; Lewis, C.; Chan, S.H.; Bartel, D.P.; Fink, G.R. m6A modification of a 3′ UTR site reduces RME1 mRNA levels to promote meiosis. Nat. Commun. 2019, 10, 3414. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Thacker, D.; Lam, I.; Knop, M.; Keeney, S. Exploiting spore-autonomous fluorescent protein expression to quantify meiotic chromosome behaviors in Saccharomyces cerevisiae. Genetics 2011, 189, 423–439. [Google Scholar] [CrossRef]
- Rogers, D.W.; McConnell, E.; Ono, J.; Greig, D. Spore-autonomous fluorescent protein expression identifies meiotic chromosome mis-segregation as the principal cause of hybrid sterility in yeast. PLoS Biol. 2018, 16, e2005066. [Google Scholar] [CrossRef]
- Brengues, M.; Pintard, L.; Lapeyre, B. mRNA decay is rapidly induced after spore germination of Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 40505–40512. [Google Scholar] [CrossRef]
- Joseph-Strauss, D.; Zenvirth, D.; Simchen, G.; Barkai, N. Spore germination in Saccharomyces cerevisiae: Global gene expression patterns and cell cycle landmarks. Genome Biol. 2007, 8, R241. [Google Scholar] [CrossRef]
- Wach, A.; Brachat, A.; Pöhlmann, R.; Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994, 10, 1793–1808. [Google Scholar] [CrossRef]
- Bitter, G.A.; Egan, K.M. Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene 1984, 32, 263–274. [Google Scholar] [CrossRef]
- Miller, A.M. The yeast MATa1 gene contains two introns. EMBO J. 1984, 3, 1061–1065. [Google Scholar] [CrossRef]
- Munding, E.M.; Shiue, L.; Katzman, S.; Donohue, J.P.; Ares, M. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol. Cell 2013, 51, 338–348. [Google Scholar] [CrossRef]
- Parenteau, J.; Durand, M.; Véronneau, S.; Lacombe, A.-A.; Morin, G.; Guérin, V.; Cecez, B.; Gervais-Bird, J.; Koh, C.-S.; Brunelle, D.; et al. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol. Biol. Cell 2008, 19, 1932–1941. [Google Scholar] [CrossRef]
- Egecioglu, D.E.; Kawashima, T.R.; Chanfreau, G.F. Quality control of MATa1 splicing and exon skipping by nuclear RNA degradation. Nucleic Acids Res. 2012, 40, 1787–1796. [Google Scholar] [CrossRef]
- Hongay, C.F.; Grisafi, P.L.; Galitski, T.; Fink, G.R. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 2006, 127, 735–745. [Google Scholar] [CrossRef]
- Lardenois, A.; Liu, Y.; Walther, T.; Chalmel, F.; Evrard, B.; Granovskaia, M.; Chu, A.; Davis, R.W.; Steinmetz, L.M.; Primig, M. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc. Natl. Acad. Sci. USA 2011, 108, 1058–1063. [Google Scholar] [CrossRef]
- Galitski, T.; Saldanha, A.J.; Styles, C.A.; Lander, E.S.; Fink, G.R. Ploidy regulation of gene expression. Science 1999, 285, 251–254. [Google Scholar] [CrossRef]
- McClure, A.W.; Jacobs, K.C.; Zyla, T.R.; Lew, D.J. Mating in wild yeast: Delayed interest in sex after spore germination. Mol. Biol. Cell 2018, 29, 3119–3127. [Google Scholar] [CrossRef] [PubMed]
- Zid, B.M.; O’Shea, E.K. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 2014, 514, 117–121. [Google Scholar] [CrossRef]
- Sinturel, F.; Navickas, A.; Wery, M.; Descrimes, M.; Morillon, A.; Torchet, C.; Benard, L. Cytoplasmic Control of Sense-Antisense mRNA Pairs. Cell Rep. 2015, 12, 1853–1864. [Google Scholar] [CrossRef] [PubMed]
- Deutschbauer, A.M.; Williams, R.M.; Chu, A.M.; Davis, R.W. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2002, 99, 15530–15535. [Google Scholar] [CrossRef] [PubMed]
- Kloimwieder, A.; Winston, F. A Screen for Germination Mutants in Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2011, 1, 143–149. [Google Scholar] [CrossRef][Green Version]
- Vanácová, S.; Wolf, J.; Martin, G.; Blank, D.; Dettwiler, S.; Friedlein, A.; Langen, H.; Keith, G.; Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005, 3, e189. [Google Scholar] [CrossRef]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Wyers, F.; Rougemaille, M.; Badis, G.; Rousselle, J.-C.; Dufour, M.-E.; Boulay, J.; Régnault, B.; Devaux, F.; Namane, A.; Séraphin, B.; et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 2005, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-M.; Futcher, B.; Leatherwood, J. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS ONE 2011, 6, e26804. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Shichino, Y.; Tanaka, H.; Hiriart, E.; Touat-Todeschini, L.; Vavasseur, A.; Ding, D.-Q.; Hiraoka, Y.; Verdel, A.; Yamamoto, M. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol. 2012, 2, 120014. [Google Scholar] [CrossRef] [PubMed]
- Kilchert, C.; Wittmann, S.; Passoni, M.; Shah, S.; Granneman, S.; Vasiljeva, L. Regulation of mRNA Levels by Decay-Promoting Introns that Recruit the Exosome Specificity Factor Mmi1. Cell Rep. 2015, 13, 2504–2515. [Google Scholar] [CrossRef] [PubMed]
- Clancy, M.J.; Shambaugh, M.E.; Timpte, C.S.; Bokar, J.A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002, 30, 4509–4518. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Berchowitz, L.E.; Gajadhar, A.S.; van Werven, F.J.; De Rosa, A.A.; Samoylova, M.L.; Brar, G.A.; Xu, Y.; Xiao, C.; Futcher, B.; Weissman, J.S.; et al. A developmentally regulated translational control pathway establishes the meiotic chromosome segregation pattern. Genes Dev. 2013, 27, 2147–2163. [Google Scholar] [CrossRef][Green Version]
- Jin, L.; Zhang, K.; Xu, Y.; Sternglanz, R.; Neiman, A.M. Sequestration of mRNAs Modulates the Timing of Translation during Meiosis in Budding Yeast. Mol. Cell. Biol. 2015, 35, 3448–3458. [Google Scholar] [CrossRef][Green Version]
- Jin, L.; Zhang, K.; Sternglanz, R.; Neiman, A.M. Predicted RNA Binding Proteins Pes4 and Mip6 Regulate mRNA Levels, Translation, and Localization during Sporulation in Budding Yeast. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef]
- Nair, R.R.; Zabezhinsky, D.; Gelin-Licht, R.; Haas, B.J.; Dyhr, M.C.; Sperber, H.S.; Nusbaum, C.; Gerst, J.E. Multiplexed mRNA assembly into ribonucleoprotein particles plays an operon-like role in the control of yeast cell physiology. Elife 2021, 10, e66050. [Google Scholar] [CrossRef]
- Roth, A.F.; Nelson, B.; Boone, C.; Davis, N.G. Asg7p-Ste3p inhibition of pheromone signaling: Regulation of the zygotic transition to vegetative growth. Mol. Cell. Biol. 2000, 20, 8815–8825. [Google Scholar] [CrossRef][Green Version]
- Robertson, C.G.; Clark-Cotton, M.R.; Lew, D.J. Mechanisms that ensure monogamous mating in Saccharomyces cerevisiae. Mol. Biol. Cell 2021, 32, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bortz, E.; Zhong, H.; Leeuw, T.; Leberer, E.; Vershon, A.K.; Hirsch, J.P. Localization and signaling of G(beta) subunit Ste4p are controlled by a-factor receptor and the a-specific protein Asg7p. Mol. Cell. Biol. 2000, 20, 8826–8835. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heasley, L.R.; Singer, E.; Cooperman, B.J.; McMurray, M.A. Saccharomyces spores are born prepolarized to outgrow away from spore-spore connections and penetrate the ascus wall. Yeast Chichester Engl. 2021, 38, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.G.; Bardwell, L.; Kron, S.J.; Thorner, J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996, 10, 2831–2848. [Google Scholar] [CrossRef] [PubMed]
- Tedford, K.; Kim, S.; Sa, D.; Stevens, K.; Tyers, M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 1997, 7, 228–238. [Google Scholar] [CrossRef]
- Pi, H.; Chien, C.T.; Fields, S. Transcriptional activation upon pheromone stimulation mediated by a small domain of Saccharomyces cerevisiae Ste12p. Mol. Cell. Biol. 1997, 17, 6410–6418. [Google Scholar] [CrossRef] [PubMed]
- Bardwell, L.; Cook, J.G.; Zhu-Shimoni, J.X.; Voora, D.; Thorner, J. Differential regulation of transcription: Repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 15400–15405. [Google Scholar] [CrossRef]
- Olson, K.A.; Nelson, C.; Tai, G.; Hung, W.; Yong, C.; Astell, C.; Sadowski, I. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell. Biol. 2000, 20, 4199–4209. [Google Scholar] [CrossRef] [PubMed]
- Houser, J.R.; Ford, E.; Nagiec, M.J.; Errede, B.; Elston, T.C. Positive roles for negative regulators in the mating response of yeast. Mol. Syst. Biol. 2012, 8, 586. [Google Scholar] [CrossRef]
- Johnson, P.R.; Swanson, R.; Rakhilina, L.; Hochstrasser, M. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 1998, 94, 217–227. [Google Scholar] [CrossRef]
- Paul, B.; Montpetit, B. Altered RNA processing and export lead to retention of mRNAs near transcription sites and nuclear pore complexes or within the nucleolus. Mol. Biol. Cell 2016, 27, 2742–2756. [Google Scholar] [CrossRef] [PubMed]
- Naro, C.; Jolly, A.; Di Persio, S.; Bielli, P.; Setterblad, N.; Alberdi, A.J.; Vicini, E.; Geremia, R.; De la Grange, P.; Sette, C. An Orchestrated Intron Retention Program in Meiosis Controls Timely Usage of Transcripts during Germ Cell Differentiation. Dev. Cell 2017, 41, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Yeom, K.-H.; Pan, Z.; Lin, C.-H.; Lim, H.Y.; Xiao, W.; Xing, Y.; Black, D.L. Tracking pre-mRNA maturation across subcellular compartments identifies developmental gene regulation through intron retention and nuclear anchoring. Genome Res. 2021, 31, 1106–1119. [Google Scholar] [CrossRef] [PubMed]
- Hibbs, M.A.; Hess, D.C.; Myers, C.L.; Huttenhower, C.; Li, K.; Troyanskaya, O.G. Exploring the functional landscape of gene expression: Directed search of large microarray compendia. Bioinformatics 2007, 23, 2692–2699. [Google Scholar] [CrossRef] [PubMed]


| Genes with Retained Introns (49) | Genes with No Obvious Intron Retention 1 (23) | Ambiguous Genes (18) |
|---|---|---|
| AMA1, APE2, ARP2, COF1, COX5B, DBP2, DCN1, DYN2, ECM33, ERV1, GCR1, GPI15, HNT1, HNT2, HOP2, LSM2, MCM21, MND1, MOB1, MRK1, MRPL44, MUD1, NSP1, PCH2, PFY1, PMI40, POP8, QCR10, QCR9, REC107, RPL17A, RPL22B, RPS17A, RPS18B, RUB1, SCS22, SMD2, STO1, TAD3, UBC13, UBC5, UBC9, VMA10, VMA9, YML6, YOP1, YRA1, YSC84, YSF3 | ACT1, BIG1, BUD25, COX42DMC1, GIM5, GLC7, LSB3, LSM7, MMS2, MOB2, MPT5, PHO85, PRE3, RAD14, RFA2, RIM1, SAE3, SRC1, TAN1, TUB1, UBC4, UBC8 1 | ARP9, CIN2, CPT1, EPT1, HFM1, IMD4, MEI4, MTR22, NMD2, REC102, REC114, SPO1, SPO22, SPT14, TUB3, UBC12, VPS75, YBR220C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeager, R.; Bushkin, G.G.; Singer, E.; Fu, R.; Cooperman, B.; McMurray, M. Post-Transcriptional Control of Mating-Type Gene Expression during Gametogenesis in Saccharomyces cerevisiae. Biomolecules 2021, 11, 1223. https://doi.org/10.3390/biom11081223
Yeager R, Bushkin GG, Singer E, Fu R, Cooperman B, McMurray M. Post-Transcriptional Control of Mating-Type Gene Expression during Gametogenesis in Saccharomyces cerevisiae. Biomolecules. 2021; 11(8):1223. https://doi.org/10.3390/biom11081223
Chicago/Turabian StyleYeager, Randi, G. Guy Bushkin, Emily Singer, Rui Fu, Benjamin Cooperman, and Michael McMurray. 2021. "Post-Transcriptional Control of Mating-Type Gene Expression during Gametogenesis in Saccharomyces cerevisiae" Biomolecules 11, no. 8: 1223. https://doi.org/10.3390/biom11081223
APA StyleYeager, R., Bushkin, G. G., Singer, E., Fu, R., Cooperman, B., & McMurray, M. (2021). Post-Transcriptional Control of Mating-Type Gene Expression during Gametogenesis in Saccharomyces cerevisiae. Biomolecules, 11(8), 1223. https://doi.org/10.3390/biom11081223






