Therapy Approaches for Stargardt Disease
Abstract
1. Introduction
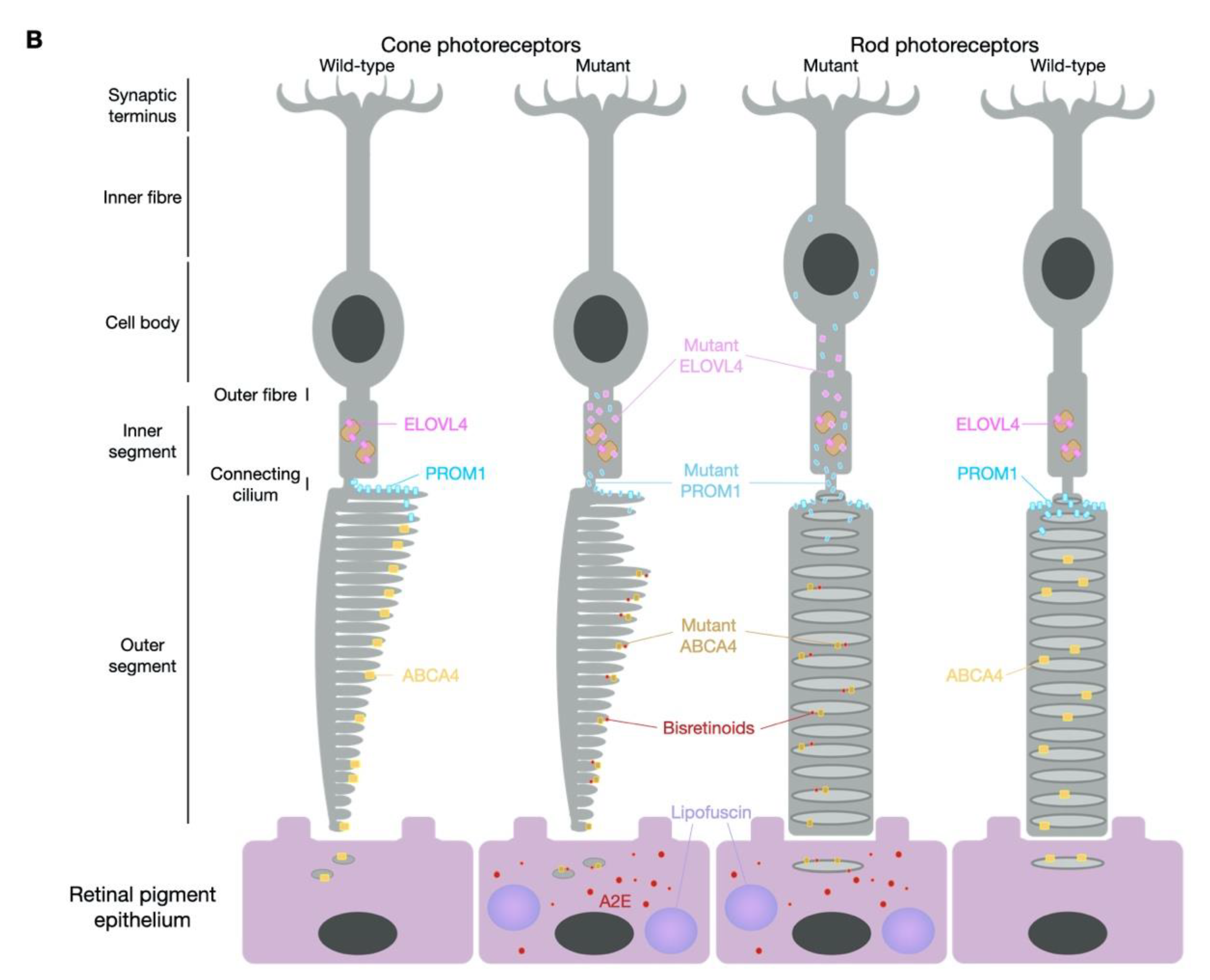
1.1. STGD1—ABCA4
1.2. STGD3—ELOVL4 (Allelic to the Former STGD2)
1.3. STGD4—PROM1
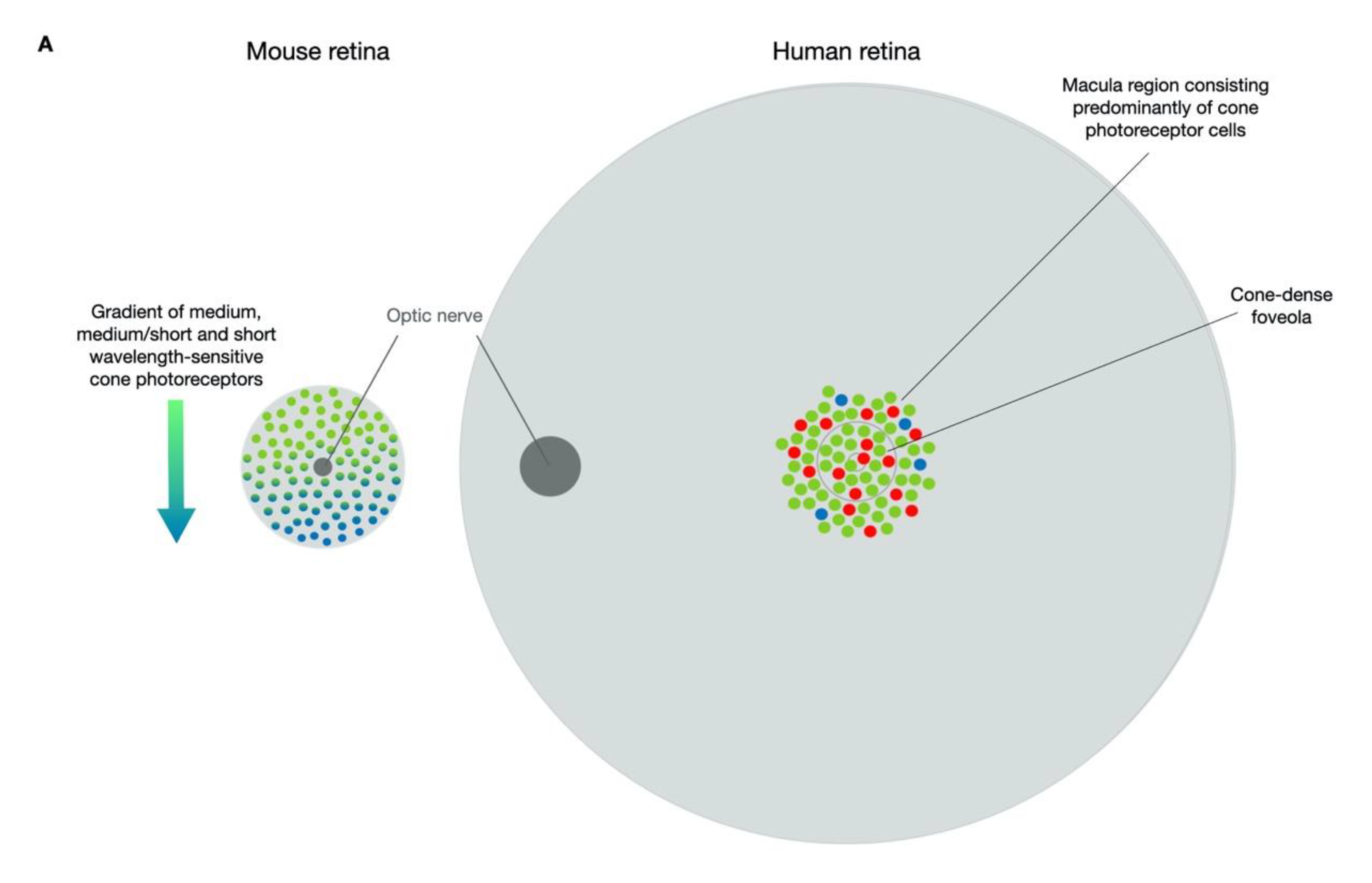
2. Models of Stargardt Disease
2.1. Mouse Models
2.2. In Vitro Models
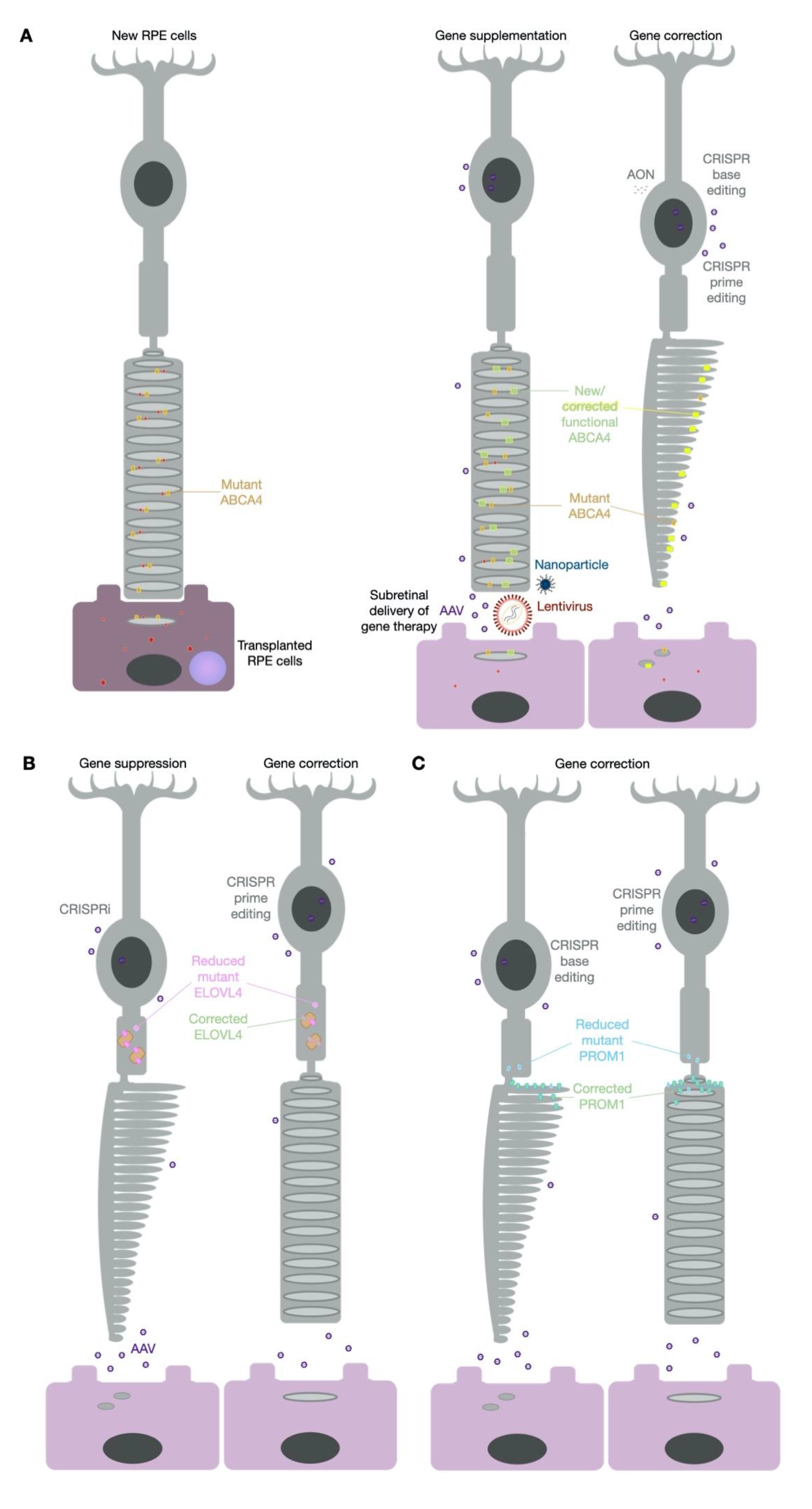
3. Gene Therapy for Stargardt Disease
3.1. Lentiviral Vectors
3.2. Adeno-Associated Viral Vectors
3.3. Nanoparticles
3.4. Anti-Sense Oligonucleotides
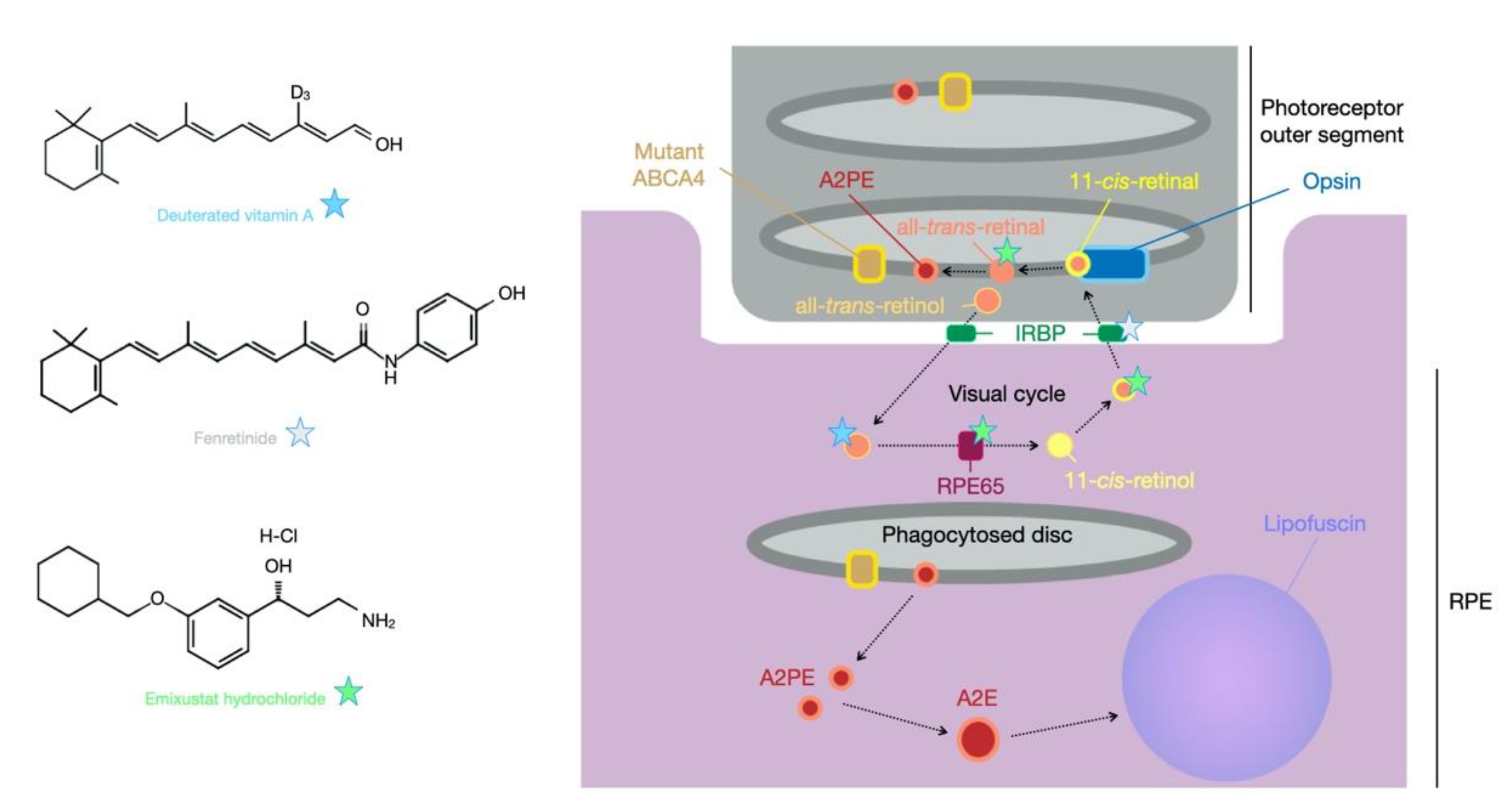
4. Small Molecule Therapy for Stargardt Disease
4.1. Pharmaceutical Interventions
4.2. Dietary Supplementation
5. Cell Replacement Therapy for Stargardt Disease
6. Future Therapy Prospects—CRISPR
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cremers, F.P.M.; Lee, W.; Collin, R.W.J.; Allikmets, R. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations. Prog. Retin. Eye Res. 2020, 100861. [Google Scholar] [CrossRef] [PubMed]
- Cornish, K.S.; Ho, J.; Downes, S.; Scott, N.W.; Bainbridge, J.; Lois, N. The Epidemiology of Stargardt Disease in the United Kingdom. Ophthalmol. Retin. 2017, 1, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Birtel, J.; Eisenberger, T.; Gliem, M.; Müller, P.L.; Herrmann, P.; Betz, C.; Zahnleiter, D.; Neuhaus, C.; Lenzner, S.; Holz, F.J.; et al. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Fry, L.E.; McClements, M.E.; MacLaren, R.E. Analysis of Pathogenic Variants Correctable with CRISPR Base Editing among Patients with Recessive Inherited Retinal Degeneration. JAMA Ophthalmol. 2021, 139. [Google Scholar] [CrossRef] [PubMed]
- Quazi, F.; Lenevich, S.; Molday, R.S. ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat. Commun. 2012, 3, 925. [Google Scholar] [CrossRef] [PubMed]
- Beharry, S.; Zhong, M.; Molday, R.S. N-retinylidene-phosphatidylethanolamine is the preferred retinoid substrate for the photoreceptor-specific ABC transporter ABCA4 (ABCR). J. Biol. Chem. 2004, 279, 53972–53979. [Google Scholar] [CrossRef]
- Sparrow, J.R.; Wu, Y.; Kim, C.Y.; Zhou, J. Phospholipid meets all-trans-retinal: The making of RPE bisretinoids. J. Lipid Res. 2010, 51, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Tsybovsky, Y.; Wang, B.; Quazi, F.; Molday, R.S.; Palczewski, K. Posttranslational modifications of the photoreceptor-specific ABC transporter ABCA4. Biochemistry 2011, 50, 6855–6866. [Google Scholar] [CrossRef] [PubMed]
- Sisk, R.A.; Leng, T. Multimodal imaging and multifocal electroretinography demonstrate autosomal recessive stargardt disease may present like occult macular dystrophy. Retina 2014, 34, 1567–1575. [Google Scholar] [CrossRef]
- Molday, R.S.; Zhang, K. Defective lipid transport and biosynthesis in recessive and dominant Stargardt macular degeneration. Prog. Lipid Res. 2010, 49, 476–492. [Google Scholar] [CrossRef][Green Version]
- Fritsche, L.G.; Fleckenstein, M.; Fiebig, B.S.; Schmitz-Valckenberg, S.; Bindewald-Wittich, A.; Keilhauer, C.N.; Renner, A.B.; Mackensen, F.; Mößner, A.; Pauleikhoff, D.; et al. A subgroup of age-related macular degeneration is associated with mono-allelic sequence variants in the ABCA4 gene. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2112–2118. [Google Scholar] [CrossRef]
- Lindner, M.; Lindner, M.; Lambertus, S.; Maushcitz, M.M.; Bax, N.B.; Kersten, E.; Lüning, A.; Nadal, J.; Schmitz-Valckenberg, S.; Schmid, M.; et al. Differential Disease Progression in Atrophic Age-Related Macular Degeneration and Late-Onset Stargardt DiseaseDisease Progression in AMD and Late-Onset STGD1. Invest. Ophth. Vis. Sci. 2017, 58, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Agbaga, M.-P.; Brush, R.S.; Mandal, M.N.A.; Henry, K.; Elliott, M.H.; Anderson, R.E. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc. Natl. Acad. Sci. USA 2008, 105, 12843–12848. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Kniazeva, M.; Han, M.; Li, W.; Yu, Z.; Yang, Z.; Li, Y.; Metzker, M.L.; Allikments, R.; Zack, D.J.; et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat. Genet. 2001, 27, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Logan, S.; Agbaga, M.-P.; Chan, M.D.; Brush, R.S.; Anderson, R.E. Endoplasmic reticulum microenvironment and conserved histidines govern ELOVL4 fatty acid elongase activity. J. Lipid Res. 2014, 55, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Dejos, C.; Kuny, S.; Han, W.H.; Capel, H.; Lemieux, H.; Sauvé, Y. Photoreceptor-induced RPE phagolysosomal maturation defects in Stargardt-like Maculopathy (STGD3). Sci. Rep. UK 2018, 8, 5944. [Google Scholar] [CrossRef]
- Aldahmesh, M.A.; Mohamed, J.Y.; Alkuraya, H.S.; Verma, I.C.; Puri, R.D.; Alaiya, A.A.; Rizzo, W.B.; Alkuraya, F.S. Recessive Mutations in ELOVL4 Cause Ichthyosis, Intellectual Disability, and Spastic Quadriplegia. Am. J. Hum. Genet. 2011, 89, 745–750. [Google Scholar] [CrossRef]
- Sompallae, R.; Hofmann, O.; Maher, C.A.; Gedye, C.; Behren, A.; Vitezic, M.; Daub, C.O.; Devalle, S.; Caballero, O.L.; Carninci, P.; et al. Comprehensive Promoter Landscape Identifies a Novel Promoter for CD133 in Restricted Tissues, Cancers, and Stem Cells. Front. Genet. 2013, 4, 209. [Google Scholar] [CrossRef] [PubMed]
- Li, Z. CD133: A stem cell biomarker and beyond. Exp. Hematol. Oncol. 2013, 2, 17. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, Y.; Lillo, C.; Chien, J.; Yu, Z.; Michaelides, M.; Klein, M.; Howes, K.A.; Li, Y.; Kaminoh, Y.; et al. Mutant Prominin 1 Found in Patients with Macular Degeneration Disrupts Photoreceptor Disk Morphogenesis in Mice. J. Clin. Investig. 2008, 118, 2908–2916. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Yin, J.; Winborn, C.S.; Zhang, Q.; Yue, J.; Chaum, E. Prominin-1 Is a Novel Regulator of Autophagy in the Human Retinal Pigment EpitheliumProm1 Negatively Regulates mTOR Signaling. Investig. Ophthalmol. Vis. Sci. 2017, 58, 2366–2387. [Google Scholar] [CrossRef]
- Permanyer, J.; Navarro, R.; Friedman, J.; Pomares, E.; Castro-Navarro, J.; Marfany, G.; Swaroop, A.; Gonzàlez-Duarte, R. Autosomal Recessive Retinitis Pigmentosa with Early Macular Affectation Caused by Premature Truncation in PROM1. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2656–2663. [Google Scholar] [CrossRef] [PubMed]
- Cehajic-Kapetanovic, J.; Birtel, J.; McClements, M.E.; Shanks, M.E.; Clouston, P.; Downes, S.M.; Issa, P.C.; MacLaren, R.E. Clinical and Molecular Characterization of PROM1-Related Retinal Degeneration. JAMA Netw. Open 2019, 2, e195752. [Google Scholar] [CrossRef]
- Michaelides, M.; Gaillard, M.-C.; Escher, P.; Tiab, L.; Bedell, M.; Borruat, F.-X.; Barthelmes, D.; Carmona, R.; Zhang, K.; White, E.; et al. The PROM1 Mutation p.R373C Causes an Autosomal Dominant Bull’s Eye Maculopathy Associated with Rod, Rod-Cone, and Macular Dystrophy. Investig. Ophthalmol. Vis. Sci. 2010, 51, 4771–4780. [Google Scholar] [CrossRef] [PubMed]
- Radu, R.A.; Yuan, Q.; Hu, J.; Peng, J.H.; Lloyd, M.; Nusinowitz, S.; Bok, D.; Travis, G.H. Accelerated Accumulation of Lipofuscin Pigments in the RPE of a Mouse Model for ABCA4-Mediated Retinal Dystrophies Following Vitamin A Supplementation. Investig. Ophthalmol. Vis. Sci. 2008, 49, 3821–3829. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Mata, N.L.; Azarian, S.M.; Tzekov, R.T.; Birch, D.G.; Travis, G.H. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt’s disease from the phenotype in abcr knockout mice. Cell 1999, 98, 13–23. [Google Scholar] [CrossRef]
- Sparrow, J.R.; Blonska, A.; Flynn, E.; Duncker, T.; Greenberg, J.P.; Secondi, R.; Ueda, K.; Delori, F.C. Quantitative Fundus Autofluorescence in Mice: Correlation with HPLC Quantitation of RPE Lipofuscin and Measurement of Retina Outer Nuclear Layer Thickness. Investig. Ophthalmol. Vis. Sci. 2013, 54, 2812–2820. [Google Scholar] [CrossRef] [PubMed]
- Issa, P.C.; Barnard, A.R.; Singh, M.S.; Carter, E.; Jiang, Z.; Radu, R.A.; Schraermeyer, U.; MacLaren, R.E. Fundus Autofluorescence in the Abca4−/− Mouse Model of Stargardt Disease—Correlation with Accumulation of A2E, Retinal Function and Histology. Investig. Ophthalmol. Vis. Sci. 2013. [Google Scholar] [CrossRef]
- Zhang, N.; Tsybovsky, Y.; Kolesnikov, A.V.; Rozanowska, M.; Swider, M.; Schwartz, S.B.; Stone, E.M.; Palczewska, G.; Maeda, A.; Kefalov, V.J.; et al. Protein Misfolding and the Pathogenesis of ABCA4-Associated Retinal Degenerations. Hum. Mol. Genet. 2015, 24, 3220–3237. [Google Scholar] [CrossRef]
- Molday, L.L.; Wahl, D.; Sarunic, M.V.; Molday, R.S. Localization and functional characterization of the p.Asn965Ser (N965S) ABCA4 variant in mice reveal pathogenic mechanisms underlying Stargardt macular degeneration. Hum. Mol. Genet. 2017, 27, 295–306. [Google Scholar] [CrossRef]
- Raz-Prag, D.; Ayyagari, R.; Fariss, R.N.; Mandal, M.N.A.; Vasireddy, V.; Majchrzak, S.; Webber, A.L.; Bush, R.A.; Salem, N.; Petrukhin, K.; et al. Haploinsufficiency Is Not the Key Mechanism of Pathogenesis in a Heterozygous Elovl4 Knockout Mouse Model of STGD3 Disease. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3603–3611. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, W.; Chen, Y.; Cameron, D.J.; Wang, C.; Karan, G.; Yang, Z.; Zhao, Y.; Pearson, E.; Chen, H.; Deng, C.; et al. Elovl4 Haploinsufficiency Does Not Induce Early Onset Retinal Degeneration in Mice. Vis. Res. 2007, 47, 714–722. [Google Scholar] [CrossRef] [PubMed]
- McMahon, A.; Jackson, S.N.; Woods, A.S.; Kedzierski, W. A Stargardt disease-3 mutation in the mouse Elovl4 gene causes retinal deficiency of C32–C36 acyl phosphatidylcholines. FEBS Lett. 2007, 581, 5459–5463. [Google Scholar] [CrossRef]
- Vasireddy, V.; Jablonski, M.M.; Khan, N.W.; Wang, X.F.; Sahu, P.; Sparrow, J.R.; Ayyagari, R. Elovl4 5-Bp Deletion Knock-in Mouse Model for Stargardt-like Macular Degeneration Demonstrates Accumulation of ELOVL4 and Lipofuscin. Exp. Eye Res. 2009, 89, 905–912. [Google Scholar] [CrossRef][Green Version]
- Karan, G.; Lillo, C.; Yang, Z.; Cameron, D.J.; Locke, K.G.; Zhao, Y.; Thirumalaichary, S.; Li, C.; Birch, D.G.; Vollmer-Snarr, H.R.; et al. Lipofuscin Accumulation, Abnormal Electrophysiology, and Photoreceptor Degeneration in Mutant ELOVL4 Transgenic Mice: A Model for Macular Degeneration. Proc. Natl. Acad. Sci. USA 2005, 102, 4164–4169. [Google Scholar] [CrossRef] [PubMed]
- Hopiavuori, B.R.; Anderson, R.E.; Agbaga, M.-P. ELOVL4: Very long-chain fatty acids serve an eclectic role in mammalian health and function. Prog. Retin. Eye Res. 2019, 69, 137–158. [Google Scholar] [CrossRef] [PubMed]
- Dellett, M.; Sasai, N.; Nishide, K.; Becker, S.; Papadaki, V.; Limb, G.A.; Moore, A.T.; Kondo, T.; Ohnuma, S.-I. Genetic Background and Light-Dependent Progression of Photoreceptor Cell Degeneration in Prominin-1 Knockout Mice. Investig. Ophthalmol. Vis. Sci. 2014, 56, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Garces, F.; Jiang, K.; Molday, L.L.; Stöhr, H.; Weber, B.H.; Lyons, C.J.; Maberley, D.; Molday, R.S. Correlating the Expression and Functional Activity of ABCA4 Disease Variants with the Phenotype of Patients With Stargardt Disease. Investig. Ophthalmol. Vis. Sci. 2018, 59, 2305–2315. [Google Scholar] [CrossRef] [PubMed]
- Sangermano, R.; Khan, M.; Cornelis, S.S.; Richelle, V.; Albert, S.; Garanto, A.; Elmelik, D.; Qamar, R.; Lugtenberg, D.; van den Born, L.I.; et al. ABCA4 Midigenes Reveal the Full Splice Spectrum of All Reported Noncanonical Splice Site Variants in Stargardt Disease. Genome Res. 2018, 28, 100–110. [Google Scholar] [CrossRef]
- Claassen, J.N.; Zhang, D.; Chen, S.-C.; Moon, S.Y.; Lamey, T.; Thompson, J.A.; McLaren, T.; Roach, J.N.D.; McLenachan, S.; Chen, F.K. Generation of the Induced Pluripotent Stem Cell Line from a Patient with Autosomal Recessive ABCA4-Mediated Stargardt Macular Dystrophy. Stem Cell Res. 2018, 34, 101352. [Google Scholar] [CrossRef]
- Ścieżyńska, A.; Soszyńska, M.; Komorowski, M.; Podgórska, A.; Krześniak, N.; Nogowska, A.; Smolińska, M.; Szulborski, K.; Szaflik, J.P.; Noszczyk, B.; et al. Molecular Analysis of the ABCA4 Gene Mutations in Patients with Stargardt Disease Using Human Hair Follicles. Int. J. Mol. Sci. 2020, 21, 3430. [Google Scholar] [CrossRef]
- Lane, A.; Jovanovic, K.; Shortall, C.; Ottaviani, D.; Panes, A.B.; Schwarz, N.; Guarascio, R.; Hayes, M.J.; Palfi, A.; Chadderton, N.; et al. Modeling and Rescue of RP2 Retinitis Pigmentosa Using IPSC-Derived Retinal Organoids. Stem Cell Rep. 2020, 15, 67–79. [Google Scholar] [CrossRef]
- Kallman, A.; Capowski, E.E.; Wang, J.; Kaushik, A.M.; Jansen, A.D.; Edwards, K.L.; Chen, L.; Berlinicke, C.A.; Phillips, M.J.; Pierce, E.A.; et al. Investigating Cone Photoreceptor Development Using Patient-Derived NRL Null Retinal Organoids. Commun. Biol. 2020, 3, 82. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.-L.; Lei, X.-L.; Han, F.; He, K.-W.; Jin, S.-Q.; Zhang, Y.-Y.; Jin, Z.-B. Patient-Specific Retinal Organoids Recapitulate Disease Features of Late-Onset Retinitis Pigmentosa. Front. Cell Dev. Biol. 2020, 8, 128. [Google Scholar] [CrossRef]
- Deng, W.-L.; Gao, M.-L.; Lei, X.-L.; Lv, J.-N.; Zhao, H.; He, K.-W.; Xia, X.-X.; Li, L.-Y.; Chen, Y.-C.; Li, Y.-P.; et al. Gene Correction Reverses Ciliopathy and Photoreceptor Loss in IPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients. Stem Cell Rep. 2018, 10, 1267–1281. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-C.; Wang, M.-L.; Chen, S.-J.; Kuo, J.-C.; Wang, W.-J.; Nguyen, P.N.N.; Wahlin, K.J.; Lu, J.-F.; Tran, A.A.; Shi, M.; et al. Morphological and Molecular Defects in Human Three-Dimensional Retinal Organoid Model of X-Linked Juvenile Retinoschisis. Stem Cell Rep. 2019, 13, 906–923. [Google Scholar] [CrossRef] [PubMed]
- Buskin, A.; Zhu, L.; Chichagova, V.; Basu, B.; Mozaffari-Jovin, S.; Dolan, D.; Droop, A.; Collin, J.; Bronstein, R.; Mehrotra, S.; et al. Disrupted Alternative Splicing for Genes Implicated in Splicing and Ciliogenesis Causes PRPF31 Retinitis Pigmentosa. Nat. Commun. 2018, 9, 4234. [Google Scholar] [CrossRef] [PubMed]
- Kruczek, K.; Qu, Z.; Gentry, J.; Fadl, B.R.; Gieser, L.; Hiriyanna, S.; Batz, Z.; Samant, M.; Samanta, A.; Chu, C.J.; et al. Gene Therapy of Dominant CRX-Leber Congenital Amaurosis Using Patient Stem Cell-Derived Retinal Organoids. Stem Cell Rep. 2021, 16, 252–263. [Google Scholar] [CrossRef]
- Cuevas, E.; Holder, D.L.; Alshehri, A.H.; Tréguier, J.; Lakowski, J.; Sowden, J.C. NRL−/− gene edited human embryonic stem cells generate rod-deficient retinal organoids enriched in S-cone-like photoreceptors. Stem Cells 2021, 39, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.; Bennett, J.; Wellman, J.A.; Chung, D.C.; Yu, Z.-F.; Tillman, A.; Wittes, J.; Pappas, J.; Elci, O.; McCague, S.; et al. Efficacy and Safety of Voretigene Neparvovec (AAV2-HRPE65v2) in Patients with RPE65-Mediated Inherited Retinal Dystrophy: A Randomised, Controlled, Open-Label, Phase 3 Trial. Lancet 2017, 390, 849–860. [Google Scholar] [CrossRef]
- Xue, K.; Jolly, J.K.; Barnard, A.R.; Rudenko, A.; Salvetti, A.P.; Patrício, M.I.; Edwards, T.L.; Groppe, M.; Orlans, H.O.; Tolmachova, T.; et al. Beneficial Effects on Vision in Patients Undergoing Retinal Gene Therapy for Choroideremia. Nat. Med. 2018, 24, 1507–1512. [Google Scholar] [CrossRef] [PubMed]
- Cehajic-Kapetanovic, J.; Xue, K.; de la Camara, C.M.-F.; Nanda, A.; Davies, A.; Wood, L.J.; Salvetti, A.P.; Fischer, M.D.; Aylward, J.W.; Barnard, A.R.; et al. Initial Results from a First-in-Human Gene Therapy Trial on X-Linked Retinitis Pigmentosa Caused by Mutations in RPGR. Nat. Med. 2020, 26, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Grieger, J.C.; Samulski, R.J. Packaging capacity of adeno-associated virus serotypes: Impact of larger genomes on infectivity and postentry steps. J. Virol. 2005, 79, 9933–9944. [Google Scholar] [CrossRef]
- Balaggan, K.S.; Binley, K.; Esapa, M.; Iqball, S.; Askham, Z.; Kan, O.; Tschernutter, M.; Bainbridge, J.W.B.; Naylor, S.; Ali, R.R. Stable and Efficient Intraocular Gene Transfer Using Pseudotyped EIAV Lentiviral Vectors. J. Gene Med. 2006, 8, 275–285. [Google Scholar] [CrossRef]
- Binley, K.; Widdowson, P.; Loader, J.; Kelleher, M.; Iqball, S.; Ferrige, G.; de Belin, J.; Carlucci, M.; Angell-Manning, D.; Hurst, F.; et al. Transduction of Photoreceptors with EIAV Lentiviral Vectors; Safety and Biodistribution of StarGenTM for Stargardt Disease. Investig. Ophthalmol. Vis. Sci. 2013. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Kim, S.-R.; Binley, K.; Pata, I.; Doi, K.; Mannik, J.; Zernant-Rajang, J.; Kan, O.; Iqball, S.; Naylor, S.; et al. Correction of the Disease Phenotype in the Mouse Model of Stargardt Disease by Lentiviral Gene Therapy. Gene Ther. 2008, 15, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, T.; Meschede, I.P.; Burden, J.J.; Bailly, M.; Seabra, M.C.; Futter, C.E. Rod disc renewal occurs by evagination of the ciliary plasma membrane that makes cadherin-based contacts with the inner segment. Proc. Natl. Acad. Sci. USA 2015, 112, 15922–15927. [Google Scholar] [CrossRef]
- Puppo, A.; Cesi, G.; Marrocco, E.; Piccolo, P.; Jacca, S.; Shayakhmetov, D.M.; Parks, R.J.; Davidson, B.L.; Colloca, S.; Brunetti-Pierri, N.; et al. Retinal Transduction Profiles by High-Capacity Viral Vectors. Gene Ther. 2014. [Google Scholar] [CrossRef] [PubMed]
- Nuzbrokh, Y.; Kassotis, A.S.; Ragi, S.D.; Jauregui, R.; Tsang, S.H. Treatment-Emergent Adverse Events in Gene Therapy Trials for Inherited Retinal Diseases: A Narrative Review. Ophthalmol. Ther. 2020, 36, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Allocca, M.; Doria, M.; Petrillo, M.; Colella, P.; Garcia-Hoyos, M.; Gibbs, D.; Kim, S.R.; Maguire, A.; Rex, T.S.; Vicino, U.D.; et al. Serotype-Dependent Packaging of Large Genes in Adeno-Associated Viral Vectors Results in Effective Gene Delivery in Mice. J. Clin. Investig. 2008, 118, 1955–1964. [Google Scholar] [CrossRef]
- Dong, B.; Nakai, H.; Xiao, W. Characterization of Genome Integrity for Oversized Recombinant AAV Vector. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 87–92. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, H.; Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Yue, Y.; Duan, D. Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome > or = 8.2 kb. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 75–79. [Google Scholar] [CrossRef]
- McClements, M.E.; MacLaren, R.E. Adeno-associated Virus (AAV) Dual Vector Strategies for Gene Therapy Encoding Large Transgenes. Yale J. Biol. Med. 2016, 90, 611–623. [Google Scholar] [CrossRef]
- McClements, M.E.; Issa, P.C.; Blouin, V.; MacLaren, R.E. A fragmented adeno-associated viral dual vector strategy for treatment of diseases caused by mutations in large genes leads to expression of hybrid transcripts. J. Genet. Syndr. Gene Ther. 2016, 7, 311. [Google Scholar] [CrossRef] [PubMed]
- Tornabene, P.; Trapani, I. Can Adeno-Associated Viral Vectors Deliver Effectively Large Genes? Hum. Gene Ther. 2020, 31, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Trapani, I.; Colella, P.; Sommella, A.; Iodice, C.; Cesi, G.; de Simone, S.; Marrocco, E.; Rossi, S.; Giunti, M.; Palfi, A.; et al. Effective Delivery of Large Genes to the Retina by Dual AAV Vectors. EMBO Mol. Med. 2014, 6, 194–211. [Google Scholar] [CrossRef] [PubMed]
- McClements, M.E.; Barnard, A.R.; Singh, M.S.; Issa, P.C.; Jiang, Z.; Radu, R.A.; MacLaren, R.E. An AAV Dual Vector Strategy Ameliorates the Stargardt Phenotype in Adult Abca4-/- Mice. Hum. Gene Ther. 2018, 30, 590–600. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, W.; Zhang, Y.; Zidon, T.; Ritchie, T.; Engelhardt, J.F. Concatamerization of adeno-associated virus circular genomes occurs through intermolecular recombination. J. Virol. 1999, 73, 9468–9477. [Google Scholar] [CrossRef] [PubMed]
- Reich, S.J.; Auricchio, A.; Hildinger, M.; Glover, E.; Maguire, A.M.; Wilson, J.M.; Bennett, J. Efficient Trans-Splicing in the Retina Expands the Utility of Adeno-Associated Virus as a Vector for Gene Therapy. Hum. Gene Ther. 2003, 14, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Colella, P.; Trapani, I.; Cesi, G.; Sommella, A.; Manfredi, A.; Puppo, A.; Iodice, C.; Rossi, S.; Simonelli, F.; Giunti, M.; et al. Efficient Gene Delivery to the Cone-Enriched Pig Retina by Dual AAV Vectors. Gene Ther. 2014, 21, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Schnepp, B.C.; Jensen, R.L.; Chen, C.-L.; Johnson, P.R.; Clark, K.R. Characterization of adeno-associated virus genomes isolated from human tissues. J. Virol. 2005, 79, 14793–14803. [Google Scholar] [CrossRef] [PubMed]
- Straus, S.E.; Sebring, E.D.; Rose, J.A. Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc. Natl. Acad. Sci. USA 1976, 73, 742–746. [Google Scholar] [CrossRef]
- Duan, D.; Yan, Z.; Yue, Y.; Engelhardt, J.F. Structural analysis of adeno-associated virus transduction circular intermediates. Virology 1999, 261, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Yue, Y.; Lai, Y.; Duan, D. A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol. Ther. J. Am. Soc. Gene Ther. 2008, 16, 124–130. [Google Scholar] [CrossRef]
- Ghosh, A.; Yue, Y.; Duan, D. Efficient transgene reconstitution with hybrid dual AAV vectors carrying the minimized bridging sequences. Hum. Gene Ther. 2011, 22, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Trapani, I.; Toriello, E.; de Simone, S.; Colella, P.; Iodice, C.; Polishchuk, E.V.; Sommella, A.; Colecchi, L.; Rossi, S.; Simonelli, F.; et al. Improved dual AAV vectors with reduced expression of truncated proteins are safe and effective in the retina of a mouse model of Stargardt disease. Hum. Mol. Genet. 2015, 24, 6811–6825. [Google Scholar] [CrossRef]
- Dyka, F.M.; Molday, L.L.; Chiodo, V.A.; Molday, R.S.; Hauswirth, W.W. Dual ABCA4-AAV Vector Treatment Reduces Pathogenic Retinal A2E Accumulation in a Mouse Model of Autosomal Recessive Stargardt Disease. Hum. Gene Ther. 2019, 30, 1361–1370. [Google Scholar] [CrossRef]
- Tornabene, P.; Trapani, I.; Minopoli, R.; Centrulo, M.; Lupo, M.; de Simone, S.; Tiberi, P.; Dell’Aquila, F.; Marrocco, E.; Iodice, C.; et al. Intein-Mediated Protein Trans-Splicing Expands Adeno-Associated Virus Transfer Capacity in the Retina. Sci. Transl. Med. 2019, 11, eaav4523. [Google Scholar] [CrossRef] [PubMed]
- Dyka, F.M.; Boye, S.L.; Chiodo, V.A.; Hauswirth, W.W.; Boye, S.E. Dual adeno-associated virus vectors result in efficient in vitro and in vivo expression of an oversized gene, MYO7A. Hum. Gene Ther. Methods 2014, 25, 166–177. [Google Scholar] [CrossRef]
- Li, J.; Sun, W.; Wang, B.; Xiao, X.; Liu, X.-Q. Protein trans-splicing as a means for viral vector-mediated in vivo gene therapy. Hum. Gene Ther. 2008, 19, 958–964. [Google Scholar] [CrossRef] [PubMed]
- McClements, M.E.; Barnard, A.R.; Issa, P.C.; MacLaren, R.E. Assessment of AAV Dual Vector Safety in the Abca4−/− Mouse Model of Stargardt Disease. Transl. Vis. Sci. Technol. 2020, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Bucher, K.; Rodríguez-Bocanegra, E.; Dauletbekov, D.; Fischer, M.D. Immune responses to retinal gene therapy using adeno-associated viral vectors—Implications for treatment success and safety. Prog. Retin. Eye Res. 2020, 100915. [Google Scholar] [CrossRef]
- Chandler, L.C.; McClements, M.E.; Yusuf, I.H.; de la Camara, C.M.-F.; MacLaren, R.E.; Xue, K. Characterizing the Cellular Immune Response to Subretinal AAV Gene Therapy in the Murine Retina. Mol. Ther. Methods Clin. Dev. 2021. [Google Scholar] [CrossRef]
- Han, Z.; Conley, S.M.; Makkia, R.S.; Cooper, M.J.; Naash, M.I. DNA nanoparticle-mediated ABCA4 delivery rescues Stargardt dystrophy in mice. J. Clin. Investig. 2012, 122, 3221–3226. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Schur, R.M.; Sears, A.E.; Gao, S.-Q.; Vaidya, A.; Sun, W.; Maeda, A.; Kern, T.; Palczewski, K.; Lu, Z.-R. Non-Viral Gene Therapy for Stargardt Disease with ECO/PRHO-ABCA4 Self-Assembled Nanoparticles. Mol. Ther. 2020, 28, 293–303. [Google Scholar] [CrossRef]
- Sun, D.; Maeno, H.; Gujrati, M.; Schur, R.; Maeda, A.; Maeda, T.; Palczewski, K.; Lu, Z. Self-Assembly of a Multifunctional Lipid With Core–Shell Dendrimer DNA Nanoparticles Enhanced Efficient Gene Delivery at Low Charge Ratios into RPE Cells. Macromol. Biosci. 2015, 15, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Gujrati, M.; Malamas, A.; Shin, T.; Jin, E.; Sun, Y.; Lu, Z.-R. Multifunctional Cationic Lipid-Based Nanoparticles Facilitate Endosomal Escape and Reduction-Triggered Cytosolic siRNA Release. Mol. Pharmaceut. 2014, 11, 2734–2744. [Google Scholar] [CrossRef]
- Sun, D.; Sun, W.; Gao, S.-Q.; Wei, C.; Naderi, A.; Schilb, A.L.; Scheidt, J.; Lee, S.; Kern, T.S.; Palczewski, K.; et al. Formulation and Efficacy of ECO/PRHO-ABCA4-SV40 Nanoparticles for Nonviral Gene Therapy of Stargardt Disease in a Mouse Model. J. Control. Release 2021, 330, 329–340. [Google Scholar] [CrossRef]
- Xue, K.; MacLaren, R.E. Antisense oligonucleotide therapeutics in clinical trials for the treatment of inherited retinal diseases. Expert Opin. Investig. Drugs 2020, 29, 1163–1170. [Google Scholar] [CrossRef]
- Tomkiewicz, T.Z.; Suárez-Herrera, N.; Cremers, F.P.M.; Collin, R.W.J.; Garanto, A. Antisense Oligonucleotide-Based Rescue of Aberrant Splicing Defects Caused by 15 Pathogenic Variants in ABCA4. Int. J. Mol. Sci. 2021, 22, 4621. [Google Scholar] [CrossRef] [PubMed]
- Cideciyan, A.V.; Jacobson, S.G.; Drack, A.V.; Ho, A.C.; Charng, J.; Garafalo, A.V.; Roman, A.J.; Sumaroka, A.; Han, I.C.; Hochstedler, M.D.; et al. Effect of an Intravitreal Antisense Oligonucleotide on Vision in Leber Congenital Amaurosis Due to a Photoreceptor Cilium Defect. Nat. Med. 2018, 25, 225–228. [Google Scholar] [CrossRef]
- Cideciyan, A.V.; Jacobson, S.G.; Ho, A.C.; Garafalo, A.V.; Roman, A.J.; Sumaroka, A.; Krishnan, A.K.; Swider, M.; Schwartz, M.R.; Girach, A. Durable Vision Improvement after a Single Treatment with Antisense Oligonucleotide Sepofarsen: A Case Report. Nat. Med. 2021, 27, 785–789. [Google Scholar] [CrossRef]
- Deák, F.; Anderson, R.E.; Fessler, J.L.; Sherry, D.M. Novel Cellular Functions of Very Long Chain-Fatty Acids: Insight from ELOVL4 Mutations. Front. Cell. Neurosci. 2019, 13, 428. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.K.; Rohrschneider, K.; Strom, T.M.; Glöckle, N.; Kohl, S.; Wissinger, B.; Weisschuh, N. Homozygosity Mapping and Whole-Genome Sequencing Reveals a Deep Intronic PROM1 Mutation Causing Cone–Rod Dystrophy by Pseudoexon Activation. Eur. J. Hum. Genet. 2016, 24, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Albert, S.; Garanto, A.; Sangermano, R.; Khan, M.; Bax, N.M.; Hoyng, C.B.; Zernant, J.; Lee, W.; Allikmets, R.; Collin, R.W.J.; et al. Identification and Rescue of Splice Defects Caused by Two Neighboring Deep-Intronic ABCA4 Mutations Underlying Stargardt Disease. Am. J. Hum. Genet. 2018, 102, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Garanto, A.; Duijkers, L.; Tomkiewicz, T.Z.; Collin, R.W.J. Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease. Genes 2019, 10, 452. [Google Scholar] [CrossRef]
- Khan, M.; Arno, G.; Fakin, A.; Parfitt, D.A.; Dhooge, P.P.A.; Albert, S.; Bax, N.M.; Duijkers, L.; Niblock, M.; Hau, K.L.; et al. Detailed Phenotyping and Therapeutic Strategies for Intronic ABCA4 Variants in Stargardt Disease. Mol. Ther. Nucleic Acids 2020, 21, 412–427. [Google Scholar] [CrossRef]
- Garanto, A.; Chung, D.C.; Duijkers, L.; Corral-Serrano, J.C.; Messchaert, M.; Xiao, R.; Bennett, J.; Vandenberghe, L.H.; Collin, R.W.J. In Vitro and in Vivo Rescue of Aberrant Splicing in CEP290 -Associated LCA by Antisense Oligonucleotide Delivery. Hum. Mol. Genet. 2016, 25, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Merkle, T.; Merz, S.; Reautschnig, P.; Blaha, A.; Li, Q.; Vogel, P.; Wettengel, J.; Li, J.B.; Stafforst, T. Precise RNA Editing by Recruiting Endogenous ADARs with Antisense Oligonucleotides. Nat. Biotechnol. 2019, 37, 133–138. [Google Scholar] [CrossRef]
- George, C.X.; Samuel, C.E. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. USA 1999, 96, 4621–4626. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.; Gommans, W.M. Identification of a selective nuclear import signal in adenosine deaminases acting on RNA. Nucleic Acids Res. 2009, 37, 5822–5829. [Google Scholar] [CrossRef]
- Wang, A.L.; Carroll, R.C.; Nawy, S. Down-Regulation of the RNA Editing Enzyme ADAR2 Contributes to RGC Death in a Mouse Model of Glaucoma. PLoS ONE 2014, 9, e91288. [Google Scholar] [CrossRef]
- Qu, L.; Yi, Z.; Zhu, S.; Wang, C.; Cao, Z.; Zhou, Z.; Yuan, P.; Yu, Y.; Tian, F.; Liu, Z.; et al. Programmable RNA Editing by Recruiting Endogenous ADAR Using Engineered RNAs. Nat. Biotechnol. 2019, 37, 1059–1069. [Google Scholar] [CrossRef]
- Katrekar, D.; Chen, G.; Meluzzi, D.; Ganesh, A.; Worlikar, A.; Shih, Y.-R.; Varghese, S.; Mali, P. In Vivo RNA Editing of Point Mutations via RNA-Guided Adenosine Deaminases. Nat. Methods 2019, 16, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Fry, L.E.; Peddle, C.F.; Barnard, A.R.; McClements, M.E.; MacLaren, R.E. RNA Editing as a Therapeutic Approach for Retinal Gene Therapy Requiring Long Coding Sequences. Int. J. Mol. Sci. 2020, 21, 777. [Google Scholar] [CrossRef]
- Maeda, A.; Golczak, M.; Maeda, T.; Palczewski, K. Limited roles of Rdh8, Rdh12, and Abca4 in all-trans-retinal clearance in mouse retina. Investig. Ophthalmol. Vis. Sci. 2009, 50, 5435–5443. [Google Scholar] [CrossRef]
- Kubota, R.; Al-Fayoumi, S.; Mallikaarjun, S.; Patil, S.; Bavik, C.; Chandler, J.W. Phase 1, dose-ranging study of emixustat hydrochloride (ACU-4429), a novel visual cycle modulator, in healthy volunteers. Retina 2014, 34, 603–609. [Google Scholar] [CrossRef]
- Zhang, J.; Kiser, P.D.; Badiee, M.; Palczewska, G.; Dong, Z.; Golczak, M.; Tochtrop, G.P.; Palczewski, K. Molecular Pharmacodynamics of Emixustat in Protection against Retinal Degeneration. J. Clin. Investig. 2015, 125, 2781–2794. [Google Scholar] [CrossRef] [PubMed]
- Kubota, R.; Birch, D.G.; Gregory, J.K.; Koester, J.M. Randomised study evaluating the pharmacodynamics of emixustat hydrochloride in subjects with macular atrophy secondary to Stargardt disease. Brit. J. Ophthalmol. 2020, 1–6. [Google Scholar] [CrossRef]
- Zhou, J.; Jang, Y.P.; Kim, S.R.; Sparrow, J.R. Complement activation by photooxidation products of A2E, a lipofuscin constituent of the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2006, 103, 16182–16187. [Google Scholar] [CrossRef] [PubMed]
- Radu, R.A.; Hu, J.; Yuan, Q.; Welch, D.L.; Makshanoff, J.; Lloyd, M.; McMullen, S.; Travis, G.H.; Bok, D. Complement System Dysregulation and Inflammation in the Retinal Pigment Epithelium of a Mouse Model for Stargardt Macular Degeneration. J. Biol. Chem. 2011, 286, 18593–18601. [Google Scholar] [CrossRef]
- Armento, A.; Ueffing, M.; Clark, S.J. The complement system in age-related macular degeneration. Cell. Mol. Life Sci. 2021, 1–19. [Google Scholar] [CrossRef]
- Jaffe, G.J.; Westby, K.; Csaky, K.G.; Monés, J.; Pearlman, J.A.; Patel, S.S.; Joondeph, B.C.; Randolph, J.; Masonson, H.; Rezaei, K.A. C5 Inhibitor Avacincaptad Pegol for Geographic Atrophy Due to Age-Related Macular Degeneration A Randomized Pivotal Phase 2/3 Trial. Ophthalmology 2021, 128, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Yehoshua, Z.; de Amorim Garcia Filho, C.A.; Nunes, R.P.; Gregori, G.; Penha, F.M.; Moshfeghi, A.A.; Zhang, K.; Sadda, S.; Feuer, W.; Rosenfeld, P.J. Systemic Complement Inhibition with Eculizumab for Geographic Atrophy in Age-Related Macular Degeneration the COMPLETE Study. Ophthalmology 2014, 121, 693–701. [Google Scholar] [CrossRef]
- Kaufman, Y.; Ma, L.; Washington, I. Deuterium Enrichment of Vitamin A at the C20 Position Slows the Formation of Detrimental Vitamin A Dimers in Wild-type Rodents. J. Biol. Chem. 2011, 286, 7958–7965. [Google Scholar] [CrossRef] [PubMed]
- Issa, P.C.; Barnard, A.R.; Herrmann, P.; Washington, I.; MacLaren, R.E. Rescue of the Stargardt phenotype in Abca4 knockout mice through inhibition of vitamin A dimerization. Proc. Natl. Acad. Sci. USA 2015, 112, 8415–8420. [Google Scholar] [CrossRef] [PubMed]
- Scholl, H.P.; Tsang, S.H.; Kay, C.N.; Conner, T.B.; Gorin, M.B.; Bernstain, P.S.; Lam, B.L.; Strecker, Z.; Zaremba, T.; DeBartolomeo, G.; et al. Stargardt disease ALK-001 phase 2 clinical trial: 12-month interim data. Investig. Ophthalmol. Vis. Sci. 2019, 60, 1336. [Google Scholar]
- Radu, R.A.; Han, Y.; Bui, T.V.; Nusinowitz, S.; Bok, D.; Lichter, J.; Widder, K.; Travis, G.H.; Mata, N.L. Reductions in Serum Vitamin A Arrest Accumulation of Toxic Retinal Fluorophores: A Potential Therapy for Treatment of Lipofuscin-Based Retinal Diseases. Investig. Ophthalmol. Vis. Sci. 2005, 46, 4393–4401. [Google Scholar] [CrossRef]
- Mata, N.L.; Lichter, J.B.; Vogel, R.; Han, Y.; Bui, T.V.; Singerman, L.J. Investigation of oral fenretinide for treatment of geographic atrophy in age-related macular degeneration. Retina 2013, 33, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Dobri, N.; Qin, Q.; Kong, J.; Yamamoto, K.; Liu, Z.; Moiseyev, G.; Ma, J.; Allikmets, R.; Sparrow, J.R.; Petrukhin, K. A1120, a Nonretinoid RBP4 Antagonist, Inhibits Formation of Cytotoxic Bisretinoids in the Animal Model of Enhanced Retinal LipofuscinogenesisA1120 in the Animal Model of Retinal Lipofuscinogenesis. Investig Ophthalmol. Vis. Sci. 2013, 54, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Neuringer, M. Infant vision and retinal function in studies of dietary long-chain polyunsaturated fatty acids: Methods, results, and implications. Am. J. Clin. Nutr. 2000, 71, 256S–267S. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, I.M.; Sieving, P.A. Investigation of the effect of dietary docosahexaenoic acid (DHA) supplementation on macular function in subjects with autosomal recessive Stargardt macular dystrophy. Ophthalmic Genet. 2018, 39, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Dornstauder, B.; Suh, M.; Kuny, S.; Gaillard, F.; MacDonald, I.M.; Clandinin, M.T.; Sauvé, Y. Dietary Docosahexaenoic Acid Supplementation Prevents Age-Related Functional Losses and A2E Accumulation in the RetinaDietary DHA Delays Retinal Degeneration. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2256–2265. [Google Scholar] [CrossRef]
- Piccardi, M.; Fadda, A.; Martelli, F.; Marangoni, D.; Magli, A.; Minnella, A.M.; Bertelli, M.; Marco, S.D.; Bisti, S.; Falsini, B. Antioxidant Saffron and Central Retinal Function in ABCA4-Related Stargardt Macular Dystrophy. Nutrients 2019, 11, 2461. [Google Scholar] [CrossRef] [PubMed]
- D’Cruz, P.M.; Yasumura, D.; Weir, J.; Matthes, M.T.; Abderrahim, H.; LaVail, M.M.; Vollrath, D. Mutation of the Receptor Tyrosine Kinase Gene Mertk in the Retinal Dystrophic RCS Rat. Hum. Mol. Genet. 2000, 9, 645–651. [Google Scholar] [CrossRef]
- Lu, B.; Malcuit, C.; Wang, S.; Girman, S.; Francis, P.; Lemieux, L.; Lanza, R.; Lund, R. Long-Term Safety and Function of RPE from Human Embryonic Stem Cells in Preclinical Models of Macular Degeneration. Stem Cells 2009, 27, 2126–2135. [Google Scholar] [CrossRef]
- Schwartz, S.D.; Regillo, C.D.; Lam, B.L.; Eliott, D.; Rosenfeld, P.J.; Gregori, N.Z.; Hubschman, J.-P.; Davis, J.L.; Heilwell, G.; Spirn, M.; et al. Human Embryonic Stem Cell-Derived Retinal Pigment Epithelium in Patients with Age-Related Macular Degeneration and Stargardt’s Macular Dystrophy: Follow-up of Two Open-Label Phase 1/2 Studies. Lancet 2015, 385, 509–516. [Google Scholar] [CrossRef]
- Mehat, M.S.; Sundaram, V.; Ripamonti, C.; Robson, A.G.; Smith, A.J.; Borooah, S.; Robinson, M.; Rosenthal, A.N.; Innes, W.; Weleber, R.G.; et al. Transplantation of Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells in Macular Degeneration. Ophthalmology 2018, 125, 1765–1775. [Google Scholar] [CrossRef]
- Weiss, J.N.; Levy, S. Stem Cell Ophthalmology Treatment Study (SCOTS): Bone Marrow-Derived Stem Cells in the Treatment of Stargardt Disease. Medicines 2021, 8, 10. [Google Scholar] [CrossRef]
- Parmar, V.M.; Parmar, T.; Arai, E.; Perusek, L.; Maeda, A. A2E-associated cell death and inflammation in retinal pigmented epithelial cells from human induced pluripotent stem cells. Stem Cell Res. 2018, 27, 95–104. [Google Scholar] [CrossRef]
- Lee, J.; Bayarsaikhan, D.; Bayarsaikhan, G.; Kim, J.-S.; Schwarzbach, E.; Lee, B. Recent advances in genome editing of stem cells for drug discovery and therapeutic application. Pharm. Ther. 2020, 209, 107501. [Google Scholar] [CrossRef]
- Kantor, A.; McClements, M.E.; Peddle, C.F.; Fry, L.E.; Salman, A.; Cehajic-Kapetanovic, J.; Xue, K.; MacLaren, R.E. CRISPR Genome Engineering for Retinal Diseases. Prog. Mol. Biol. Transl. 2021. [Google Scholar] [CrossRef]
- Peddle, C.F.; Fry, L.E.; McClements, M.E.; MacLaren, R.E. CRISPR Interference–Potential Application in Retinal Disease. Int. J. Mol. Sci. 2020, 21, 2329. [Google Scholar] [CrossRef]
- Quinn, J.; Musa, A.; Kantor, A.; McClements, M.E.; Cehajic-Kapetanovic, J.; MacLaren, R.E.; Xue, K. Genome-Editing Strategies for Treating Human Retinal Degenerations. Hum. Gene Ther. 2020. [Google Scholar] [CrossRef]
- Kantor, A.; McClements, M.E.; MacLaren, R.E. CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int. J. Mol. Sci. 2020, 21, 6240. [Google Scholar] [CrossRef]
- First CRISPR therapy dosed. Nat. Biotechnol. 2020, 38, 382. [CrossRef] [PubMed]
- Maeder, M.L.; Stefanidakis, M.; Wilson, C.J.; Baral, R.; Barrera, L.A.; Bounoutas, G.S.; Bumcrot, D.; Chao, H.; Ciulla, D.M.; DaSilva, J.A.; et al. Development of a Gene-Editing Approach to Restore Vision Loss in Leber Congenital Amaurosis Type 10. Nature medicine 2019, 25, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Imani, S.; Cheng, J.; Shasaltaneh, M.D.; Wei, C.; Yang, L.; Fu, S.; Zou, H.; Khan, M.A.; Zhang, X.; Chen, H.; et al. Genetic Identification and Molecular Modeling Characterization Reveal a Novel PROM1 Mutation in Stargardt4-like Macular Dystrophy. Oncotarget 2017, 9, 122–141. [Google Scholar] [CrossRef] [PubMed]
- Salles, M.V.; Motta, F.L.; da Silva, E.D.; Teixeira, P.V.L.; Costa, K.A.; Filippelli-Silva, R.; Martin, R.; Pesquero, J.B.; Sallum, J.M.F. PROM1 Gene Variations in Brazilian Patients with Macular Dystrophy. Ophthalmic Genet 2017, 38, 39–42. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, S.; Zhang, C.; Gao, N.; Li., M.; Wang, D.; Wang, D.; Liu, D.; Liu, H.; Ong, S.; et al. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 2020, 18, e3000686. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Topkar, V.V.; Zheng, Z.; Joung, J.K. Broadening the Targeting Range of Staphylococcus Aureus CRISPR-Cas9 by Modifying PAM Recognition. Nat. Biotechnol. 2015, 33, 1293–1298. [Google Scholar] [CrossRef]
- Suh, S.; Choi, E.H.; Leinonen, H.; Foik, A.T.; Newby, G.A.; Yeh, W.-H.; Dong, Z.; Kiser, P.D.; Lyon, D.C.; Liu, D.R.; et al. Restoration of Visual Function in Adult Mice with an Inherited Retinal Disease via Adenine Base Editing. Nat. Biomed. Eng. 2020. [Google Scholar] [CrossRef] [PubMed]
- McCullough, K.T.; Boye, S.L.; Fajardo, D.; Calabro, K.; Peterson, J.J.; Strang, C.E.; Chakraborty, D.; Gloskowski, S.; Haskett, S.; Samuelsson, S.; et al. Somatic Gene Editing of GUCY2D by AAV-CRISPR/Cas9 Alters Retinal Structure and Function in Mouse and Macaque. Hum. Gene Ther. 2019, 30, 571–589. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.M.; Yeh, W.-H.; Pendse, N.; Davis, J.R.; Hennessey, E.; Butcher, R.; Koblan, L.W.; Comander, J.; Liu, Q.; Liu, D.R. Cytosine and Adenine Base Editing of the Brain, Liver, Retina, Heart and Skeletal Muscle of Mice via Adeno-Associated Viruses. Nat. Biomed. Eng. 2020, 4, 97–110. [Google Scholar] [CrossRef]
- Chung, S.H.; Mollhoff, I.N.; Nguyen, U.; Nguyen, A.; Stucka, N.; Tieu, E.; Manna, S.; Maleppat, R.K.; Zhang, P.; Nguyen, E.L.; et al. Factors Impacting Efficacy of AAV-Mediated CRISPR-Based Genome Editing for Treatment of Choroidal Neovascularization. Mol. Ther. Methods Clin. Dev. 2020, 17, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, K.; Panda, S.; Gonzales-Rojsa, R.; Chong, A.; Bugay, V.; Park, H.Y.; Brenner, R.; Murthy, N.; Lee, H.Y. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018, 2, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.L.; Gliem, M.; Mangold, E.; Bolz, H.J.; Finger, R.P.; McGuinness, M.; Betz, C.; Jiang, Z.; Weber, B.H.F.; MacLaren, R.E.; et al. Monoallelic ABCA4 Mutations Appear Insufficient to Cause Retinopathy: A Quantitative Autofluorescence Study. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8179–8186. [Google Scholar] [CrossRef]
- Jin, S.; Fei, H.; Zhu, Z.; Luo, Y.; Liu, J.; Gao, S.; Zhang, F.; Chen, Y.-H.; Wang, Y.; Gao, C. Rationally Designed APOBEC3B Cytosine Base Editors with Improved Specificity. Mol. Cell 2020, 79, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 337, 1–21. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Kweon, J.; Yoon, J.-K.; Jang, A.-H.; Shin, H.R.; See, J.-E.; Jang, G.; Kim, J.-I.; Kim, Y. Engineered Prime Editors with PAM Flexibility. Mol. Ther. 2021. [Google Scholar] [CrossRef] [PubMed]
| Model Type | Details | Structural Features | Functional Features | Strengths/Limitations |
|---|---|---|---|---|
| Mouse | ||||
| STGD1 | Abca4 KO [25,26] | Absence of Abca4 expression. On an albino background, loss of outer nuclear layer (ONL) structure at 11 months. Pigmented mice show no loss in structure. Lipofuscin granule accumulation in the RPE. | Abca4 KO models exhibit increased autofluorescence compared to age-matched wild-type mice that correlates to accumulation of bisretinoids/A2E/lipofuscin. | Easy detection of ABCA4 protein following gene supplementation. Assessment of pharmaceutical, dietary and gene therapy efficacy achievable by reduction in autofluorescence and associated build-up of bisretinoids/A2E/lipofuscin. However, the KO genotype and absence of Abca4 does not reflect typical human disease. |
| STGD1 | Leu451Pro and Ala1038Val (PV/PV) [29] Asn965Ser [30] | Reduced expression of Abca4 with mislocalisation within the photoreceptor cells. | Models exhibit increased autofluorescence compared to age-matched wild-type mice that correlates to accumulation of bisretinoids/A2E/lipofuscin. | Efficacy evident in these models would be more relevant to human disease and achieved by rescue of bisretinoid/A2E/lipofuscin build-up and the associated autofluorescence phenotype. |
| STGD3 | Elovl4 KO [31,32] | Normal retinal structure. | Normal retinal function. | The KO is of limited value as it can only be reared as a heterozygous model and offers no clear features of retinal disease. |
| STGD3 | ELOVL4 5-bp deletion knock-in [33,34,35,36] | Accumulation of ELOVL4 at 4 months with progressive loss of ONL and, in particular, cones at 6–18 months. | Abnormal ERG and accumulation of lipofuscin. | Rescue of retinal structure and function. Transgenic models are more representative of human disease both in genotype and phenotype. |
| STGD4 | Rd19 | Progressive loss of ONL beginning at 2 months of age. | Normal cone ERG but abnormal rod a-wave responses. | This naturally occurring model has yet to be used in pre-clinical studies. |
| STGD4 | Prom1 KO [37] | Extensive loss of ONL beginning at 2 weeks of age. | Abnormal ERG. | Loss of retinal structure and function begins early; therefore, treatment intervention may not be provided in time to observe efficacy. Rearing in the dark could be applied to slow the rate of degeneration. |
| STGD4 | PROM1 Arg373Cys knock-in [20] | Mislocalisation of PROM1with abnormal outer segment morphology and degeneration. | Abnormal rod and cone ERG by 3 months of age. | The knock-in better reflects the human state and offers an opportunity to assess treatment efficacy through correction of structural and functional changes. |
| In Vitro | ||||
| Immortalised cell lines | Wild-type | Lack of native retinal gene expression and absence of specialised retinal structures. | Enables expression and localisation assessments plus downstream isolation and functional assays. | Exogenous delivery of retinal genes of interest is required but basic assessments of vectors and downstream functional assays are achievable. |
| Induced pluripotent stem cells (iPSCs) | Patient-specific genotype | Cells can be differentiated to better reflect photoreceptor cell morphology and gene expression profiles. | Functional outputs could be achieved by expression profile analysis and downstream protein isolation and functional assays. | These will be particularly useful for future gene-editing techniques in assessing mutation-specific therapies. Editing efficiencies and protein outputs could be compared to cells from control donors. |
| Fibroblasts | Patient-specific genotype | Some retinal gene expression may be evident, as for ABCA4 [41]. | Functional outputs could be achieved by expression profile analysis and downstream protein isolation and functional assays. | The use of these will likely be supplementary to preliminary pre-clinical assessments of new therapies as expression of retinal genes will be limited. However, being patient-derived, they will have the added benefit of being useful for gene-editing strategies. |
| Hair follicles | Patient-specific genotype | Some retinal gene expression may be evident, as for ABCA4 [41]. | As for fibroblast samples, functional outputs could be achieved by expression profile analysis and downstream protein isolation and functional assays. | As for fibroblast samples, the use of these will likely be supplementary to other preliminary pre-clinical assessments but being patient-derived they will have the added benefit of being useful for gene-editing therapies. |
| Retinal organoids | Patient-specific genotype | Structural differences may be evident and include protein mislocalisation [42,43,44,45,46,47,48,49]. | As for other patient-derived samples, functional outputs could be achieved by expression profile analysis and downstream protein isolation and functional assays. | Changes in expression profiles and protein localisation plus cell morphology could be assessed following treatment application. Retinal organoid will provide an ideal model for mutation-specific treatments. |
| Strategy | Therapeutic | Trial | Phase | Data |
|---|---|---|---|---|
| Gene-based | ||||
| Gene supplementation | Lentivirus | NCT01367444 | Terminated in 2019 due to sponsor issues, not for reasons of safety. | Yet to be peer-reviewed. |
| NCT01736592 | Follow-up of patients involved in the above trial. | |||
| Gene supplementation | Dual AAV | N/A | Trials have yet to be initiated. | |
| Gene modulation | AON | N/A | N/A | |
| Gene editing | CRISPR | N/A | N/A | |
| Pharmaceutical | ||||
| Visual cycle modulator | Emixustat hydrochloride | NCT03033108 | Phase I/IIa (completed 2021) | Delayed dark adaptation at 5 and 10 mg doses confirmed biological activity of the drug [110]. |
| NCT03772665 | Phase III (initiated 2018) | Data not yet reported. | ||
| Deuterated vitamin A | ALK-001 | NCT02230228 | Phase I (completed 2015) | Therapeutic was well tolerated. |
| NCT02402660 | Phase II (initiated 2015) | Tolerability data were reported at ARVO 2019 [118]. | ||
| NCT04239625 | Phase II (initiated 2020) | This is an extension of the above study of tolerability, safety and efficacy. | ||
| Inhibitors of retinol-binding protein | Fenretinide | No STGD trials | N/A | Adverse events in 20% of AMD patients at the high dose with no signs of efficacy. |
| A1120 | None reported | N/A | ||
| STG-001 | NCT04489511 | Phase IIa (completed 2021) | Data yet to be reported. | |
| C5 inhibition | Avacincaptad pegol | NCT03364153 | Phase IIb (initiated 2017) | No data for STGD1 patients published. |
| Eculzimab | No STGD trials | N/A | Well tolerated in AMD patients but no signs of efficacy. | |
| Dietary | ||||
| DHA | NCT00060749 | Phase I (completed 2017) | Data for 11 STGD1 patients reported no adverse events or signs of efficacy [123]. Data for STGD3 cohort yet to be published. | |
| Omega-3 fatty acids | NCT03297515 | Prospective trial completed 2021 | Data yet to be reported. | |
| Saffron | NCT01278277 | Phase I/II (initiated 2011) | No safety concerns and no significant changes in macular function of STGD1 patients after 6 months [125]. | |
| Cell replacement | ||||
| ESC-RPE | MA09-hRPE | NCT01625559 | Phase I (initiated 2012) | No serious adverse events and no signs of efficacy 12 months post treatment in advanced STGD1 patients [128]. |
| NCT01345006 | Phase I/II (completed 2021) | |||
| NCT01469832 | Phase I/II (completed 2021) | Escalating dose of transplanted cells produced no serious adverse events with no signs of efficacy 12 months post treatment in 12 patients with advanced STGD1 [129]. | ||
| NCT02445612 | Long-term follow-up to Phase I/II (initiated 2015) | Data yet to be reported. | ||
| hESC-RPE | NCT02903576 | Phase I/II (completed 2020) | Data yet to be reported. | |
| NCT02941991 | Follow-up to Phase I/II (initiated 2016) | Data yet to be reported. | ||
| BMSC | NCT01920867 | Non-randomised open label (initiated 2013) | Data have been reported but issues exist regarding recruitment and study design [130]. | |
| Mutation | CRISPR Strategy | ABCA4 Total = 349 | ELOVL4 Total = 23 | PROM1 Total = 3 |
|---|---|---|---|---|
| G > A | ABE (coding strand) and RNA-ABE | 72 | 3 | 1 |
| A > G | CBE (non-coding strand) | 21 | 1 | 0 |
| T > C | CBE (coding strand) | 31 | 3 | 1 |
| C > T | ABE (non-coding strand) | 63 | 4 | 1 |
| G > T | Prime | 20 | 2 | 0 |
| G > C | Prime | 14 | 0 | 0 |
| T > A | Prime | 12 | 0 | 0 |
| A > T | Prime | 8 | 1 | 0 |
| C > A | Prime | 15 | 2 | 0 |
| A > C | Prime | 4 | 1 | 0 |
| C > G | Prime | 13 | 4 | 0 |
| T > G | Prime | 17 | 0 | 0 |
| Insert/deletion/duplication | Prime | 59 | 2 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piotter, E.; McClements, M.E.; MacLaren, R.E. Therapy Approaches for Stargardt Disease. Biomolecules 2021, 11, 1179. https://doi.org/10.3390/biom11081179
Piotter E, McClements ME, MacLaren RE. Therapy Approaches for Stargardt Disease. Biomolecules. 2021; 11(8):1179. https://doi.org/10.3390/biom11081179
Chicago/Turabian StylePiotter, Elena, Michelle E McClements, and Robert E MacLaren. 2021. "Therapy Approaches for Stargardt Disease" Biomolecules 11, no. 8: 1179. https://doi.org/10.3390/biom11081179
APA StylePiotter, E., McClements, M. E., & MacLaren, R. E. (2021). Therapy Approaches for Stargardt Disease. Biomolecules, 11(8), 1179. https://doi.org/10.3390/biom11081179





