Evolutionary and Characteristic Analysis of RING-DUF1117 E3 Ubiquitin Ligase Genes in Gossypium Discerning the Role of GhRDUF4D in Verticillium dahliae Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characterization of RDUF Gene Members in Gossypium
2.2. Phylogenetic, Gene Structure and Conserved Domain, and Motif Analysis
2.3. Chromosomal Location, Gene Synteny, and RNA-Seq Dataset Analysis
2.4. Transcription Factors (TFs) and miRNAs in Targeting GhRDUF Genes
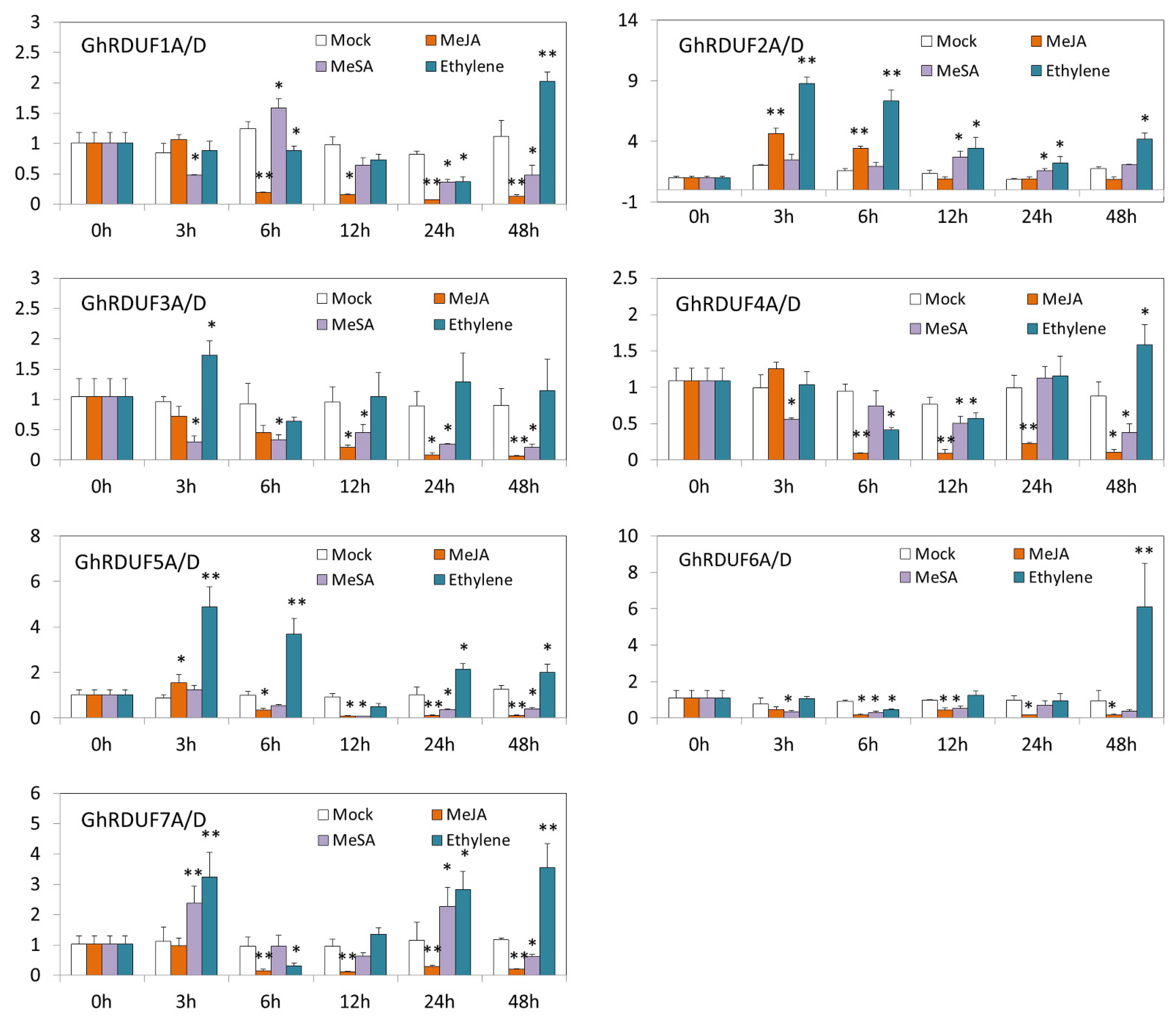
2.5. Cotton Seedlings under Hormone Treatments
2.6. GFP Vector Construction and Fluorescent Visualization
2.7. Overexpression Vector Construction and Screening of Transgenic Arabidopsis
2.8. VIGS Vector Construction and Assays Performing
2.9. V. dahliae Infection and Disease Evaluation
2.10. Reactive Oxygen Species (ROS) Analysis and Callose
2.11. RT-qPCR Analysis
3. Results
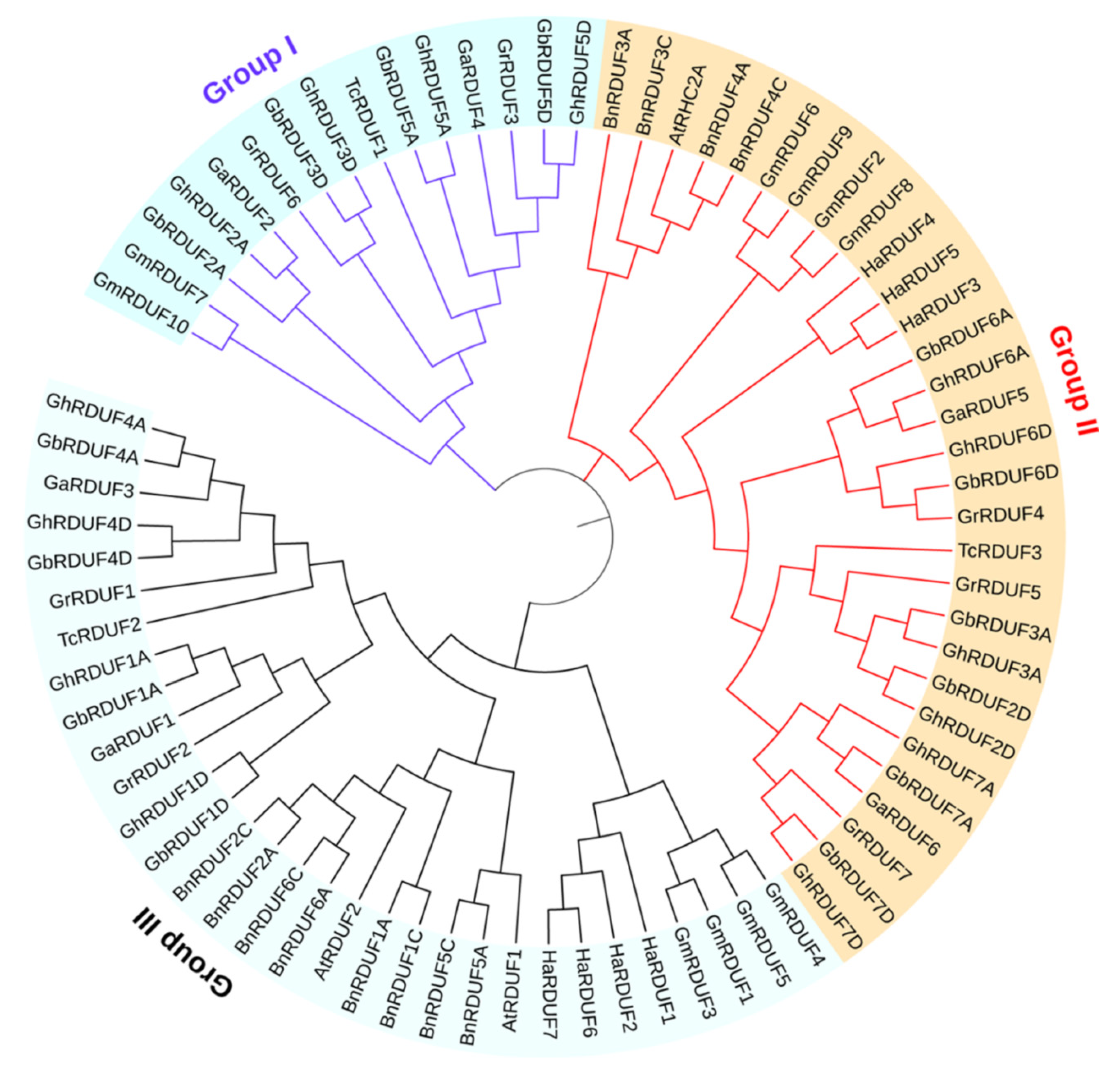
3.1. Identification and Phylogenetic Analysis of RDUF Family Genes
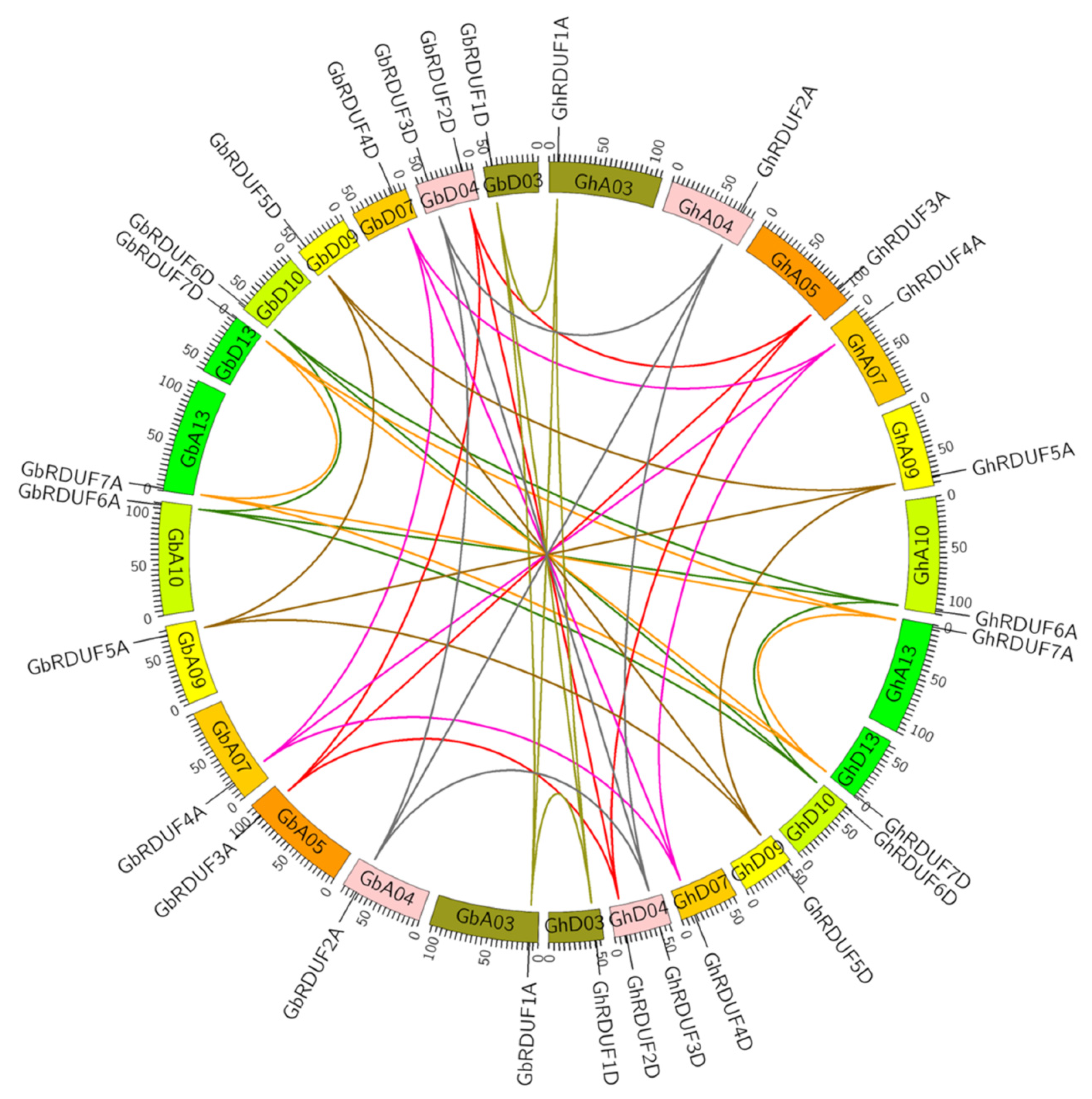
3.2. Chromosomal Localization and Gene Synteny Analysis of RDUF Genes
3.3. Conserved Structure and Domains in GhRDUF Proteins
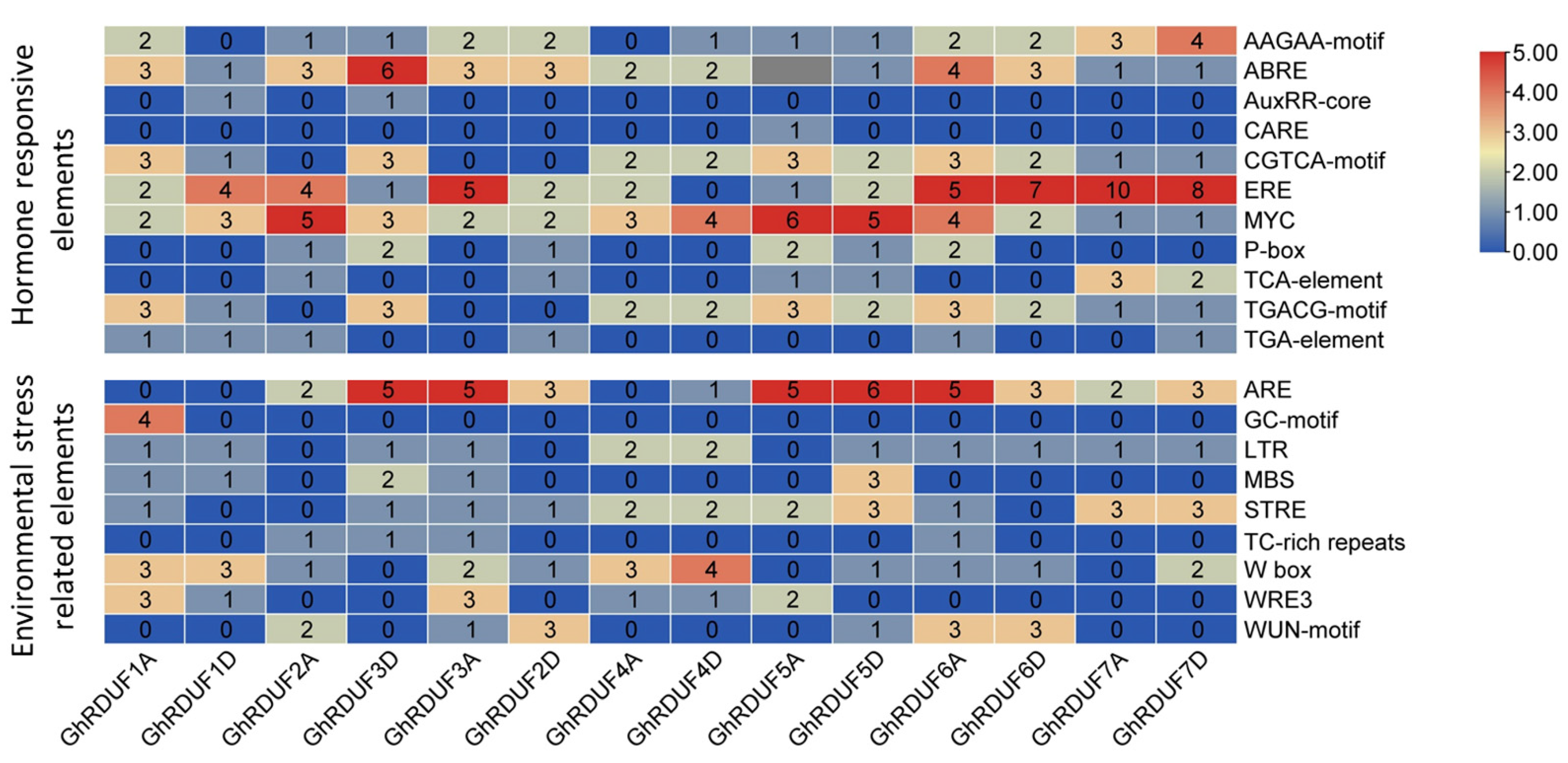
3.4. Cis-Elements in the GhRDUFs Promoter and the Targeting TFs
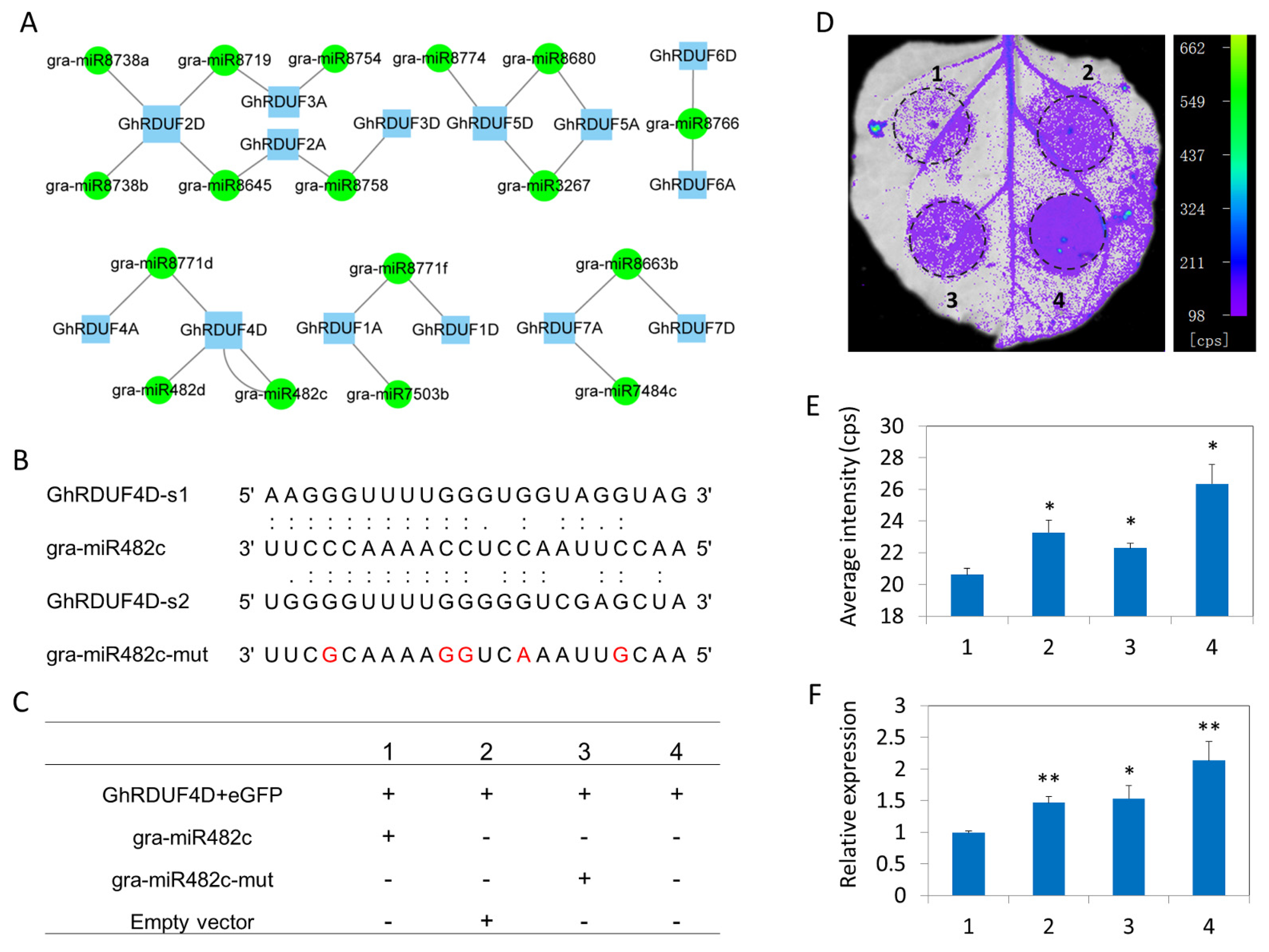
3.5. GhRDUF Targeting miRNAs Predict the Regulatory Network of GhRDUF4D and gra-miR482c
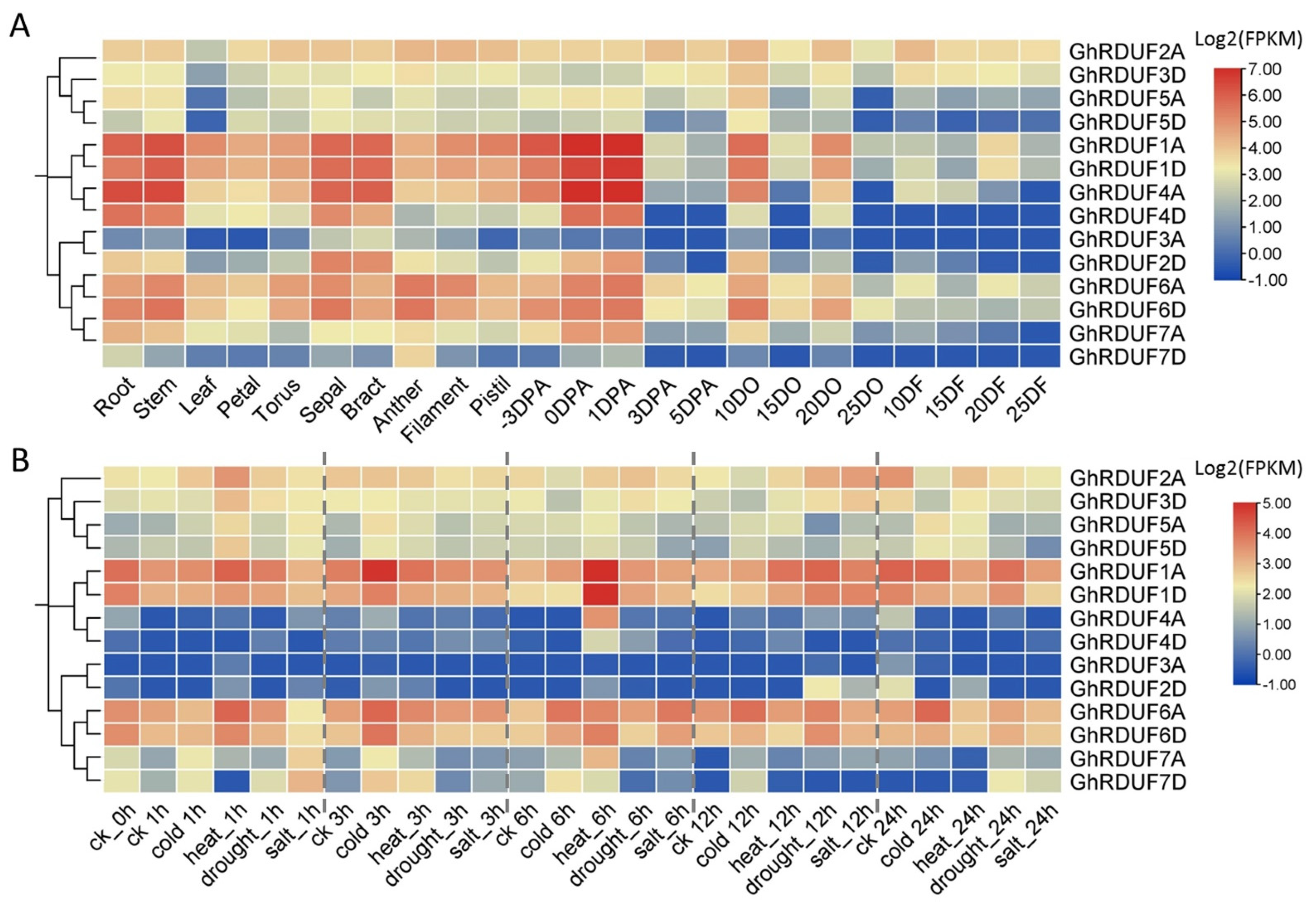
3.6. Expression Profiling of GhRDUF Genes in Upland Cotton
3.7. Overexpression of GhRDUF4D Enhanced the V. dahliae Resistance in Arabidopsis
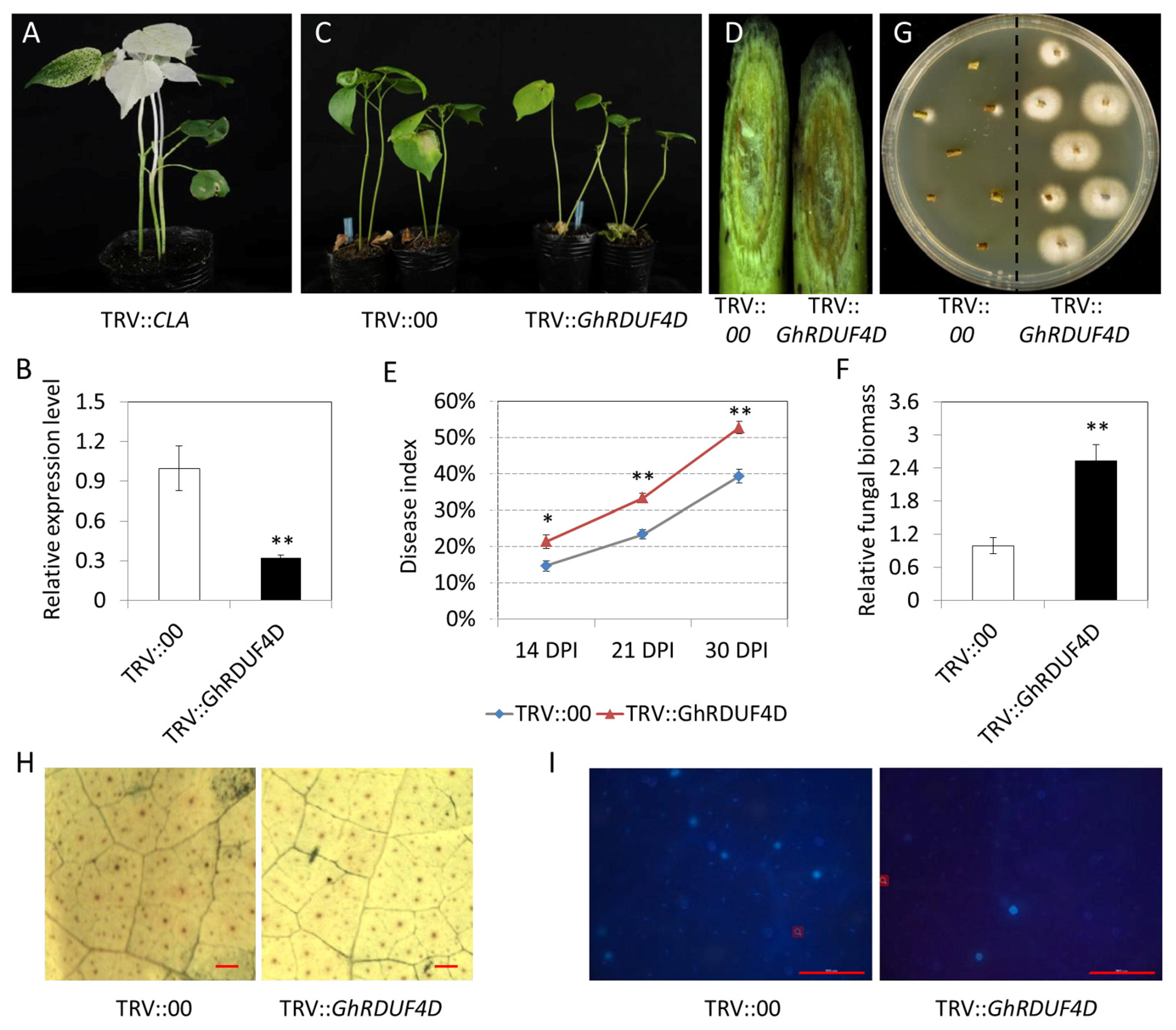
3.8. Knockdown of GhRDUF4D Compromise V. dahliae Resistance in Upland Cotton
4. Discussion
4.1. Evolution and Functional Diversification of GhRDUF Genes
4.2. RDUF Function in Plants Adapting to Abiotic Stress
4.3. GhRDUF4D Participates in Resistance to V. dahliae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Wu, Z.; Percy, R.G.; Bai, M.; Li, Y.; Frelichowski, J.E.; Hu, J.; Wang, K.; Yu, J.Z.; Zhu, Y. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 2020, 52, 516–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.L.; Liang, S.; Wang, H.Y.; Han, L.B.; Wang, F.X.; Cheng, H.Q.; Wu, X.M.; Qu, Z.L.; Wu, J.H.; Xia, G.X. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol. Plant 2015, 8, 399–411. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Zhu, L.; Zhang, X.; Guan, Q.; Xiao, S.; Min, L.; Zhang, X. GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol. 2018, 178, 876–889. [Google Scholar] [CrossRef] [Green Version]
- Shaban, M.; Miao, Y.; Ullah, A.; Khan, A.Q.; Menghwar, H.; Khan, A.H.; Ahmed, M.M.; Tabassum, M.A.; Zhu, L. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 2018, 125, 193–204. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, H.L.; Wang, X.N.; Yang, Y.H.; Zhang, C.J.; Zhu, H.Q.; Shi, L.; Tang, C.M.; Zhao, M.W. Dynamic infection of Verticillium dahliae in upland cotton. Plant Biol. 2020, 22, 90–105. [Google Scholar] [CrossRef]
- Long, L.; Xu, F.C.; Zhao, J.R.; Li, B.; Xu, L.; Gao, W. GbMPK3 overexpression increases cotton sensitivity to Verticillium dahliae by regulating salicylic acid signaling. Plant Sci. 2020, 292, 110374. [Google Scholar] [CrossRef]
- Liu, N.; Sun, Y.; Pei, Y.; Zhang, X.; Wang, P.; Li, X.; Li, F.; Hou, Y. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wu, L.; Wang, X.; Chen, B.; Zhao, J.; Cui, J.; Li, Z.; Yang, J.; Wu, L.; Wu, J.; et al. The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 2019, 20, 309–322. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Jin, Y.; Gong, Q.; Li, Z.; Zhao, L.; Han, X.; Zhou, J.; Li, F.; Yang, Z. Mechanismal analysis of resistance to Verticillium dahliae in upland cotton conferred by overexpression of RPL18A-6 (Ribosomal Protein L18A-6). Ind. Crop. Prod. 2019, 141, 111742. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, L.; Wassan, G.M.; He, X.; Shaban, M.; Zhang, L.; Zhu, L.; Zhang, X. GbSOBIR1 confers Verticillium wilt resistance by phosphorylating the transcriptional factor GbbHLH171 in Gossypium barbadense. Plant Biotechnol. J. 2019, 17, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Zhang, Y.; Yang, J.; Zhang, M.; Ma, Q.; Wang, X.; Ma, Z. The G-protein a subunit GhGPA positively regulates Gossypium hirsutum resistance to Verticillium dahliae via induction of SA and JA signaling pathways and ROS accumulation. Crop J. 2020. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Kraft, E.; Stone, S.L.; Ma, L.; Su, N.; Gao, Y.; Lau, O.S.; Deng, X.W.; Callis, J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 2005, 139, 1597–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Dye, B.T.; Schulman, B.A. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 131–150. [Google Scholar] [CrossRef]
- Hunter, T. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 2007, 28, 730–738. [Google Scholar] [CrossRef]
- Kosarev, P.; Mayer, K.F.; Hardtke, C.S. Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 2002, 3, research0016. [Google Scholar] [CrossRef]
- Freemont, P.S. The RING finger: A novel protein sequence motif related to the zinc finger. Ann. N. Y. Acad. Sci. 1993, 684, 174–192. [Google Scholar] [CrossRef]
- Callis, J. The ubiquitination machinery of the ubiquitin system. Arabidopsis B 2014, 12, e0174. [Google Scholar] [CrossRef] [Green Version]
- Lorick, K.L.; Jensen, J.P.; Fang, S.; Ong, A.M.; Hatakeyama, S.; Weissman, A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 1999, 96, 11364–11369. [Google Scholar] [CrossRef] [Green Version]
- Freemont, P.S.; Hanson, I.M.; Trowsdale, J. A novel cysteine-rich sequence motif. Cell 1991, 64, 483–484. [Google Scholar] [CrossRef]
- Lovering, R.; Hanson, I.M.; Borden, K.L.; Martin, S.; O’Reilly, N.J.; Evan, G.I.; Rahman, D.; Pappin, D.J.; Trowsdale, J.; Freemont, P.S. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc. Natl. Acad. Sci. USA 1993, 90, 2112–2116. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Guo, Y.; Wu, C.; Yang, G.; Li, Y.; Zheng, C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008, 9, 44. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Li, M.Y.; Zhao, J.; Zhang, Y.C.; Xie, Q.J.; Chen, D.H. Genome-wide analysis of RING finger proteins in the smallest free-living photosynthetic eukaryote Ostreococus tauri. Mar. Genomics 2016, 26, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Alam, I.; Yang, Y.Q.; Wang, Y.; Zhu, M.L.; Wang, H.B.; Chalhoub, B.; Lu, Y.H. Genome-wide identification, evolution and expression analysis of RING finger protein genes in Brassica rapa. Sci. Rep. 2017, 7, 40690. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, C.; Li, Y.; Zheng, N.; Chen, H.; Zhao, Q.; Gao, T.; Guo, H.; Xie, Q. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 2007, 19, 1912–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Cui, F.; Wu, Y.; Lou, L.; Liu, L.; Tian, M.; Ning, Y.; Shu, K.; Tang, S.; Xie, Q. The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 2015, 27, 214–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, M.Y.; Zeng, N.Y.; Tong, S.W.; Li, F.W.; Zhao, K.J.; Zhang, Q.; Sun, S.S.; Lam, H.M. Expression of a RING-HC protein from rice improves resistance to Pseudomonas syringae pv. tomato DC3000 in transgenic Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 4147–4159. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.S.; Pi, L.Y.; Chen, X.; Chakrabarty, P.K.; Jiang, J.; De Leon, A.L.; Liu, G.Z.; Li, L.; Benny, U.; Oard, J.; et al. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 2006, 18, 3635–3646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Jin, L.; Huang, X.; Geng, Y.; Li, F.; Qin, Q.; Wang, R.; Ji, S.; Zhao, S.; Xie, Q.I.; et al. OsRFPH2-10, a ring-H2 finger E3 ubiquitin ligase, is involved in rice antiviral defense in the early stages of rice dwarf virus infection. Mol. Plant 2014, 7, 1057–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Han, Y.; Zhao, Q.; Li, C.; Xie, Q.; Chong, K.; Xu, Y. The E3 ligase AtRDUF1 positively regulates salt stress responses in Arabidopsis thaliana. PLoS ONE 2013, 8, e71078. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Ryu, M.Y.; Kim, W.T. Suppression of Arabidopsis RING-DUF1117 E3 ubiquitin ligases, AtRDUF1 and AtRDUF2, reduces tolerance to ABA-mediated drought stress. Biochem. Biophys. Res. Commun. 2012, 420, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Wan, J.; Czechowski, T.; Udvardi, M.; Stacey, G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant Microbe Interact. 2007, 20, 900–911. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Huang, G.; He, S.; Yang, Z.; Sun, G.; Ma, X.; Li, N.; Zhang, X.; Sun, J.; Liu, M.; et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018, 50, 796–802. [Google Scholar] [CrossRef]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Jung, S.; Cheng, C.H.; Ficklin, S.P.; Lee, T.; Zheng, P.; Jones, D.; Percy, R.G.; Main, D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014, 42, D1229–D1236. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Liu, S.; Sun, R.; Zhang, X.; Feng, Z.; Wei, F.; Zhao, L.; Zhang, Y.; Zhu, L.; Feng, H.; Zhu, H. Genome-wide analysis of OPR family genes in cotton identified a role for GhOPR9 in Verticillium dahliae resistance. Genes 2020, 11, 1134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Guo, A.; Wang, Y.; Hua, J. Evolution of PEPC gene family in Gossypium reveals functional diversification and GhPEPC genes responding to abiotic stresses. Gene 2019, 698, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Guo, M.; Wang, X.; Guo, Z.; Xu, Z.; Xu, L.; Zhao, H.; Sun, H.; Yan, C.; Yi, K. Two RING-finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Mol. Plant 2019, 12, 1060–1074. [Google Scholar] [CrossRef]
- Cao, H.; Li, X.; Wang, Z.; Ding, M.; Sun, Y.; Dong, F.; Chen, F.; Liu, L.; Doughty, J.; Li, Y.; et al. Histone H2B monoubiquitination mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 is involved in anther development by regulating tapetum degradation-related genes in rice. Plant Physiol. 2015, 168, 1389–1405. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wang, W.; Wu, Y.; Li, Q.; Zhang, G.; Shi, R.; Yang, J.; Wang, Y.; Wang, W. The involvement of wheat U-box E3 ubiquitin ligase TaPUB1 in salt stress tolerance. J. Integr. Plant Biol. 2020, 62, 631–651. [Google Scholar] [CrossRef]
- Min, H.J.; Jung, Y.J.; Kang, B.G.; Kim, W.T. CaPUB1, a hot pepper U-box E3 ubiquitin ligase, confers enhanced cold stress tolerance and decreased drought stress tolerance in transgenic rice (Oryza sativa L.). Mol. Cells 2016, 39, 250–257. [Google Scholar]
- Peng, L.; Wan, X.; Huang, K.; Pei, L.; Xiong, J.; Li, X.; Wang, J. AtPUB48 E3 ligase plays a crucial role in the thermotolerance of Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 509, 281–286. [Google Scholar] [CrossRef]
- Park, Y.C.; Lim, S.D.; Moon, J.C.; Jang, C.S. A rice really interesting new gene H2-type E3 ligase, OsSIRH2-14, enhances salinity tolerance via ubiquitin/26S proteasome-mediated degradation of salt-related proteins. Plant Cell Environ. 2019, 42, 3061–3076. [Google Scholar] [CrossRef]
- Inzé, A.; Vanderauwera, S.; Hoeberichts, F.A.; Vandorpe, M.; Van Gaever, T.; Van Breusegem, F. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 2012, 35, 308–320. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Li, X.P.; Zhang, T.Q.; Wang, Y.Y.; Wang, C.; Gao, C.Q. Overexpression of ThMYB8 mediates salt stress tolerance by directly activating stress-responsive gene expression. Plant Sci. 2021, 302, 110668. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Li, H.G.; Wang, J.J.; Su, Y.; Wang, H.L.; Feng, C.H.; Yang, Y.; Niu, M.X.; Liu, C.; Yin, W.; et al. PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol. J. 2019, 17, 2169–2183. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Yan, M.; Li, L.; He, J.; Zhou, S.; Li, Z.; Niu, C.; Bao, C.; Zhi, F.; Ma, F.; et al. The apple DNA-binding one zinc-finger protein MdDof54 promotes drought resistance. Hortic. Res. 2020, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Wang, R.; Shi, X.; Zhou, X.; Wang, G.L. A layered defense strategy mediated by rice E3 ubiquitin ligases against diverse pathogens. Mol. Plant 2016, 9, 1096–1098. [Google Scholar] [CrossRef] [Green Version]
- Berrocal-Lobo, M.; Stone, S.; Yang, X.; Antico, J.; Callis, J.; Ramonell, K.M.; Somerville, S. ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses. PLoS ONE 2010, 5, e14426. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Guo, T.; Lefebvre, M.; Scaglione, S.; Antico, C.J.; Jing, T.; Yang, X.; Shan, W.; Ramonell, K.M. Expression and regulation of ATL9, an E3 ubiquitin ligase involved in plant defense. PLoS ONE 2017, 12, e0188458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kachewar, N.R.; Gupta, V.; Ranjan, A.; Patel, H.K.; Sonti, R.V. Overexpression of OsPUB41, a Rice E3 ubiquitin ligase induced by cell wall degrading enzymes, enhances immune responses in rice and Arabidopsis. BMC Plant Biol. 2019, 19, 530. [Google Scholar] [CrossRef] [PubMed]
- Molnár, G.; Bancoş, S.; Nagy, F.; Szekeres, M. Characterisation of BRH1, a brassinosteroid-responsive RING-H2 gene from Arabidopsis thaliana. Planta 2002, 215, 127–133. [Google Scholar] [CrossRef]
- Serrano, M.; Guzmán, P. Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 2004, 167, 919–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, T.; Liu, S.; Zhang, Z.; Sun, L.; He, X.; Lindsey, K.; Zhu, L.; Zhang, X. GhCyP3 improves the resistance of cotton to Verticillium dahliae by inhibiting the E3 ubiquitin ligase activity of GhPUB17. Plant Mol. Biol. 2019, 99, 379–393. [Google Scholar] [CrossRef] [PubMed]
- González, V.M.; Müller, S.; Baulcombe, D.; Puigdomènech, P. Evolution of NBS-LRR gene copies among dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs. Mol. Plant 2015, 8, 329–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canto-Pastor, A.; Santos, B.A.M.C.; Valli, A.A.; Summers, W.; Schornack, S.; Baulcombe, D.C. Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. USA 2019, 116, 2755–2760. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Mu, X.; Liu, C.; Cai, J.; Shi, K.; Zhu, W.; Yang, Q. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015, 57, 1078–1088. [Google Scholar] [CrossRef]
- de Vries, S.; Kukuk, A.; von Dahlen, J.K.; Schnake, A.; Kloesges, T.; Rose, L.E. Expression profiling across wild and cultivated tomatoes supports the relevance of early miR482/2118 suppression for Phytophthora resistance. Proc. Biol. Sci. 2018, 285, 20172560. [Google Scholar] [CrossRef] [Green Version]
- Jiang, N.; Meng, J.; Cui, J.; Sun, G.; Luan, Y. Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. Hortic. Res. 2018, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Q.H.; Fan, L.; Liu, Y.; Xu, H.; Llewellyn, D.; Wilson, I. miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE 2013, 8, e84390. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Name | Gene Locus ID | Nucleic Acid | Amino Acid | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Location | CDS (bp) | Exons | Size (aa) | Mw (Da) | pI | Formula | Subcellular Location | ||
| GaRDUF1 | Ga01G2247.1 | Chr01: 104114537-104114537 | 1095 | 1 | 364 | 40,145.64 | 6.72 | C1709H2699N529O553S20 | cyto: 5, nucl: 3, chlo: 2 |
| GaRDUF2 | Ga04G0720.1 | Chr04: 15879489-15880598 | 1110 | 1 | 369 | 41,080.80 | 8.36 | C1780H2753N547O545S17 | nucl: 8, mito: 4, chlo: 1 |
| GaRDUF3 | Ga07G1079.1 | Chr07: 15730492-15731585 | 1023 | 2 | 340 | 38,340.47 | 5.96 | C1655H2554N498O525S16 | chlo: 6, mito: 4, nucl: 2 |
| GaRDUF4 | Ga09G1752.1 | Chr09: 75411415-75412479 | 1065 | 1 | 354 | 39,662.52 | 9.27 | C1720H2690N536O517S16 | nucl: 7, chlo: 3, mito: 3 |
| GaRDUF5 | Ga10G0274.1 | Chr10: 3699993-3701000 | 1008 | 1 | 335 | 36,635.1 | 6.15 | C1588H2470N474O489S19 | chlo: 10, nucl: 2, cyto: 1 |
| GaRDUF6 | Ga13G0217.1 | Chr13: 2143604-2144689 | 1086 | 1 | 361 | 39,945.89 | 7.03 | C1729H2687N519O529S23 | nucl: 14 |
| GrRDUF1 | Gorai.001G111900.1 | Chr01: 13043565-13045611 | 1099 | 1 | 361 | 40,055.51 | 5.77 | C1728H2685N513O552S18 | chlo: 5, mito: 5, nucl: 1 |
| GrRDUF2 | Gorai.003G129300.1 | Chr03: 38196500-38198583 | 1114 | 1 | 366 | 40,318.81 | 6.72 | C1716H2710N530O557S20 | nucl: 4, cyto: 3, mito: 3 |
| GrRDUF3 | Gorai.006G171100.1 | Chr06: 43050323-43052612 | 1078 | 1 | 354 | 39,714.48 | 9.02 | C1718H2682N534O522S17 | nucl: 10, chlo: 2, mito: 2 |
| GrRDUF4 | Gorai.011G267000.1 | Chr11: 59881340-59882767 | 953 | 2 | 313 | 34,156.21 | 8.39 | C1485H2298N448O453S15 | chlo: 9, nucl: 2, mito: 2 |
| GrRDUF5 | Gorai.012G071400.1 | Chr12: 10539979-10541968 | 1011 | 1 | 332 | 36,698.02 | 7.59 | C1580H2471N491O488S17 | nucl: 8, mito: 3, chlo: 2 |
| GrRDUF6 | Gorai.012G105100.1 | Chr12: 23596133-23598256 | 1123 | 1 | 369 | 41,122.88 | 8.36 | C1783H2759N547O545S17 | nucl: 8, mito: 4, chlo: 2 |
| GrRDUF7 | Gorai.013G021800.1 | Chr13: 1532332-1533783 | 1102 | 1 | 362 | 40,120.10 | 8.02 | C1733H2701N527O529S23 | nucl: 14 |
| GhRDUF1A | GH_A03G0566 | A03: 8775627-8776721 | 1116 | 1 | 364 | 40,144.7 | 6.72 | C1711H2704N528O552S20 | cyto: 5, nucl: 3, chlo: 2 |
| GhRDUF2A | GH_A04G1023 | A04: 72823528-72824637 | 1132 | 1 | 369 | 41,114.82 | 8.36 | C1783H2751N547O545S17 | nucl: 9, mito: 4, chlo: 1 |
| GhRDUF3A | GH_A05G3679 | A05: 96863898-96864839 | 960 | 1 | 313 | 34,110.2 | 7.59 | C1470H2308N450O457S16 | nucl: 7, mito: 4, chlo: 2 |
| GhRDUF4A | GH_A07G1092 | A07: 16689279-16690247 | 988 | 1 | 322 | 36,130.2 | 5.25 | C1566H2418N456O497S17 | chlo: 6, mito: 4, nucl: 2 |
| GhRDUF5A | GH_A09G1709 | A09: 74061366-74062430 | 1086 | 1 | 354 | 39,765.64 | 9.25 | C1727H2695N537O517S16 | mito: 7, nucl: 3.5, chlo: 3 |
| GhRDUF6A | GH_A10G2448 | A10: 111936421-111937428 | 1028 | 1 | 335 | 36,635.1 | 6.15 | C1588H2470N474O489S19 | chlo: 10, nucl: 2, cyto: 1 |
| GhRDUF7A | GH_A13G0199 | A13: 2147580-2148665 | 1107 | 1 | 361 | 39,948.85 | 7.03 | C1727H2684N520O530S23 | nucl: 14 |
| GhRDUF1D | GH_D03G1398 | D03: 45757090-45758175 | 1107 | 1 | 361 | 39,809.38 | 6.72 | C1696H2687N525O546S20 | cyto: 4, chlo: 3, nucl: 3 |
| GhRDUF2D | GH_D04G0664 | D04: 11134024-11135037 | 1034 | 1 | 337 | 37,207.58 | 8.34 | C1602H2506N498O495S17 | nucl: 9, mito: 3, chlo: 2 |
| GhRDUF3D | GH_D04G1354 | D04: 44739659-44740768 | 1132 | 1 | 369 | 41,202.03 | 8.56 | C1788H2768N550O543S17 | nucl: 9, mito: 3, chlo: 2 |
| GhRDUF4D | GH_D07G1077 | D07: 13116176-13117261 | 1107 | 1 | 361 | 40,221.74 | 5.82 | C1737H2699N515O553S18 | chlo: 6, mito: 4, nucl: 2 |
| GhRDUF5D | GH_D09G1658 | D09: 43682720-43683784 | 1086 | 1 | 354 | 39,723.49 | 8.89 | C1720H2681N533O522S17 | nucl: 9, mito: 3, chlo: 1 |
| GhRDUF6D | GH_D10G2556 | D10: 64049541-64050551 | 1031 | 1 | 336 | 36,686.15 | 6.89 | C1590H2471N477O488S19 | chlo: 8, nucl: 3, mito: 2 |
| GhRDUF7D | GH_D13G0196 | D13: 1703107-1704195 | 1110 | 1 | 362 | 40,142.16 | 8.02 | C1735H2703N529O527S23 | nucl: 14 |
| GbRDUF1A | GB_A03G0554 | A03: 8482495-8483589 | 1116 | 1 | 364 | 40,144.7 | 6.72 | C1711H2704N528O552S20 | cyto: 5, nucl: 3, chlo: 2 |
| GbRDUF2A | GB_A04G1063 | A04: 67240547-67241656 | 1132 | 1 | 369 | 41,080.8 | 8.36 | C1780H2753N547O545S17 | nucl: 8, mito: 4, chlo: 1 |
| GbRDUF3A | GB_A05G3770 | A05: 94125157-94126098 | 960 | 1 | 313 | 34,134.26 | 7.57 | C1474H2312N448O457S16 | nucl: 6, mito: 5, chlo: 2 |
| GbRDUF4A | GB_A07G1079 | A07: 17016234-17017202 | 988 | 1 | 322 | 36,188.24 | 5.17 | C1568H2420N456O499S17 | chlo: 6, mito: 4, nucl: 2 |
| GbRDUF5A | GB_A09G1832 | A09: 70138025-70139089 | 1086 | 1 | 354 | 39,716.57 | 9.27 | C1722H2692N538O517S16 | mito: 7, nucl: 3.5, chlo: 3 |
| GbRDUF6A | GB_A10G2617 | A10: 108379933-108380940 | 1028 | 1 | 335 | 36,587.06 | 6.15 | C1584H2470N474O489S19 | chlo: 9, nucl: 2, mito: 2 |
| GbRDUF7A | GB_A13G0200 | A13: 2045827-2048345 | 1312 | 2 | 428 | 46,919.93 | 8.38 | C2038H3185N603O622S26 | nucl: 14 |
| GbRDUF1D | GB_D03G1418 | D03: 45690505-45691611 | 1129 | 1 | 368 | 40,430.98 | 6.72 | C1719H2726N532O559S20 | nucl: 4, chlo: 3, cyto: 3 |
| GbRDUF2D | GB_D04G0687 | D04: 10848147-10850667 | 1049 | 2 | 342 | 37,749.2 | 8.05 | C1625H2541N503O503S18 | nucl: 9, mito: 3, chlo: 2 |
| GbRDUF3D | GB_D04G1434 | D04: 45218042-45219151 | 1132 | 1 | 369 | 41,132.92 | 8.36 | C1785H2761N547O544S17 | nucl: 8, mito: 4, chlo: 2 |
| GbRDUF4D | GB_D07G1081 | D07: 13412269-13413354 | 1107 | 1 | 361 | 40,182.70 | 5.89 | C1734H2698N516O552S18 | chlo: 5, mito: 5, nucl: 1 |
| GbRDUF5D | GB_D09G1672 | D09: 45246636-45247700 | 1086 | 1 | 354 | 39,735.54 | 8.89 | C1722H2685N533O521S17 | nucl: 9, mito: 3, chlo: 2 |
| GbRDUF6D | GB_D10G2571 | D10: 63188105-63189115 | 1031 | 1 | 336 | 36,658.1 | 6.46 | C1589H2467N475O489S19 | chlo: 8, mito: 3, nucl: 2 |
| GbRDUF7D | GB_D13G0189 | D13: 1578513-1579601 | 1110 | 1 | 362 | 40,112.13 | 8.03 | C1734H2701N529O526S23 | nucl: 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.-P.; Shen, J.-L.; Li, W.-J.; Wu, N.; Chen, C.; Hou, Y.-X. Evolutionary and Characteristic Analysis of RING-DUF1117 E3 Ubiquitin Ligase Genes in Gossypium Discerning the Role of GhRDUF4D in Verticillium dahliae Resistance. Biomolecules 2021, 11, 1145. https://doi.org/10.3390/biom11081145
Zhao Y-P, Shen J-L, Li W-J, Wu N, Chen C, Hou Y-X. Evolutionary and Characteristic Analysis of RING-DUF1117 E3 Ubiquitin Ligase Genes in Gossypium Discerning the Role of GhRDUF4D in Verticillium dahliae Resistance. Biomolecules. 2021; 11(8):1145. https://doi.org/10.3390/biom11081145
Chicago/Turabian StyleZhao, Yan-Peng, Jian-Ling Shen, Wen-Jie Li, Na Wu, Chen Chen, and Yu-Xia Hou. 2021. "Evolutionary and Characteristic Analysis of RING-DUF1117 E3 Ubiquitin Ligase Genes in Gossypium Discerning the Role of GhRDUF4D in Verticillium dahliae Resistance" Biomolecules 11, no. 8: 1145. https://doi.org/10.3390/biom11081145
APA StyleZhao, Y.-P., Shen, J.-L., Li, W.-J., Wu, N., Chen, C., & Hou, Y.-X. (2021). Evolutionary and Characteristic Analysis of RING-DUF1117 E3 Ubiquitin Ligase Genes in Gossypium Discerning the Role of GhRDUF4D in Verticillium dahliae Resistance. Biomolecules, 11(8), 1145. https://doi.org/10.3390/biom11081145






