The Identification of Zinc-Finger Protein 433 as a Possible Prognostic Biomarker for Clear-Cell Renal Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene and Protein Expression Analysis
2.1.1. UALCAN Analysis
2.1.2. TNM Plotter
2.1.3. TIMER2.0
2.1.4. Human Protein Atlas
2.2. Overall Survival Analysis
2.3. Methylation Analysis
2.4. Gene Ontology and Pathway Analysis
2.5. Statistical Analysis
3. Results
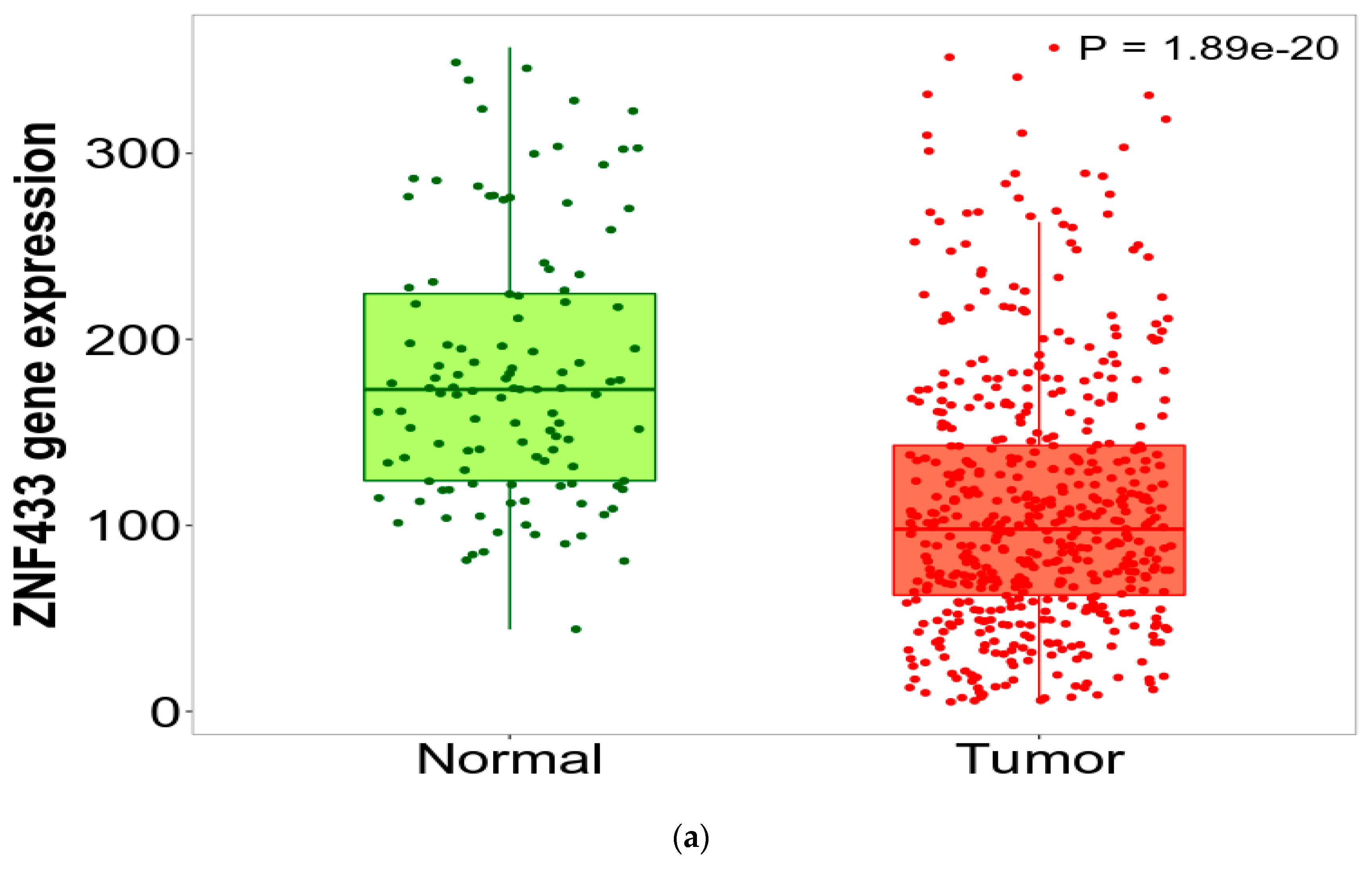
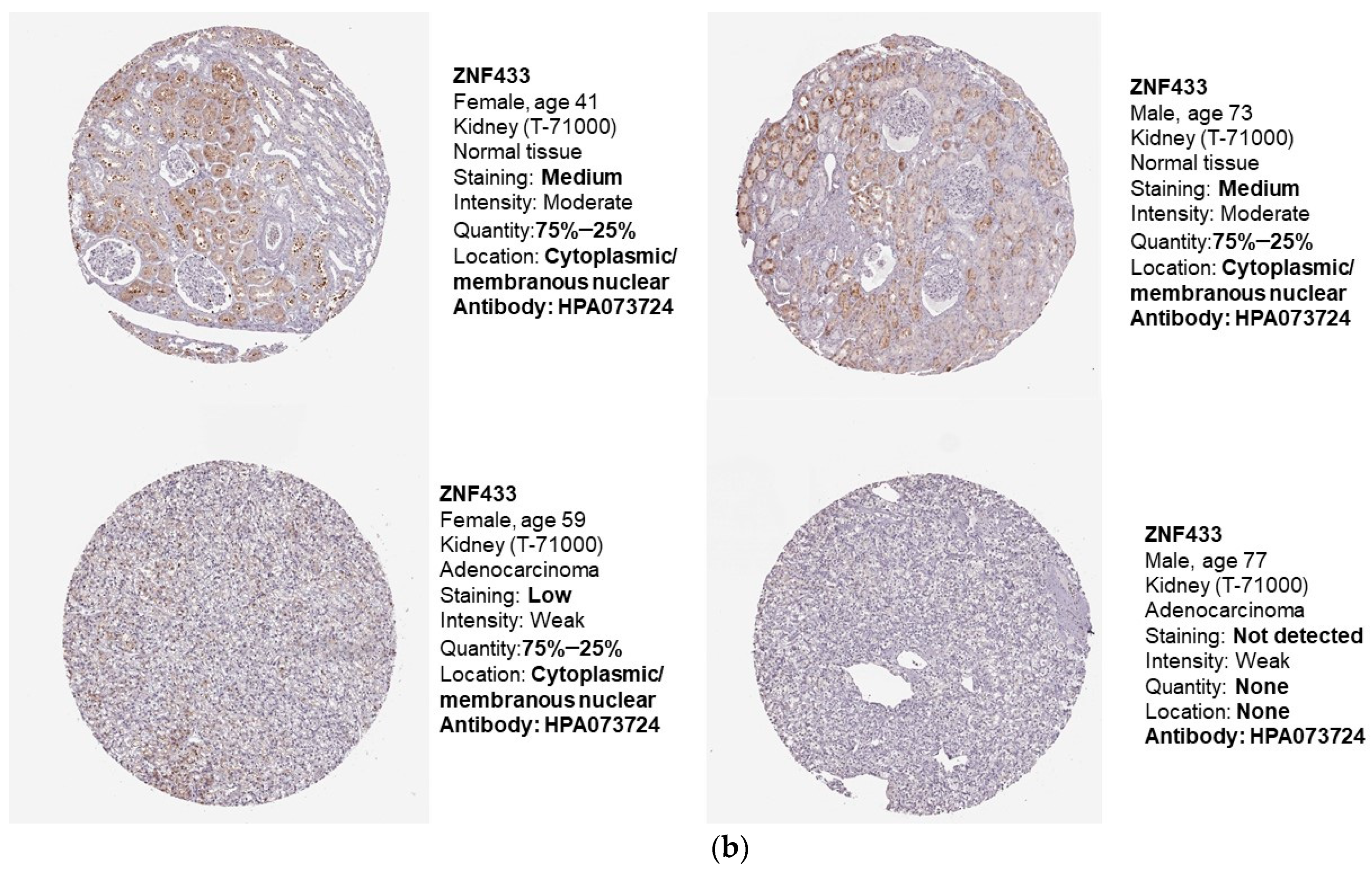
3.1. ZNF433 mRNA and Protein Expression in Normal and Clear-Cell Renal Cell Carcinoma
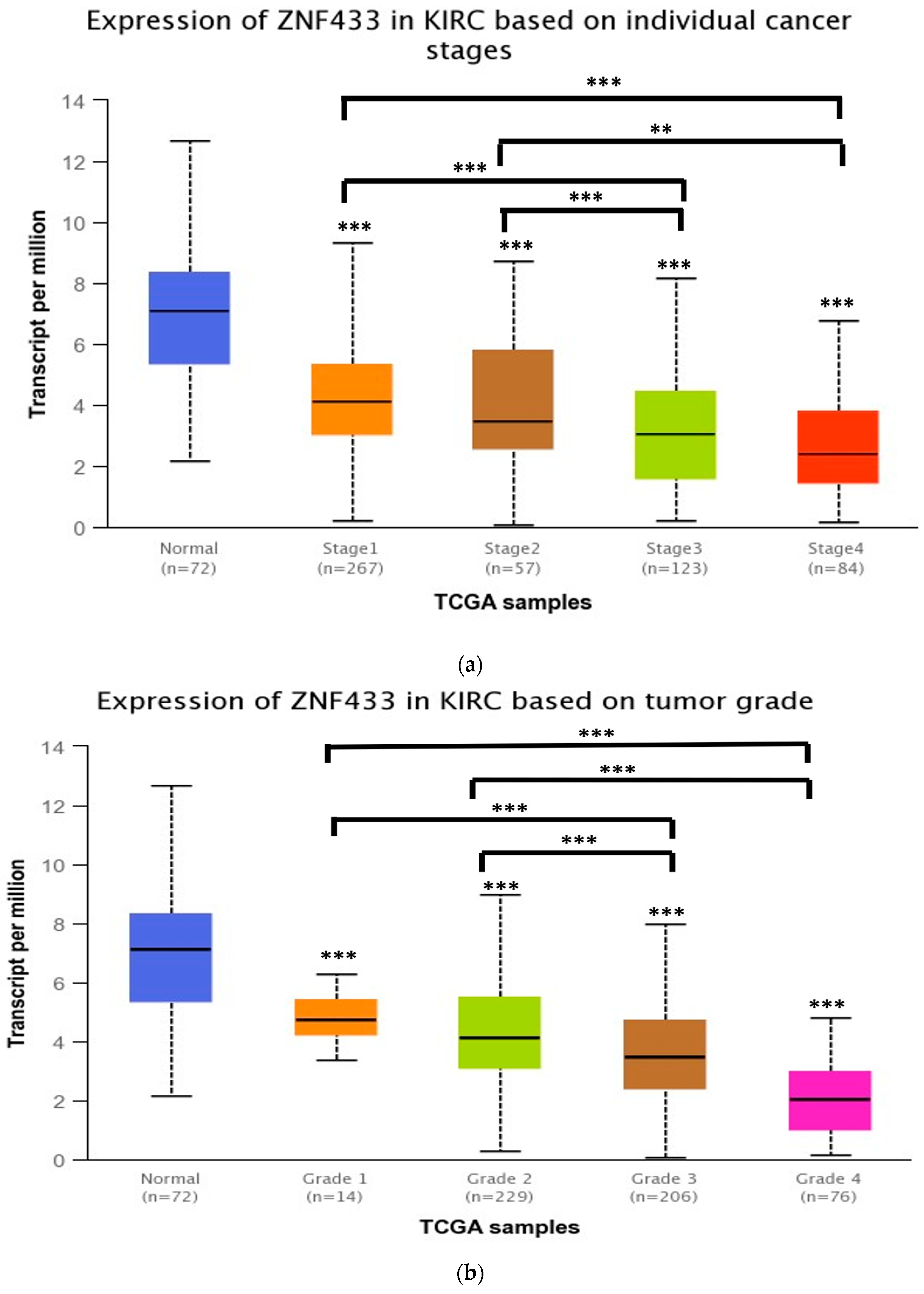
3.2. ZNF433 mRNA Expression in Association with the Clinicopathological Features of Clear-Cell Renal Cell Carcinoma
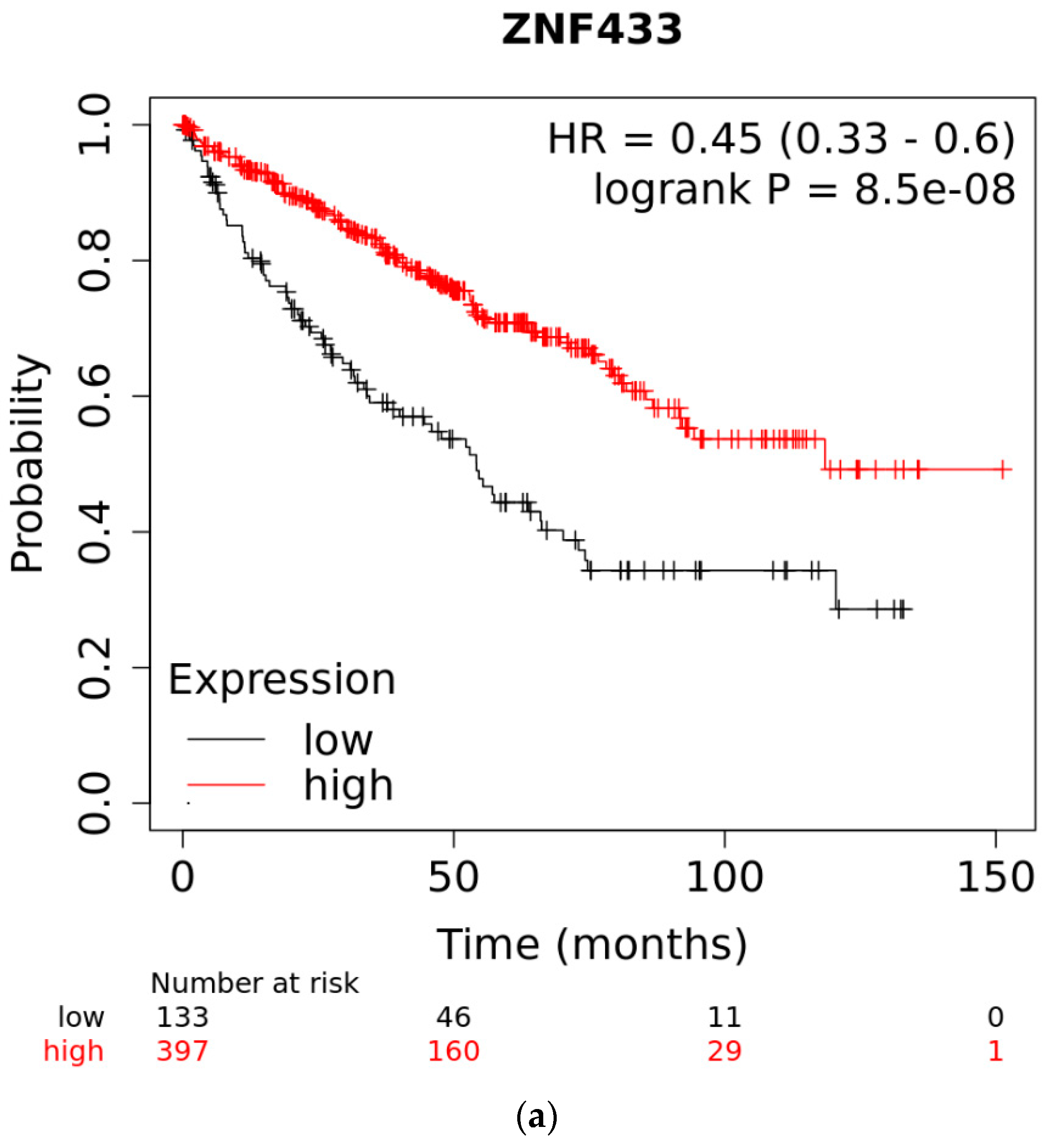
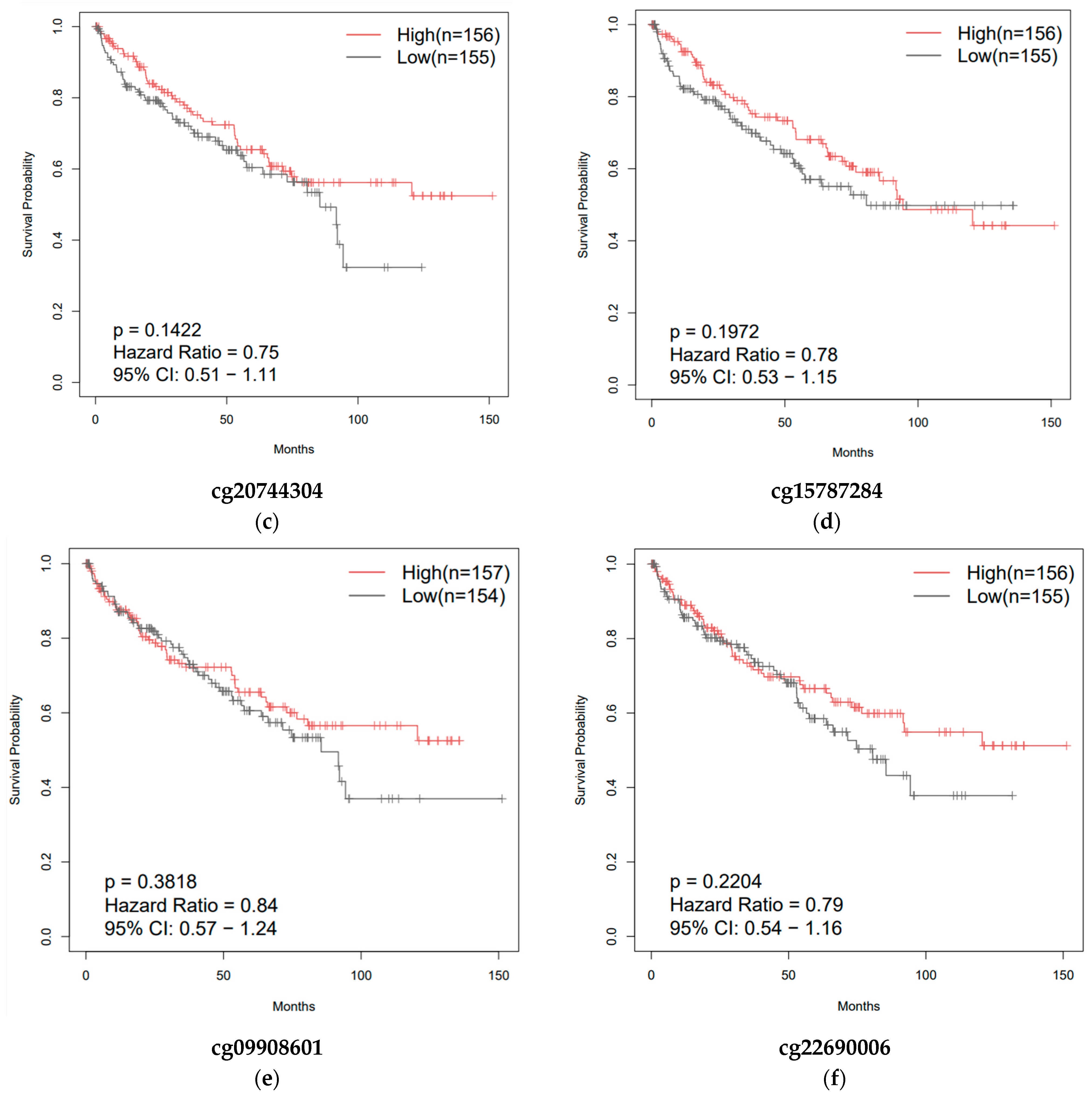
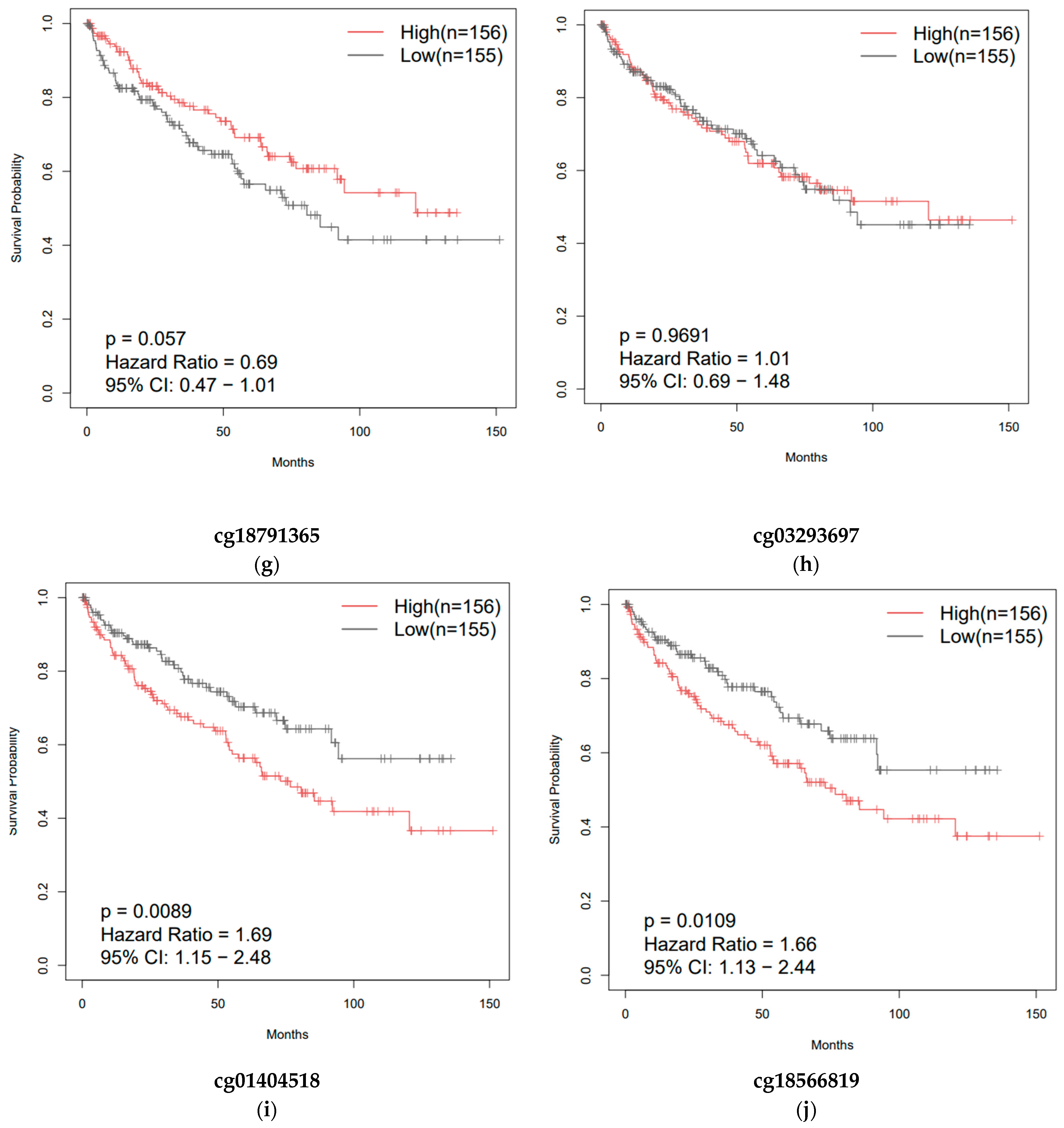
3.3. Overall Survival of Patients with Clear-Cell and Papillary Renal Cell Carcinomas as a Function of ZNF433 Expression
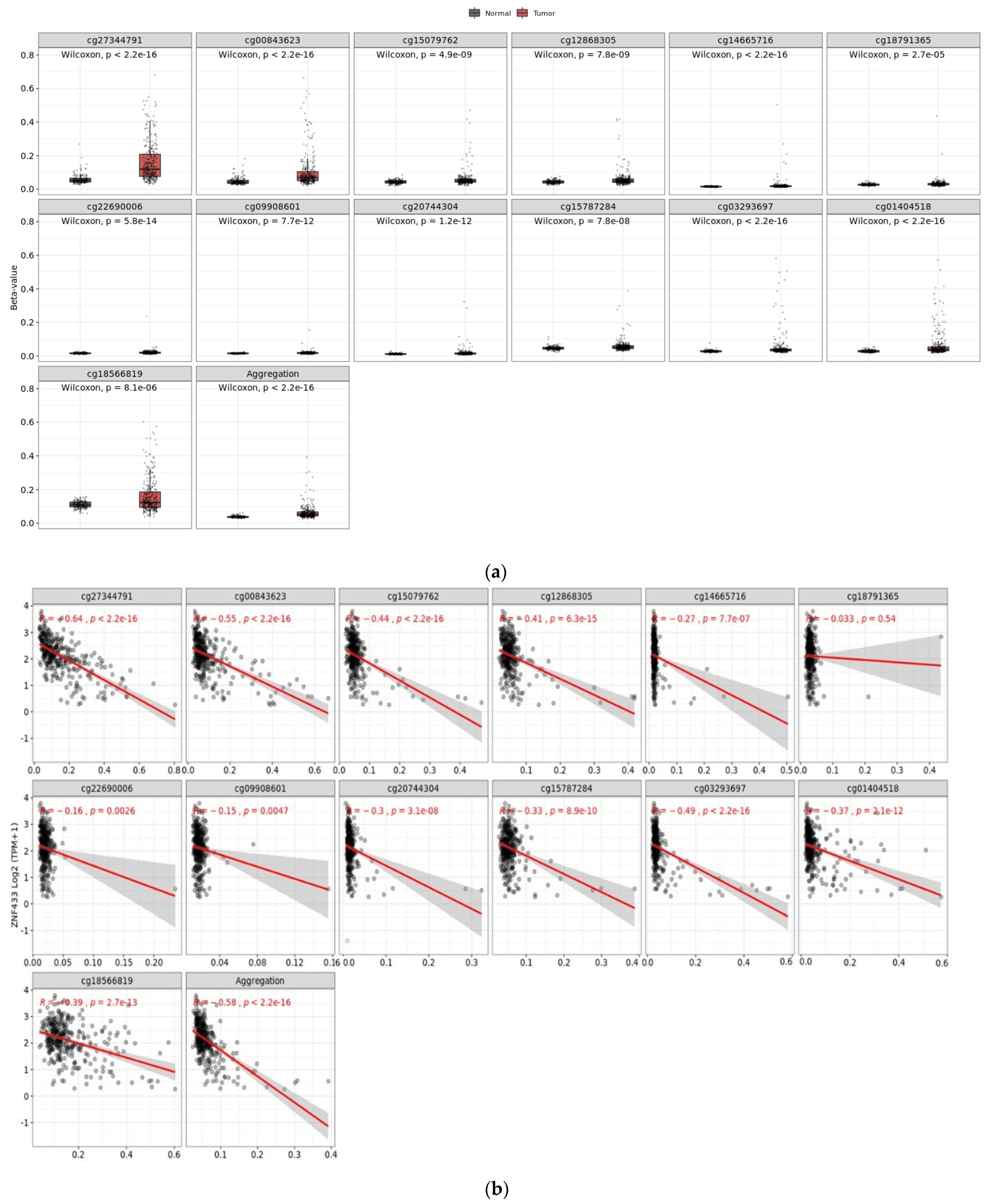
3.4. Effect of CpG Methylation on ZNF433 Gene Expression
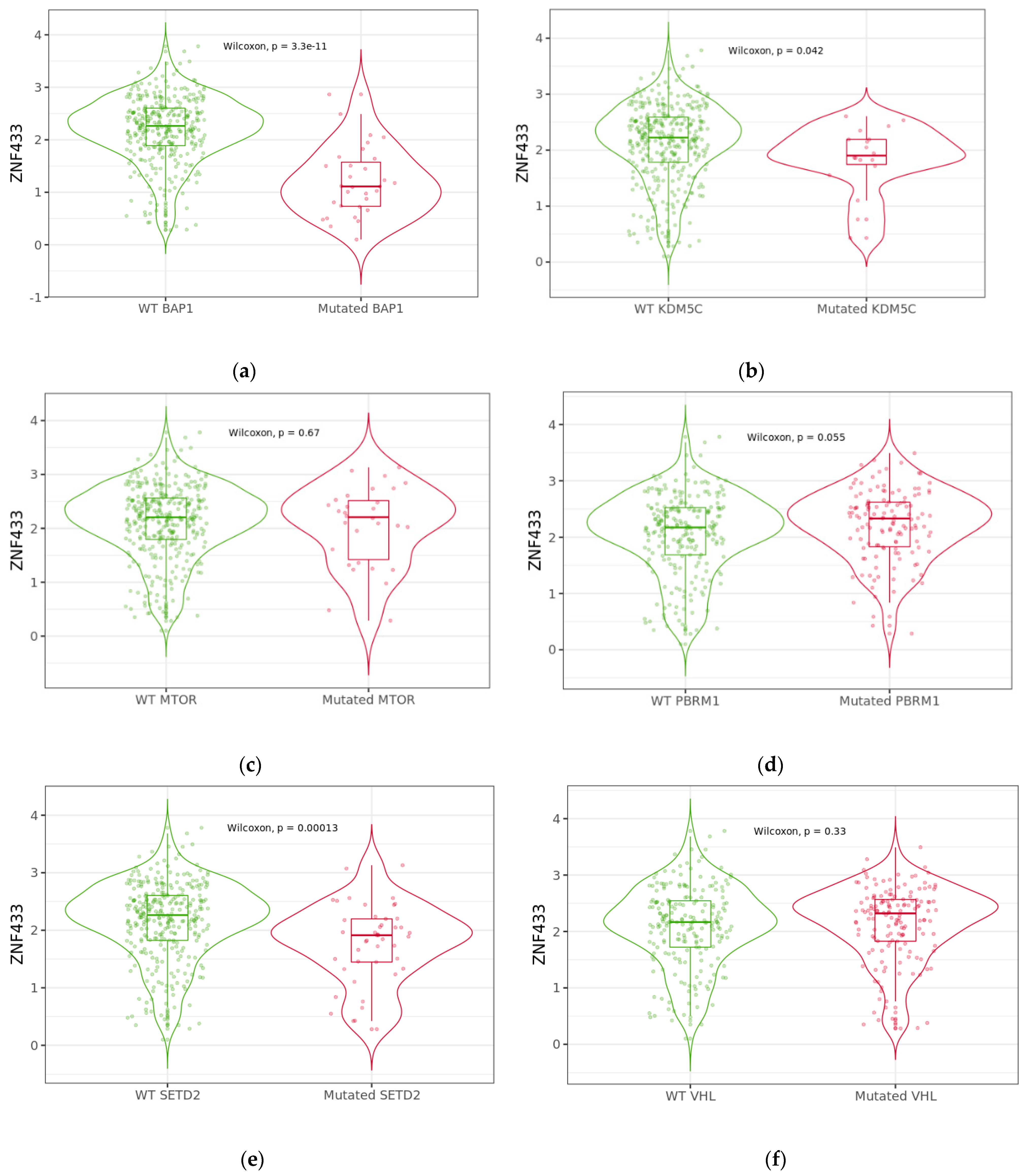
3.5. Effect of BAP1, KDM5C, MTOR, PBRM1, SETD2, and VHL Mutations on ZNF433 mRNA Expression
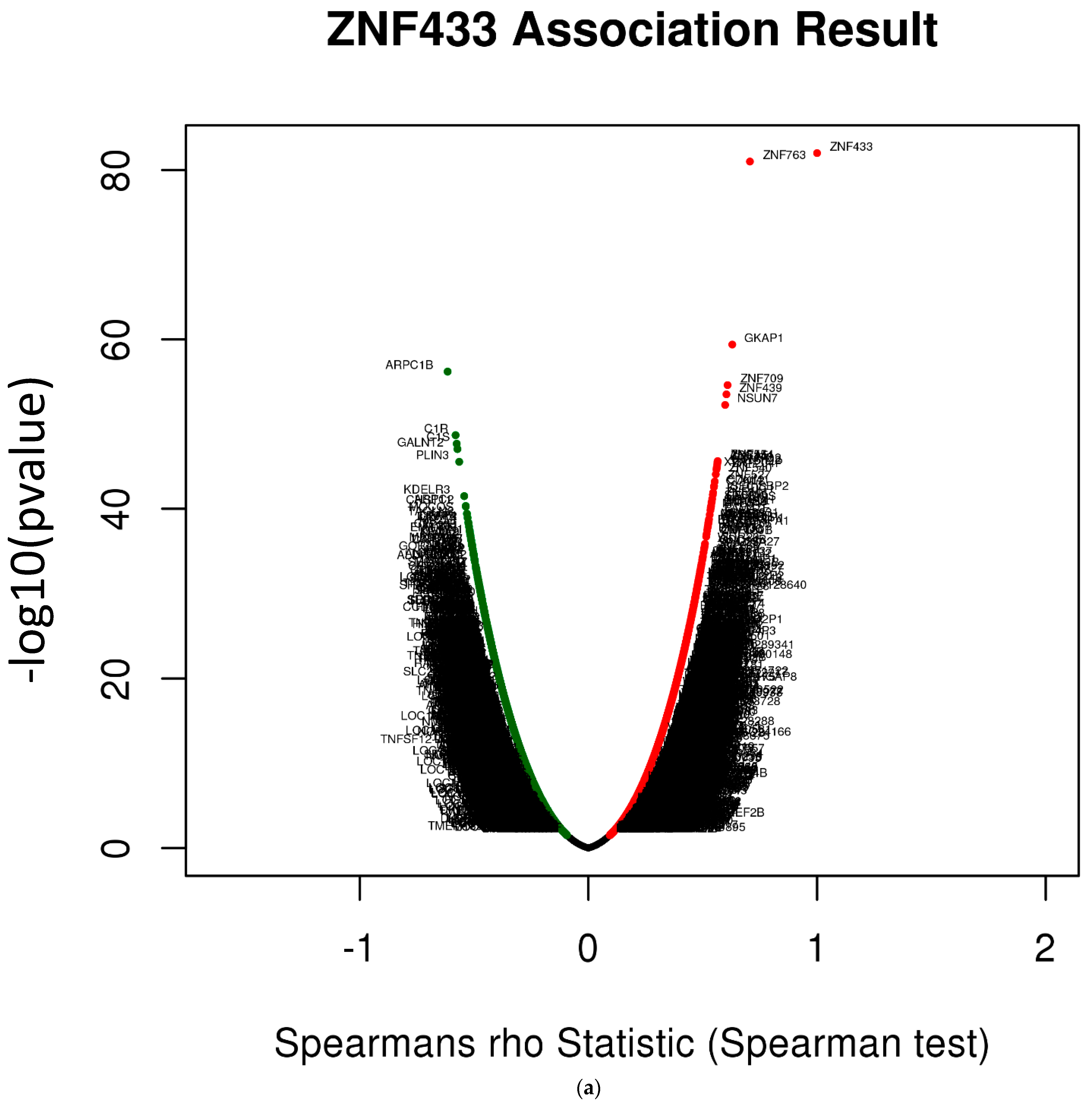
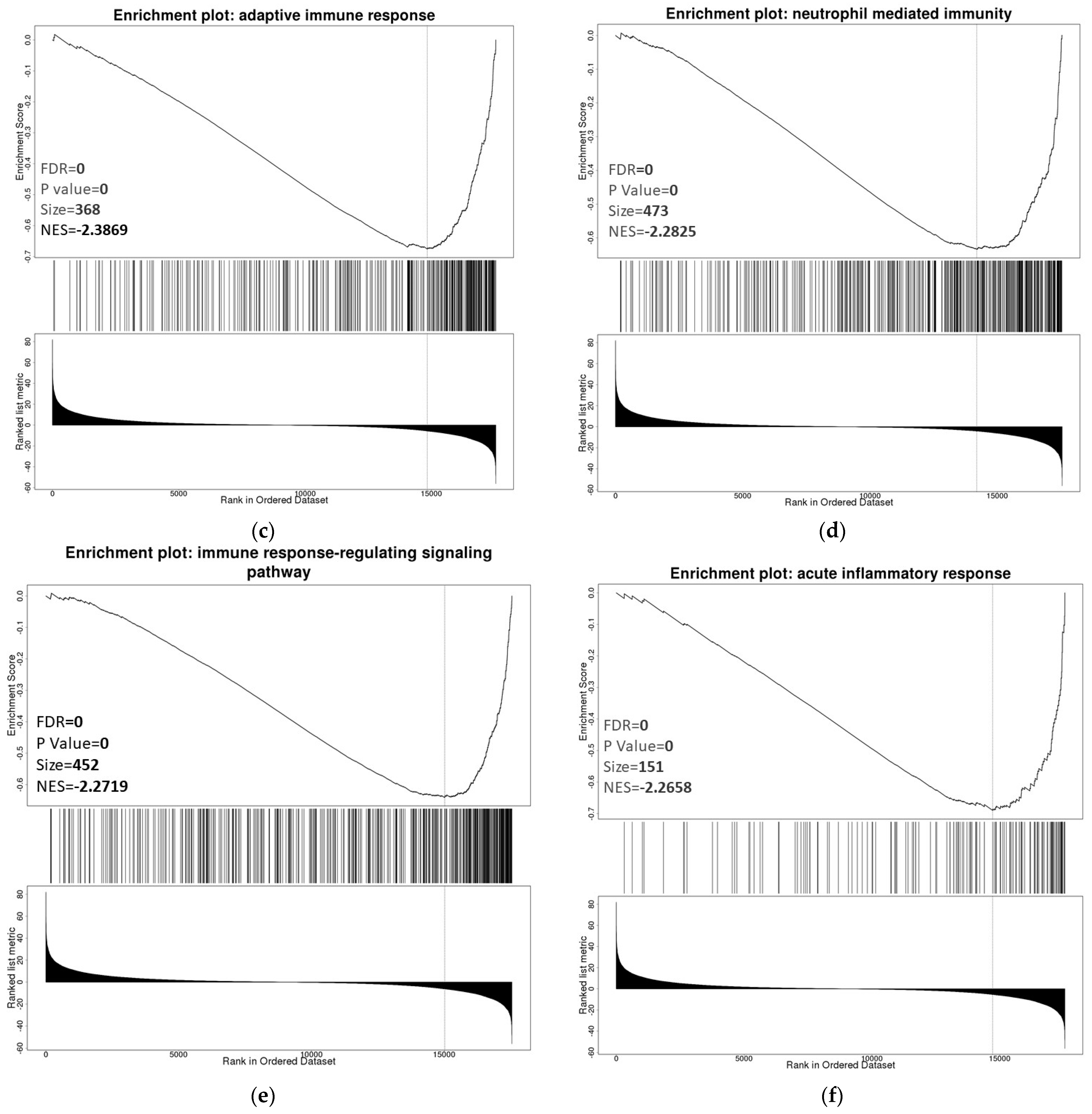
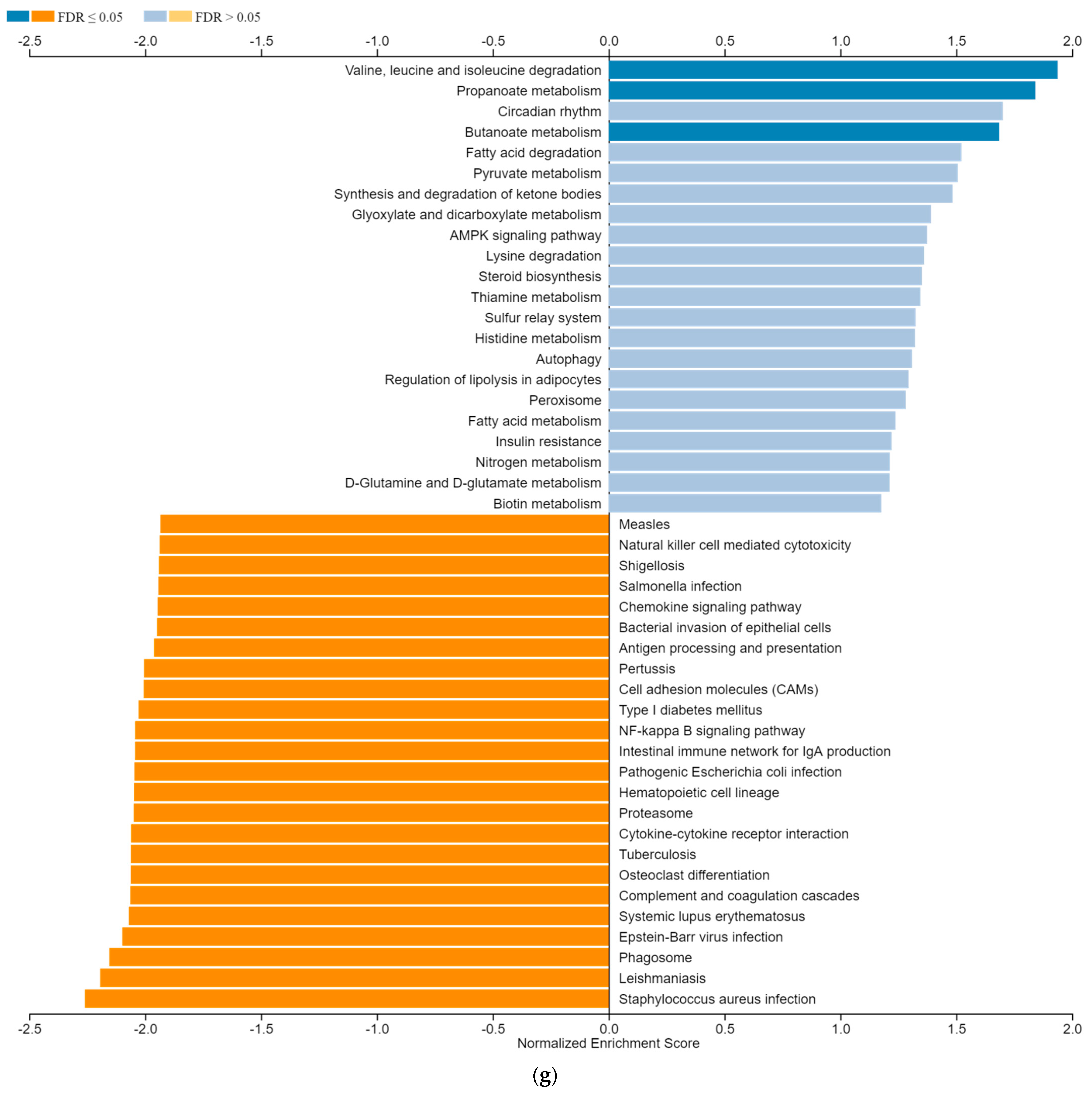
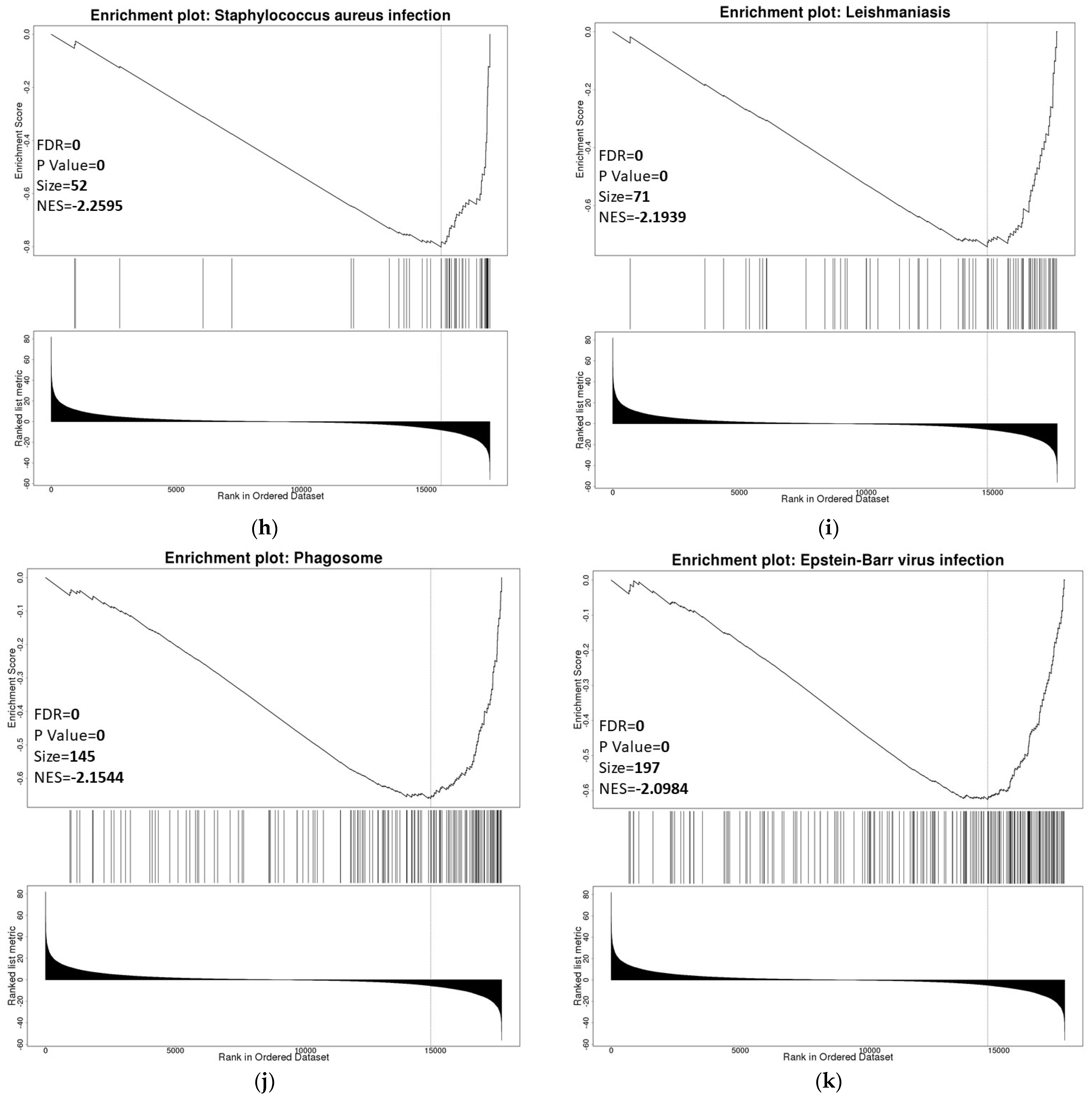
3.6. ZNF433 Pathway Enrichment, Target Gene Expression, and Target Gene Ontology
3.7. Pan-Cancer Analysis of ZNF433 Target Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eble, J.N.; Sauter, G.; Epstein, J.I.; Sesterhenn, I.A. Renal Cell Carcinoma. In World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs; IARC Press: Lyon, France, 2004; pp. 8–14. [Google Scholar]
- Dimitrieva, S.; Schlapbach, R.; Rehrauer, H. Prognostic value of cross-omics screening for kidney clear cell renal cancer survival. Biol. Direct. 2016, 11, 68. [Google Scholar] [CrossRef]
- Low, G.; Huang, G.; Fu, W.; Moloo, Z.; Girgis, S. Review of renal cell carcinoma and its common subtypes in radiology. World J. Radiol. 2016, 8, 484–500. [Google Scholar] [CrossRef]
- Powles, T.; Albiges, L.; Staehler, M.; Bensalah, K.; Dabestani, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; Lam, T.B.; et al. Updated European association of urology guidelines recommendations for the treatment of first-line metastatic clear cell renal cancer. Eur. Urol. 2017, 73, 311–315. [Google Scholar] [CrossRef]
- Huang, M.; Chen, Y.; Han, D.; Lei, Z.; Chu, X. Role of the zinc finger and SCAN domain-containing transcription factors in cancer. Am. J. Cancer Res. 2019, 9, 816–836. [Google Scholar]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef]
- Urrutia, R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003, 4, 231. [Google Scholar] [CrossRef]
- Sobocińska, J.; Molenda, S.; Machnik, M.; Oleksiewicz, U. KRAB-ZFP Transcriptional Regulators Acting as Oncogenes and Tumor Suppressors: An Overview. Int. J. Mol. Sci. 2021, 22, 2212. [Google Scholar] [CrossRef]
- Wuttke, D.S.; Foster, M.P.; Case, D.A.; Gottesfeld, J.M.; Wright, P.E. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: Determinants of affinity and sequence specificity. J. Mol. Biol. 1997, 273, 183–206. [Google Scholar] [CrossRef]
- Lupo, A.; Cesaro, E.; Montano, G.; Zurlo, D.; Izzo, P.; Costanzo, P. KRAB-zinc finger proteins: A repressor family displaying multiple biological functions. Curr. Genom. 2013, 14, 268–278. [Google Scholar] [CrossRef]
- Machnik, M.; Cylwa, R.; Kiełczewski, K.; Biecek, P.; Liloglou, T.; Mackiewicz, A.; Oleksiewicz, U. The expression signature of cancer-associated KRAB-ZNF factors identified in TCGA pan-cancer transcriptomic data. Mol. Oncol. 2019, 13, 701–724. [Google Scholar] [CrossRef]
- Sun, R.; Xiang, T.; Tang, J.; Peng, W.; Luo, J.; Li, L.; Qiu, Z.; Tan, Y.; Ye, L.; Zhang, M.; et al. 19q13 KRAB zinc-finger protein ZNF471 activates MAPK10/JNK3 signaling but is frequently silenced by promoter CpG methylation in esophageal cancer. Theranostics 2020, 10, 2243–2259. [Google Scholar] [CrossRef]
- Tao, Y.F.; Hu, S.Y.; Lu, J.; Cao, L.; Zhao, W.L.; Xiao, P.F.; Xu, L.X.; Li, Z.H.; Wang, N.N.; Du, X.J.; et al. Zinc finger protein 382 is downregulated by promoter hypermethylation in pediatric acute myeloid leukemia patients. Int. J. Mol. Med. 2014, 34, 1505–1515. [Google Scholar] [CrossRef][Green Version]
- Pei, L.; He, X.; Li, S.; Sun, R.; Xiang, Q.; Ren, G.; Xiang, T. KRAB zinc-finger protein 382 regulates epithelial-mesenchymal transition and functions as a tumor suppressor but is silenced by CpG methylation in gastric cancer. Int. J. Oncol. 2018, 53, 961–972. [Google Scholar] [CrossRef]
- Tao, C.; Luo, J.; Tang, J.; Zhou, D.; Feng, S.; Qiu, Z.; Putti, T.C.; Xiang, T.; Tao, Q.; Li, L.; et al. The tumor suppressor Zinc finger protein 471 suppresses breast cancer growth and metastasis through inhibiting AKT and Wnt/β-catenin signaling. Clin. Epigenetics 2020, 12, 173. [Google Scholar] [CrossRef]
- Zhang, J.; Wen, X.; Liu, N.; Li, Y.Q.; Tang, X.R.; Wang, Y.Q.; He, Q.M.; Yang, X.J.; Zhang, P.P.; Ma, J.; et al. Epigenetic mediated zinc finger protein 671 downregulation promotes cell proliferation and tumorigenicity in nasopharyngeal carcinoma by inhibiting cell cycle arrest. J. Exp. Clin. Cancer Res. 2017, 36, 147. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, J.; Jiang, H.; Xie, T.; Zheng, J.; Tian, Y.; Li, R.; Wang, B.; Lin, J.; Xu, A.; et al. The Tumor Suppressor Role of Zinc Finger Protein 671 (ZNF671) in Multiple Tumors Based on Cancer Single-Cell Sequencing. Front. Oncol. 2019, 9, 1214. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Bartha, Á.; Balázs, G. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER 2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, J48, W509–W514. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas–A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A.; Munkárcsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar]
- Modhukur, V.; Iljasenko, T.; Lokk, K.; Metsalu, T.; Laisk-Podar, T.; Vilo, J. MethSurv: A web tool to perform multivariate survival analysis using DNA methylation data. Epigenomics 2018, 10, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, D.; Lu, C. The SMART App: An interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin 2019, 12, 71. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions, and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Mi, H.; Thomas, P. PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar]
- Benstead-Hume, G.; Wooller, S.K.; Downs, J.A.; Pearl, F.M.G. Defining Signatures of Arm-Wise Copy Number Change and Their Associated Drivers in Kidney Cancers. Int. J. Mol. Sci. 2019, 20, 762. [Google Scholar] [CrossRef]
- Munro, D.; Ghersi, D.; Singh, M. Two critical positions in zinc finger domains are heavily mutated in three human cancer types. PLoS Comput. Biol. 2018, 14, e1006290. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, L.; Qiu, Y.; Yin, A.; Zhong, M. ZNF217 is associated with poor prognosis and enhances proliferation and metastasis in ovarian cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3038–3047. [Google Scholar]
- Zhang, C.; Xiang, T.; Li, S.; Ye, L.; Feng, Y.; Pei, L.; Li, L.; Wang, X.; Sun, R.; Ren, G.; et al. The novel 19q13 KRAB zinc-finger tumour suppressor ZNF382 is frequently methylated in oesophageal squamous cell carcinoma and antagonises Wnt/β-catenin signalling. Cell Death Dis. 2018, 9, 573. [Google Scholar] [CrossRef]
- Rousseau-Merck, M.F.; Koczan, D.; Legrand, I.; Moller, S.; Autran, S.; Thiesen, H.J. The KOX zinc finger genes: Genome-wide mapping of 368 ZNF PAC clones with zinc finger gene clusters predominantly in 23 chromosomal loci are confirmed by human sequences annotated in EnsEMBL. Cytogenet. Genome Res. 2002, 98, 147–153. [Google Scholar] [CrossRef]
- Brooks, S.A.; Brannon, A.R.; Parker, J.S.; Fisher, J.C.; Sen, O.; Kattan, M.W.; Hakimi, A.A.; Hsieh, J.J.; Choueiri, T.K.; Tamboli, P.; et al. ClearCode34: A prognostic risk predictor for localized clear cell renal cell carcinoma. Eur. Urol. 2014, 66, 77–84. [Google Scholar] [CrossRef]
- Brugarolas, J. Molecular genetics of clear-cell renal cell carcinoma. J. Clin. Oncol. 2014, 32, 1968–1976. [Google Scholar] [CrossRef]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of renal cell carcinoma: Findings and clinical implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef]
- Mehdi, A.; Riazalhosseini, Y. Epigenome Aberrations: Emerging Driving Factors of the Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2017, 18, 1774. [Google Scholar] [CrossRef]
- Peña-Llopis, S.; Vega-Rubín-de-Celis, S.; Liao, A.; Leng, N.; Pavía-Jiménez, A.; Wang, S.; Yamasaki, T.; Zhrebker, L.; Sivanand, S.; Spence, P.; et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 2012, 44, 751–759. [Google Scholar] [CrossRef]
- Wagner, E.; Carpenter, P. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell. Biol. 2012, 13, 115–126. [Google Scholar] [CrossRef]
- Dalgliesh, G.L.; Furge, K.; Greenman, C.; Chen, L.; Bignell, G.; Butler, A.; Davies, H.; Edkins, S.; Hardy, C.; Latimer, C.; et al. Systematic sequencing of renal carcinoma reveals inactivation of histone-modifying genes. Nature 2010, 463, 360–363. [Google Scholar] [CrossRef]
- Pantuck, A.J.; Seligson, D.B.; Klatte, T.; Yu, H.; Leppert, J.T.; Moore, L.; O’Toole, T.; Gibbons, J.; Belldegrun, A.S.; Figlin, R.A. Prognostic relevance of the mTOR pathway in renal cell carcinoma: Implications for molecular patient selection for targeted therapy. Cancer 2007, 109, 2257–2267. [Google Scholar] [CrossRef]
- Louie, B.H.; Kurzrock, R. BAP1: Not just a BRCA1-associated protein. Cancer Treat. Rev. 2020, 90, 102091. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Ma, N.; Zhao, L.; Yang, G.; Cao, B. KDM5c Promotes Colon Cancer Cell Proliferation through the FBXW7-c-Jun Regulatory Axis. Front. Oncol. 2020, 10, 535449. [Google Scholar] [CrossRef] [PubMed]
- Schabath, M.B.; Giuliano, A.R.; Thompson, Z.J.; Amankwah, E.K.; Gray, J.E.; Fenstermacher, D.A.; Jonathan, K.A.; Beg, A.A.; Haura, E.B. TNFRSF10B polymorphisms and haplotypes associated with increased risk of death in non-small cell lung cancer. Carcinogenesis 2013, 34, 2525–2530. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Provost, E.; Bailey, J.M.; Aldrugh, S.; Liu, S.; Iacobuzio-Donahue, C.; Leach, S.D. The tumor suppressor rpl36 restrains KRAS(G12V)-induced pancreatic cancer. Zebrafish 2014, 11, 551–559. [Google Scholar] [CrossRef]
- Zheng, X.; Wu, K.; Liao, S.; Pan, Y.; Sun, Y.; Chen, X.; Zhang, Y.; Xia, S.; Hu, Y.; Zhang, J. MicroRNA-transcription factor network analysis reveals miRNAs cooperatively suppress RORA in oral squamous cell carcinoma. Oncogenesis. 2018, 7, 79. [Google Scholar] [CrossRef]
- Khan, A.; Valli, E.; Lam, H.; Scott, D.A.; Murray, J.; Hanssen, K.M.; Eden, G.; Gamble, L.D.; Pandher, R.; Flemming, C.L.; et al. Targeting metabolic activity in high-risk neuroblastoma through Monocarboxylate Transporter 1 (MCT1) inhibition. Oncogene 2020, 39, 3555–3570. [Google Scholar] [CrossRef] [PubMed]
- Lucarelli, G.; Loizzo, D.; Franzin, R.; Battaglia, S.; Ferro, M.; Cantiello, F.; Castellano, G.; Bettocchi, C.; Ditonno, P.; Battaglia, M. Metabolomic insights into pathophysiological mechanisms and biomarker discovery in clear cell renal cell carcinoma. Exp. Rev. Mol. Diagn. 2019, 19, 397–407. [Google Scholar] [CrossRef]
- Wettersten, H.; Aboud, O.A.; Lara, P.N.J.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 7, 410–419. [Google Scholar] [CrossRef]
- Netti, G.S.; Lucarelli, G.; Spadaccino, F.; Castellano, G.; Gigante, M.; Divella, C.; Rocchetti, M.T.; Rascio, F.; Mancini, V.; Stallone, G.; et al. PTX3 modulates the immunoflogosis in tumor microenvironment and is a prognostic factor for patients with clear cell renal cell carcinoma. Aging 2020, 12, 7585–7602. [Google Scholar] [CrossRef] [PubMed]
- Beckermann, K.E.; Dudzinski, S.O.; Rathmell, J.C. Dysfunctional T cell metabolism in the tumor microenvironment. Cytokine Growth Factor Rev. 2017, 35, 7–14. [Google Scholar] [CrossRef]
- Şenbabaoğlu, Y.; Gejman, R.S.; Winer, A.G.; Liu, M.; Van Allen, E.M.; de Velasco, G.; Miao, D.; Ostrovnaya, I.; Drill, E.; Luna, A. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016, 17, 231. [Google Scholar] [CrossRef]
- Siska, P.J.; Beckermann, K.E.; Mason, F.M.; Andrejeva, G.; Greenplate, A.R.; Sendor, A.B.; Chiang, Y.C.J.; Corona, A.L.; Gemta, L.F.; Vincent, B.G.; et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2017, 2, e93411. [Google Scholar] [CrossRef] [PubMed]



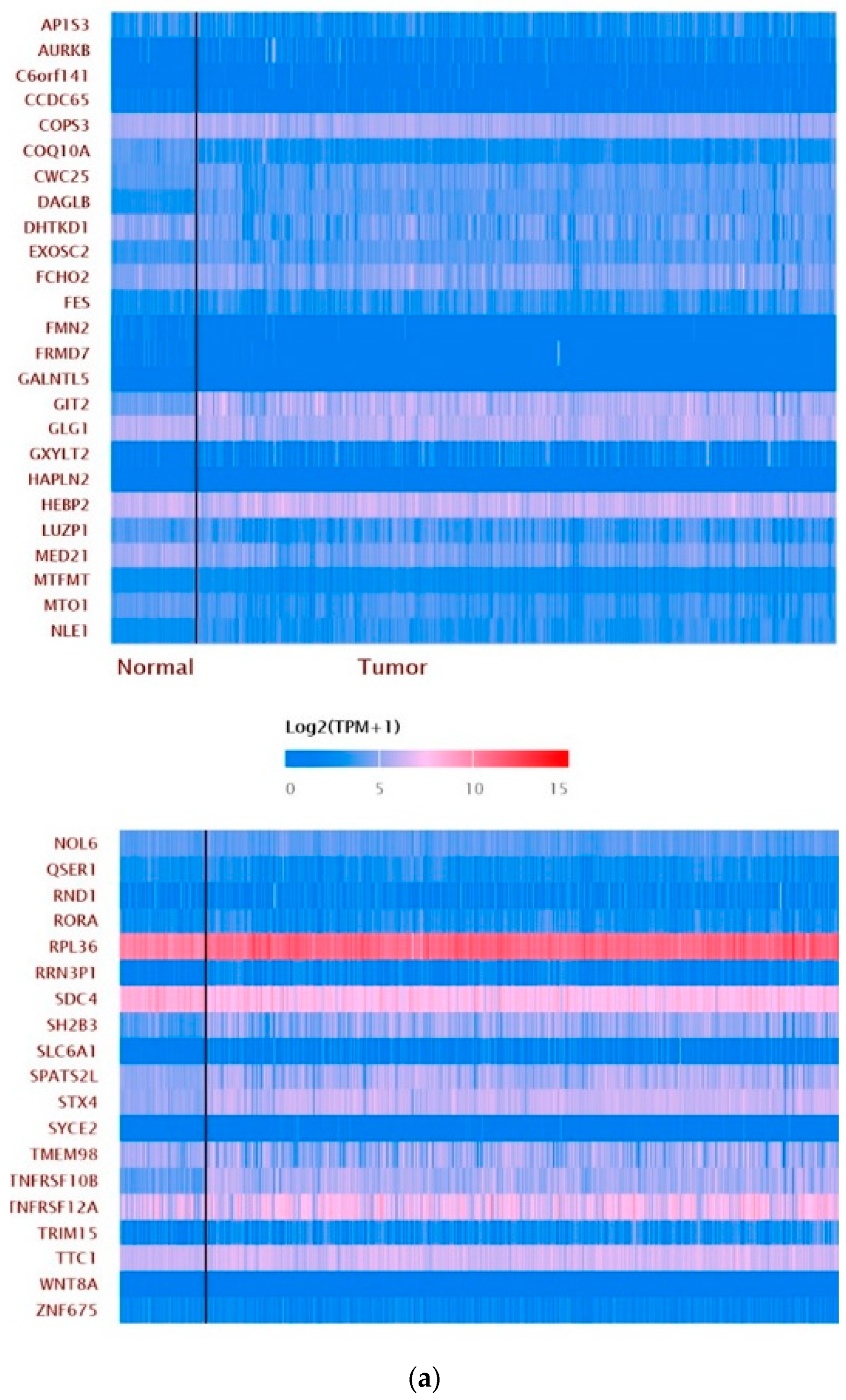
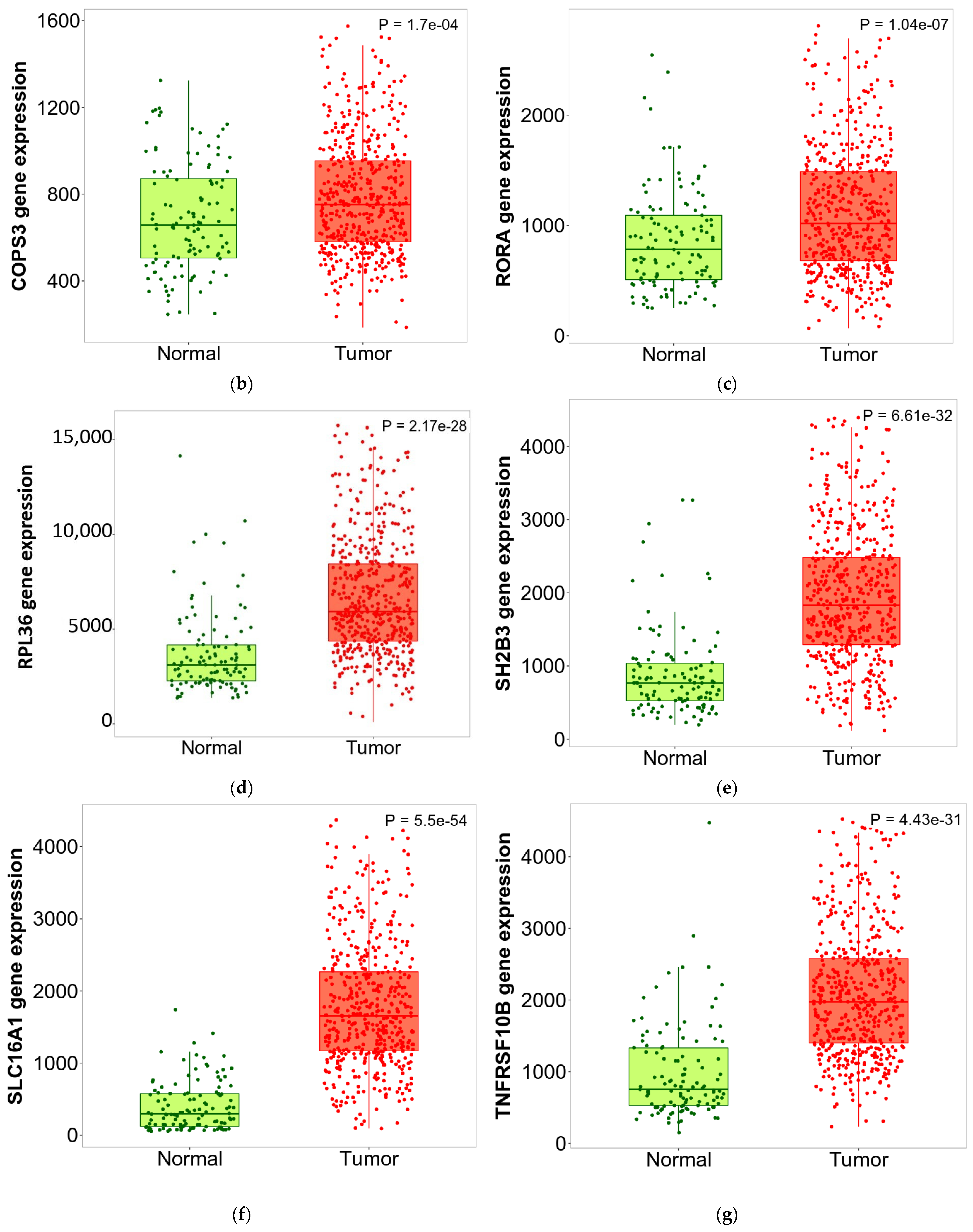
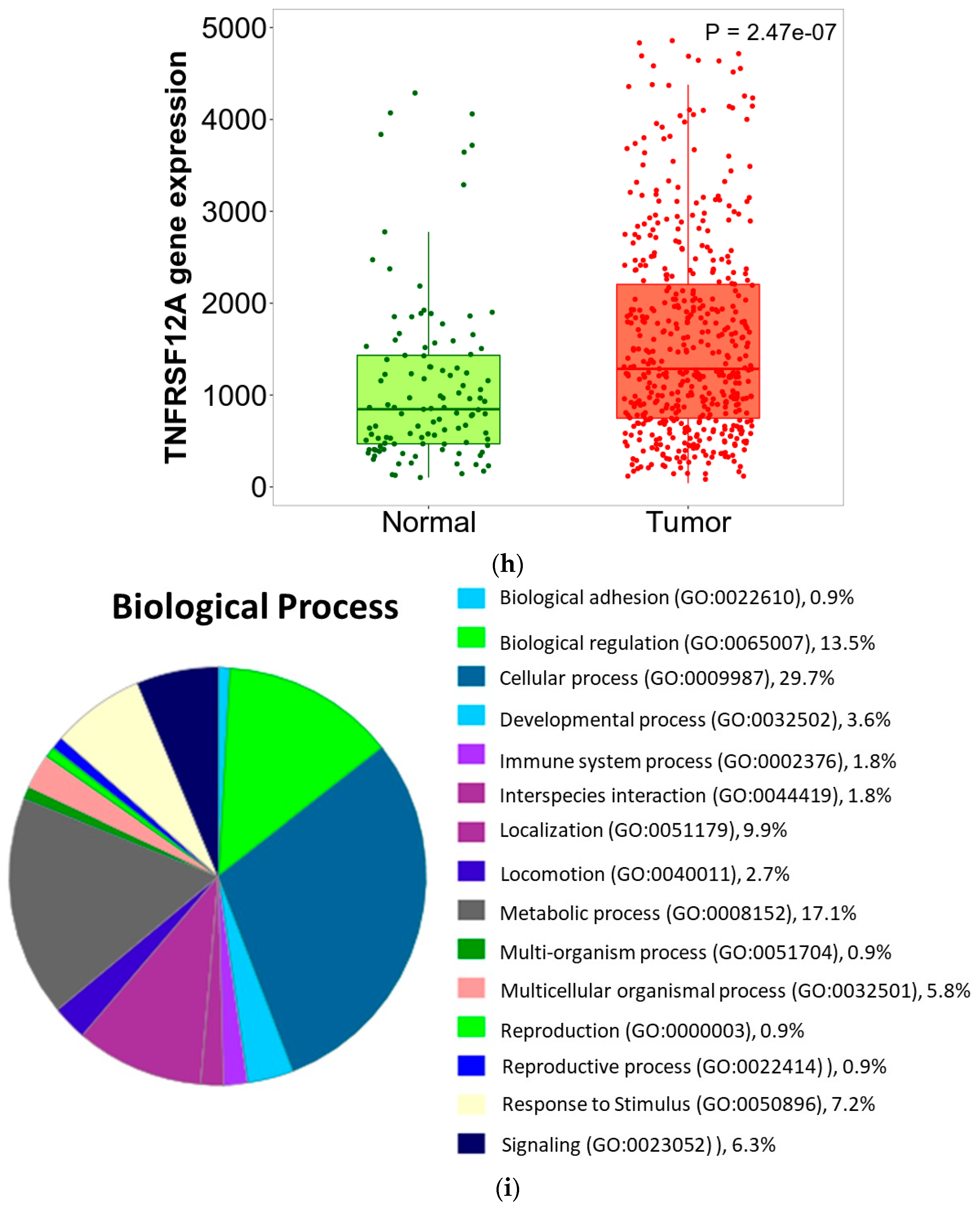
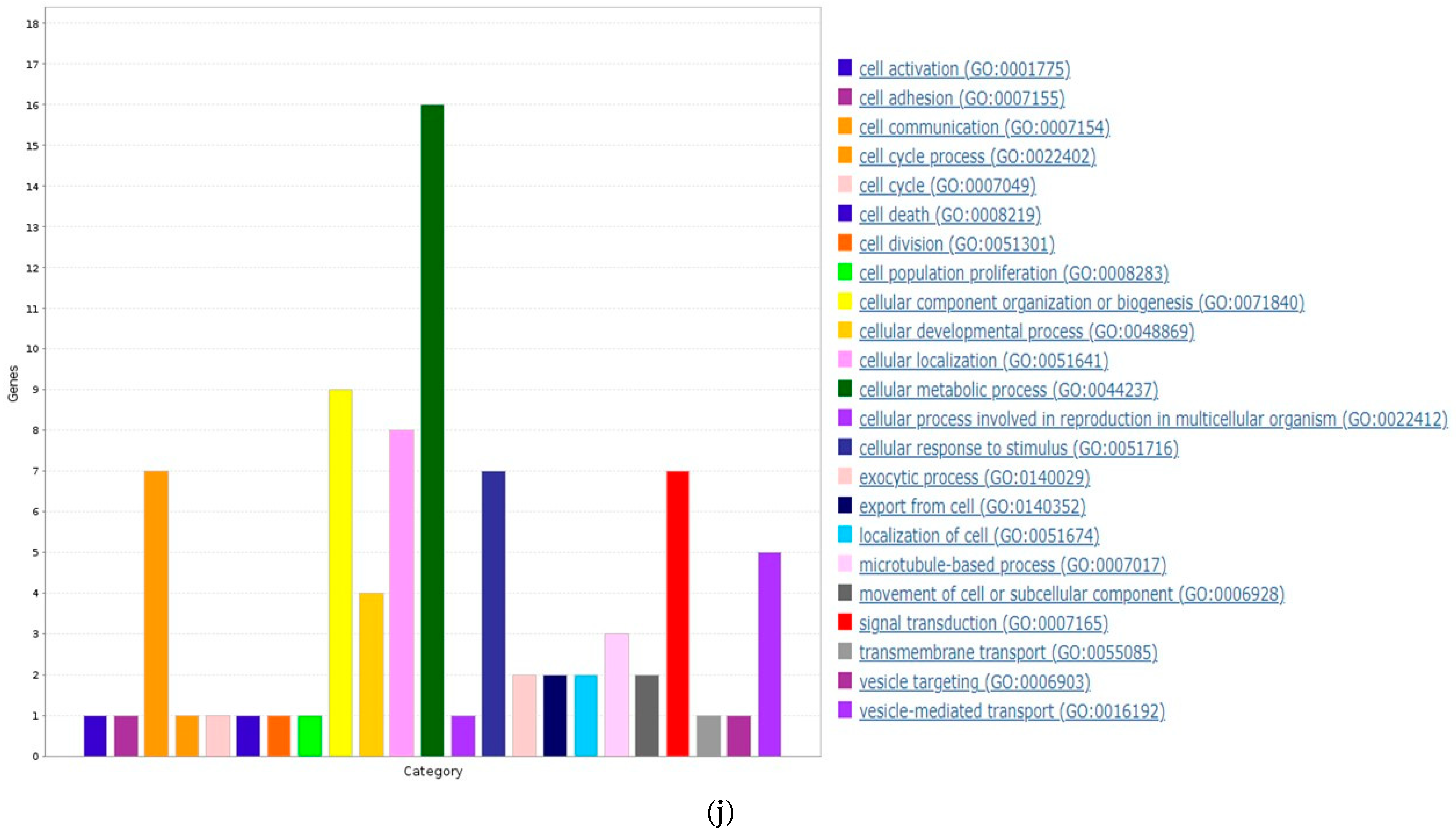
| Clinicopathological Features | N | Statistics |
|---|---|---|
| Gender | N.S. | |
| Male | 385 | |
| Female | 188 | |
| Histological Grade | p < 0.001 | |
| Normal | 72 | |
| 1 | 14 | |
| 2 | 229 | |
| 3 | 206 | |
| 4 | 76 | |
| Pathological Stage | p < 0.001 | |
| Normal | 72 | |
| 1 | 267 | |
| 2 | 57 | |
| 3 | 123 | |
| 4 | 84 | |
| Metastasis | p < 0.001 | |
| Normal | 72 | |
| N0 | 240 | |
| N1 | 16 | |
| Age | N.S. | |
| 21–40 years | 26 | |
| 41–60 years | 238 | |
| 61–80 years | 246 | |
| 81+ years | 23 |
| Clinicopathological Feature | Univariate Analysis HR (95% CI) p-Value | Multivariate Analysis HR (95% CI) p-Value |
|---|---|---|
| ZNF433 | 0.6227 (0.4593–0.8440) | 0.7390 (0.6987–1.3065) |
| p = 0.0023 | p = 0.0597 | |
| Gender | 0.9508 (0.7003–1.2910) | 0.9711 (0.7094–1.3292) |
| p = 0.7466 | p = 0.8547 | |
| Age | 1.7589 (1.2979–2.3835) | 1.5889 (1.1612–2.1742) |
| p = 0.0003 | p = 0.0038 | |
| Stage | 3.8179 (2.7869–5.2303) | 3.3766 (2.4441–4.6649) |
| p < 0.0001 | p < 0.001 | |
| Grade | 1.5172 (1.1152–2.0639) | 1.2429 (0.9088–1.6697) |
| p = 0.0079 | p = 0.1734 |
| Tumor | Normal | Change | Significance (p-Value) |
|---|---|---|---|
| BLCA.Tumor (n = 408) | BLCA.Normal (n = 19) | Upregulation | 0.016096887 |
| BRCA.Tumor (n = 1093) | BRCA.Normal (n = 112) | Downregulation | 0.446186328 |
| CESC.Tumor (n = 304) | CESC.Normal (n = 3) | Downregulation | 0.210693518 |
| CHOL.Tumor (n = 36) | CHOL.Normal (n = 9) | Upregulation | 0.005768157 |
| COAD.Tumor (n = 457) | COAD.Normal (n = 41) | Upregulation | 0.792680029 |
| ESCA.Tumor (n = 184) | ESCA.Normal (n = 11) | Downregulation | 0.164913994 |
| GBM.Tumor (n = 153) | GBM.Normal (n = 5) | Upregulation | 0.001122476 |
| HNSC.Tumor (n = 520) | HNSC.Normal (n = 44) | Downregulation | 6.12 × 10−8 |
| KICH.Tumor (n = 66) | KICH.Normal (n = 25) | Downregulation | 8.04 × 10−5 |
| KIRC.Tumor (n = 533) | KIRC.Normal (n = 72) | Downregulation | 1.13 × 10−23 |
| KIRP.Tumor (n = 290) | KIRP.Normal (n = 32) | Downregulation | 2.86 × 10−9 |
| LIHC.Tumor (n = 371) | LIHC.Normal (n = 50) | Upregulation | 7.93 × 10−11 |
| LUAD.Tumor (n = 515) | LUAD.Normal (n = 59) | Downregulation | 0.039291777 |
| LUSC.Tumor (n = 501) | LUSC.Normal (n = 51) | Downregulation | 1.16 × 10−5 |
| PAAD.Tumor (n = 178) | PAAD.Normal (n = 4) | Downregulation | 0.112226334 |
| PCPG.Tumor (n = 179) | PCPG.Normal (n = 3) | Upregulation | 0.032947137 |
| PRAD.Tumor (n = 497) | PRAD.Normal (n = 52) | Upregulation | 0.028391067 |
| READ.Tumor (n = 166) | READ.Normal (n = 10) | Downregulation | 0.101167394 |
| SKCM.Tumor (n = 103) | SKCM.Metast (n = 368) | Downregulation | 0.826588455 |
| STAD.Tumor (n = 415) | STAD.Normal (n = 35) | Downregulation | 0.433244729 |
| THCA.Tumor (n = 501) | THCA.Normal (n = 59) | Downregulation | 1.98 × 10−11 |
| UCEC.Tumor (n = 545) | UCEC.Normal (n = 35) | Upregulation | 0.015728415 |
| Gene Symbol | Official Name |
|---|---|
| ACER3 | alkaline ceramidase 3 |
| ALDOA | aldolase, fructose-bisphosphate A |
| ANGPTL6 | angiopoietin-like 6 |
| AP1S3 | adaptor related protein complex 1 subunit sigma 3 |
| ASS1P5 | argininosuccinate synthetase 1 pseudogene 5 |
| AURKB | aurora kinase B |
| BORCS6 | BLOC-1 related complex subunit 6 |
| C6orf141 | chromosome 6 open reading frame 141 |
| CCDC65 | coiled-coil domain containing 65 |
| COPS3 | COP9 signalosome subunit 3 |
| COQ10A | coenzyme Q10A |
| CROCCP3 | CROCC pseudogene 3 |
| CWC25 | CWC25 spliceosome associated protein homolog |
| CYP1B1-AS1 | CYP1B1 antisense RNA 1 |
| DAGLB | diacylglycerol lipase beta |
| DHTKD1 | dehydrogenase E1 and transketolase domain containing 1 |
| EXOSC2 | exosome component 2 |
| FABP5P3 | fatty-acid-binding protein 5 pseudogene 3 |
| FCHO2 | FCH and mu domain-containing endocytic adaptor 2 |
| FES | FES proto-oncogene, tyrosine kinase |
| FMN2 | formin 2 |
| FRMD7 | FERM domain containing 7 |
| GALNTL5 | polypeptide N-acetylgalactosaminyltransferase like 5 |
| GIT2 | GIT ArfGAP 2 |
| GLG1 | Golgi glycoprotein 1 |
| GPR1-AS | GPR1 antisense RNA |
| GXYLT2 | glucoside xylosyltransferase 2 |
| HAPLN2 | hyaluronan and proteoglycan link protein 2 |
| HEBP2 | heme binding protein 2 |
| KAT8 | lysine acetyltransferase 8 |
| LINC01235 | long intergenic non-protein coding RNA 1235 |
| LINC01641 | long intergenic non-protein coding RNA 1641 |
| LUZP1 | leucine zipper protein 1 |
| MED21 | mediator complex subunit 21 |
| MTFMT | mitochondrial methionyl-tRNA formyl transferase |
| MTND1P14 | MT-ND1 pseudogene 14 |
| MTO1 | mitochondrial tRNA translation optimization 1 |
| NLE1 | notchless homolog 1 |
| NOL6 | nucleolar protein 6 |
| NUCB1-AS1 | NUCB1 antisense RNA 1 |
| OR1X5P | olfactory receptor family 1 subfamily X member 5 pseudogene |
| QSER1 | glutamine and serine rich 1 |
| RN7SL93P | RNA, 7SL, cytoplasmic 93, pseudogene |
| RND1 | Rho family GTPase 1 |
| RNU6-1003P | RNA, U6 small nuclear 1003, pseudogene |
| RNU6-166P | RNA, U6 small nuclear 166, pseudogene |
| RORA | RAR related orphan receptor A |
| RPL32P27 | ribosomal protein L32 pseudogene 27 |
| RPL36 | ribosomal protein L36 |
| RRN3P1 | RRN3 pseudogene 1 |
| SDC4 | syndecan 4 |
| SH2B3 | SH2B adaptor protein 3 |
| SLC6A1 | solute carrier family 6-member 1 |
| SNHG30 | small nucleolar RNA host gene 30 |
| SPATS2L | spermatogenesis associated serine rich 2 like |
| STX4 | syntaxin 4 |
| SYCE2 | synaptonemal complex central element protein 2 |
| TMEM98 | transmembrane protein 98 |
| TNFRSF10B | TNF receptor superfamily member 10b |
| TNFRSF12A | TNF receptor superfamily member 12A |
| TRAV15 | T cell receptor alpha variable 15 (pseudogene) |
| TRIM15 | tripartite motif containing 15 |
| TTC1 | tetratricopeptide repeat domain 1 |
| TTLL13P | tubulin tyrosine ligase like 13, pseudogene |
| WNT8A | Wnt family member 8A |
| ZNF675 | zinc-finger protein 675 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heyliger, S.O.; Soliman, K.F.A.; Saulsbury, M.D.; Reams, R.R. The Identification of Zinc-Finger Protein 433 as a Possible Prognostic Biomarker for Clear-Cell Renal Cell Carcinoma. Biomolecules 2021, 11, 1193. https://doi.org/10.3390/biom11081193
Heyliger SO, Soliman KFA, Saulsbury MD, Reams RR. The Identification of Zinc-Finger Protein 433 as a Possible Prognostic Biomarker for Clear-Cell Renal Cell Carcinoma. Biomolecules. 2021; 11(8):1193. https://doi.org/10.3390/biom11081193
Chicago/Turabian StyleHeyliger, Simone O., Karam F. A. Soliman, Marilyn D. Saulsbury, and Romonia Renee Reams. 2021. "The Identification of Zinc-Finger Protein 433 as a Possible Prognostic Biomarker for Clear-Cell Renal Cell Carcinoma" Biomolecules 11, no. 8: 1193. https://doi.org/10.3390/biom11081193
APA StyleHeyliger, S. O., Soliman, K. F. A., Saulsbury, M. D., & Reams, R. R. (2021). The Identification of Zinc-Finger Protein 433 as a Possible Prognostic Biomarker for Clear-Cell Renal Cell Carcinoma. Biomolecules, 11(8), 1193. https://doi.org/10.3390/biom11081193






