High Throughput miRNA Screening Identifies miR-574-3p Hyperproductive Effect in CHO Cells
Abstract
:1. Introduction
2. Material and Methods
2.1. CHO-EPO and CHO-ETN Stable Clones Preparation
2.2. miRNA Mimics Screening
2.3. Stable miRNAs Expression Clones Preparation
2.4. Real-Time PCR
2.5. EPO Purification
2.6. Western Blot Analysis
2.7. Iso-Electric Focusing
2.8. siRNA Transfections
3. Results
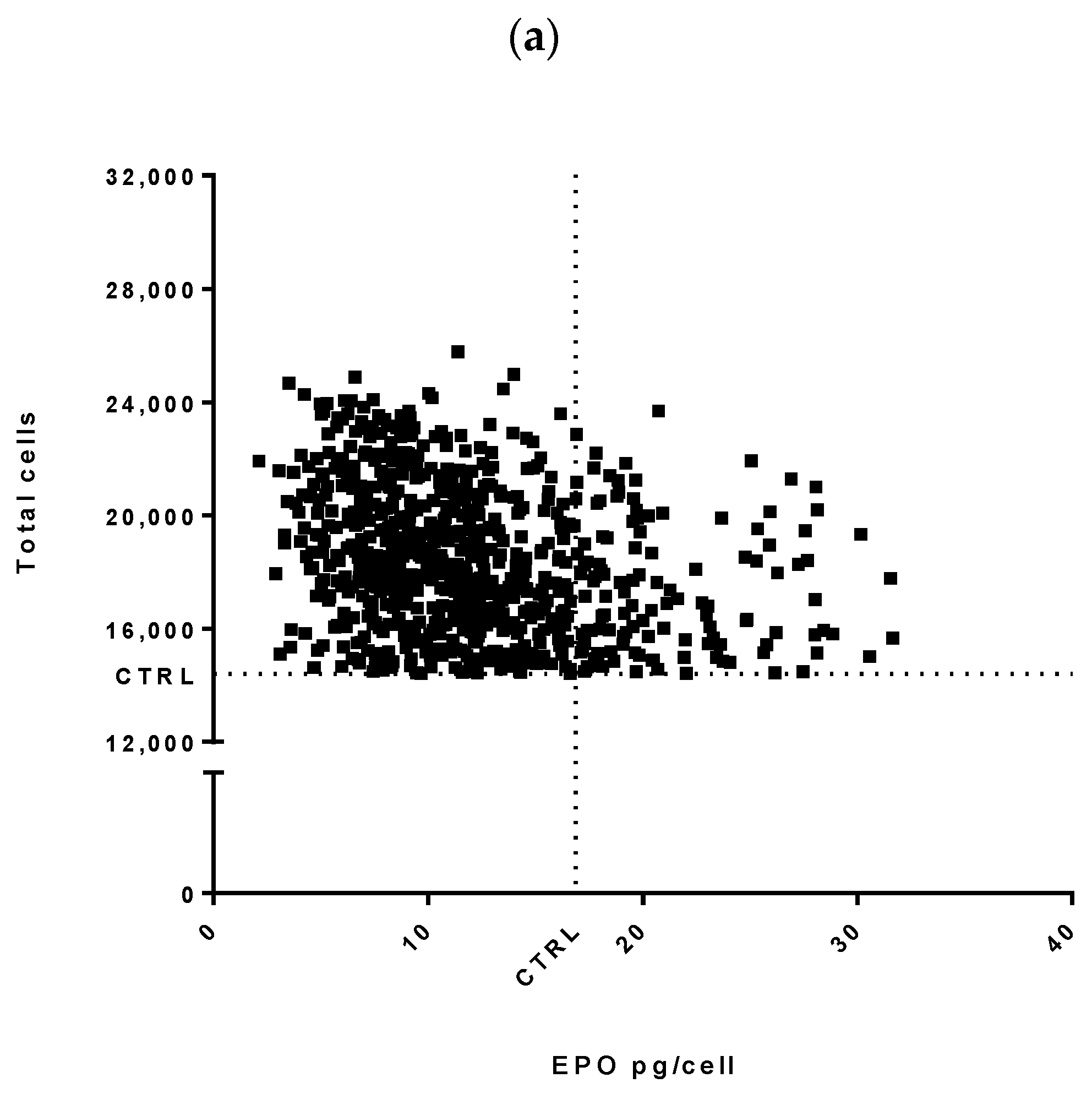
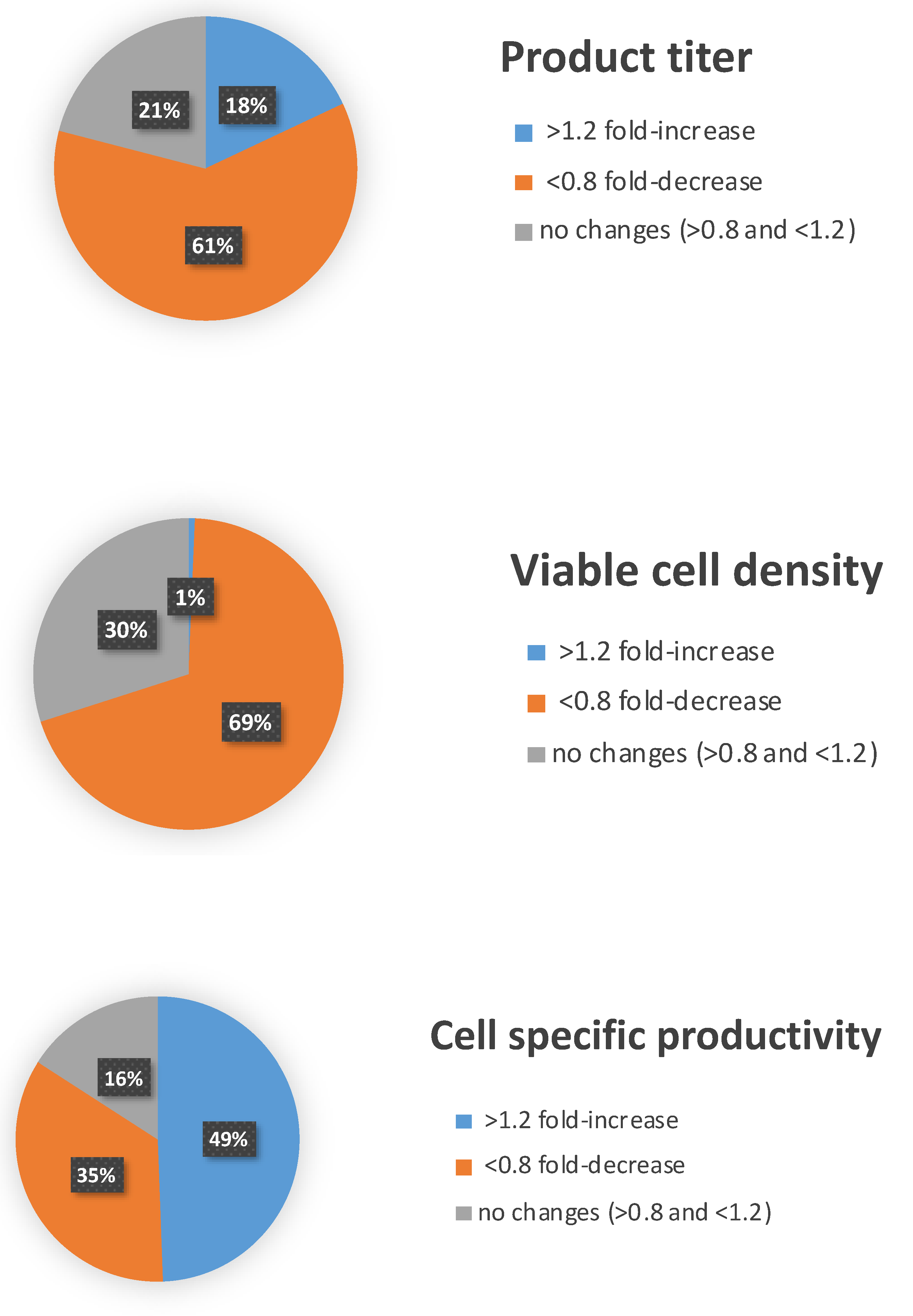
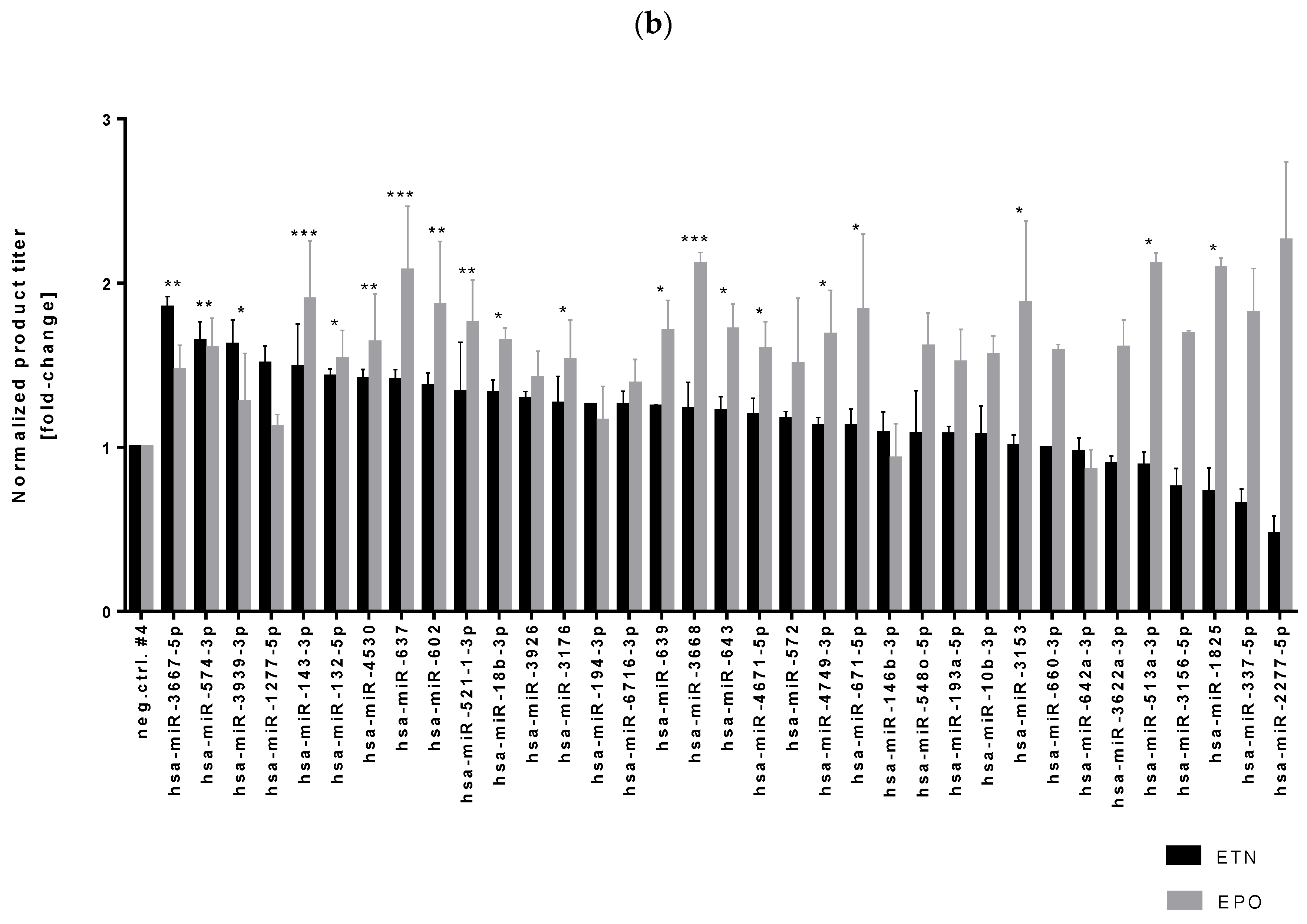
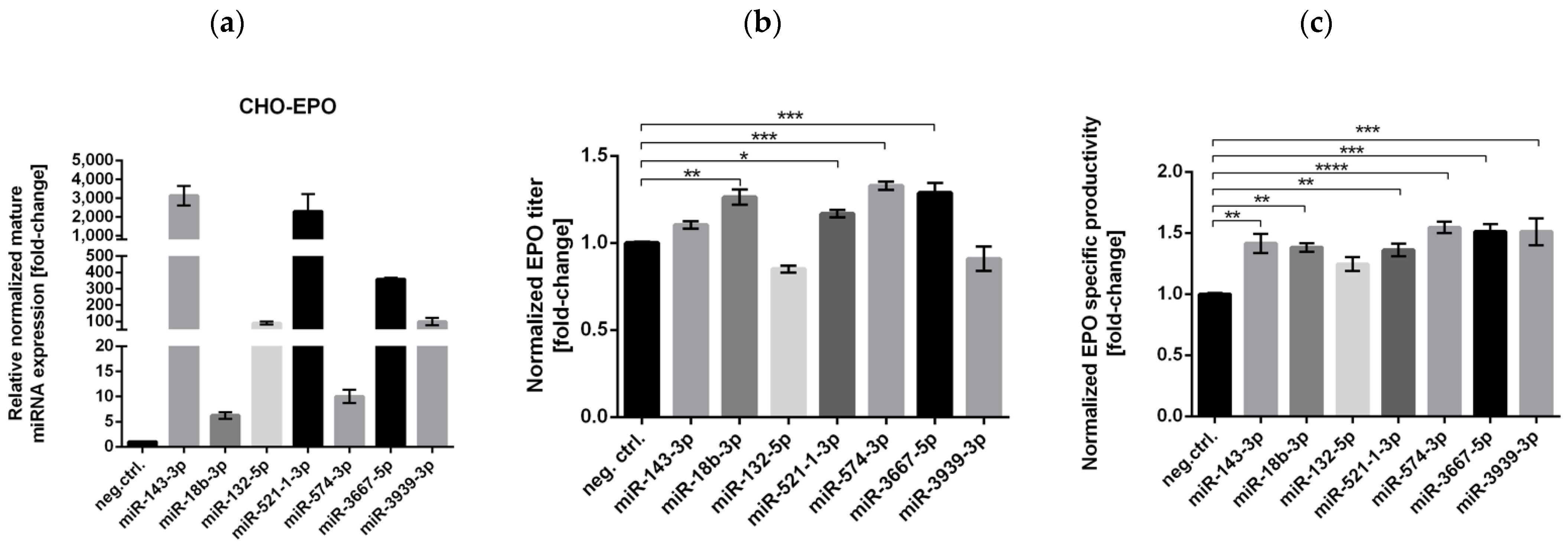
3.1. High-Throughput miRNA Mimics Screening in CHO-EPO and CHO-ETN Cells
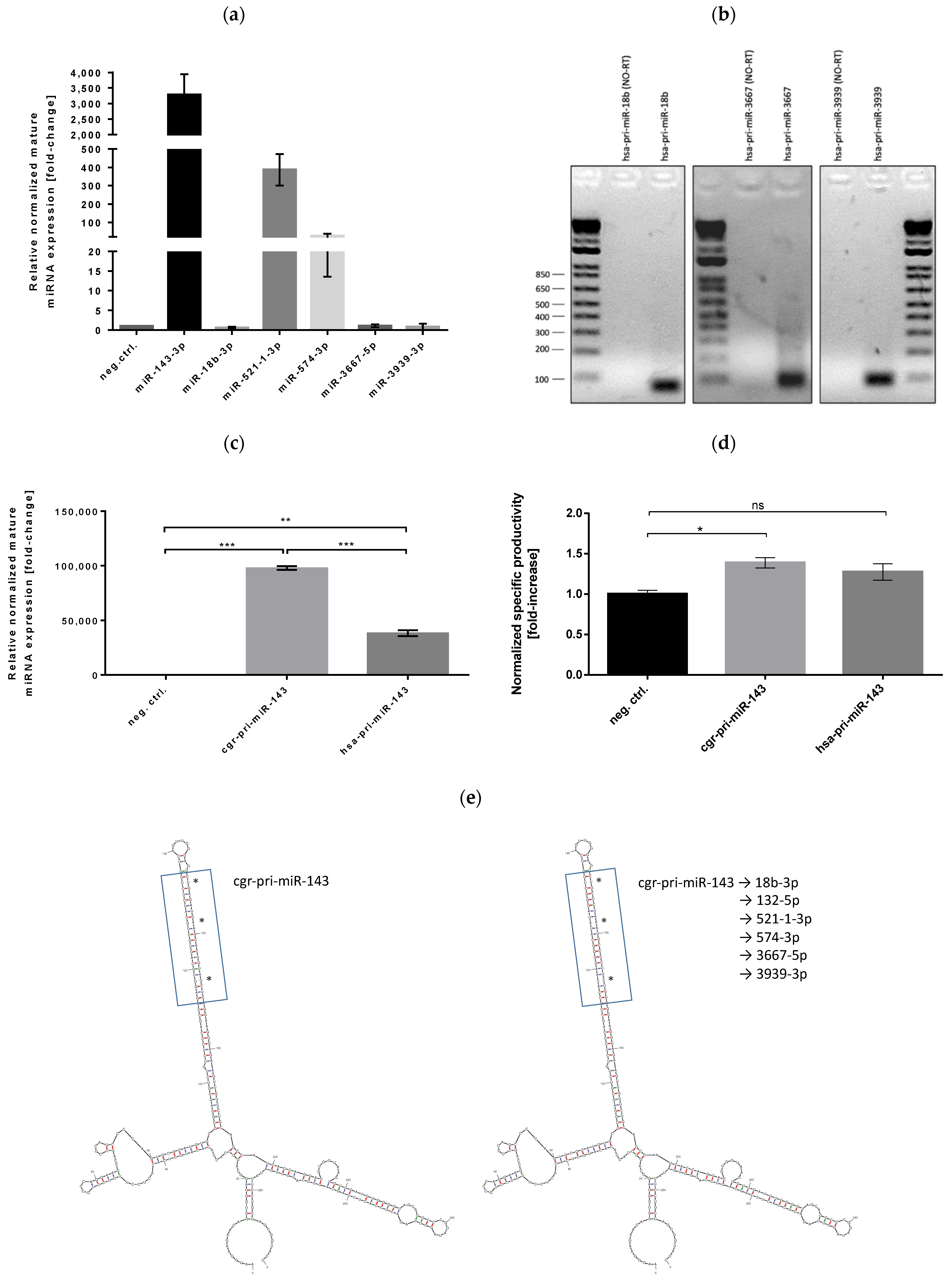
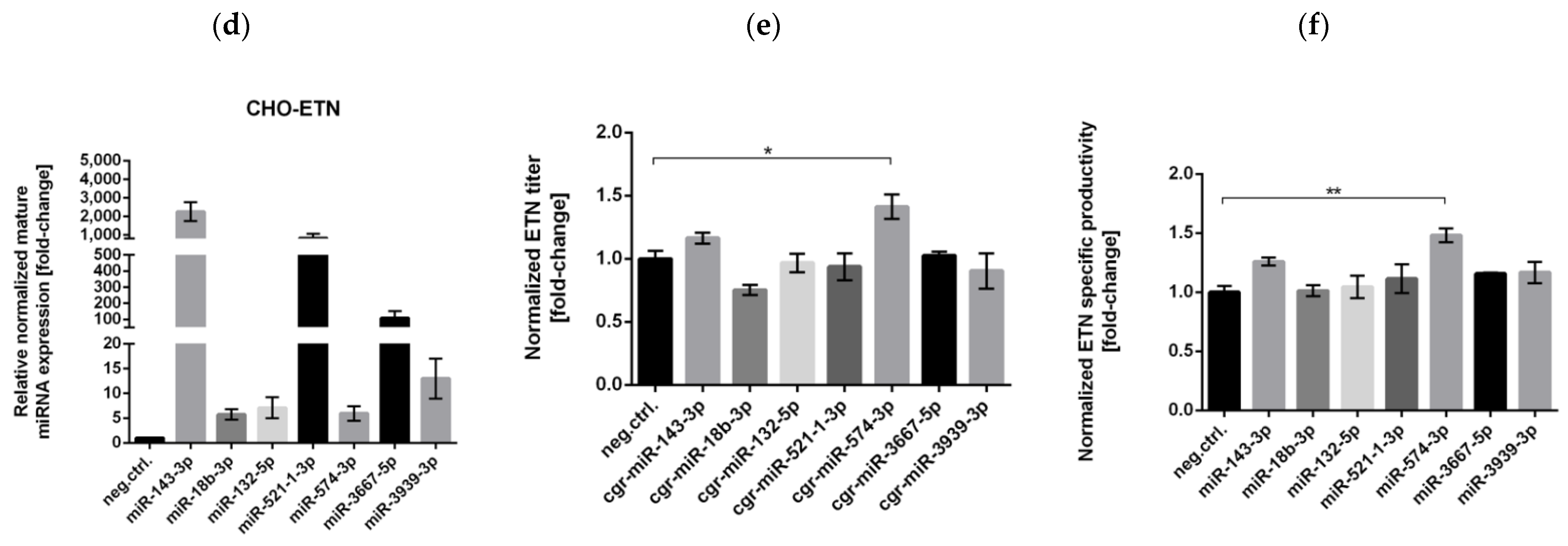
3.2. Stable Expression of Selected miRNAs in CHO Cells
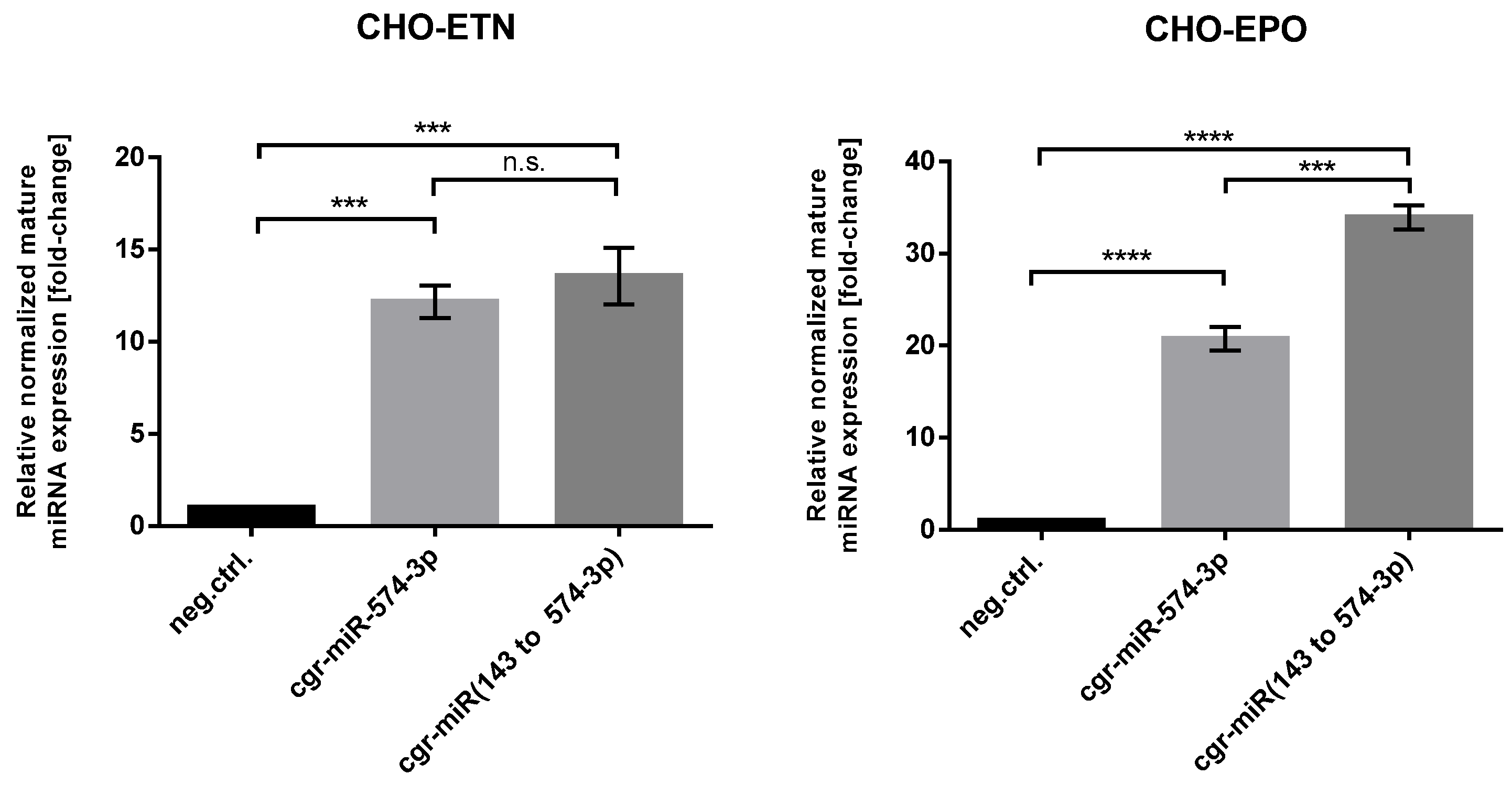
3.3. Successful Stable Expression of Seven miRNAs from Chinese Hamster pri-miR-143 Flanking Motifs in CHO Cells
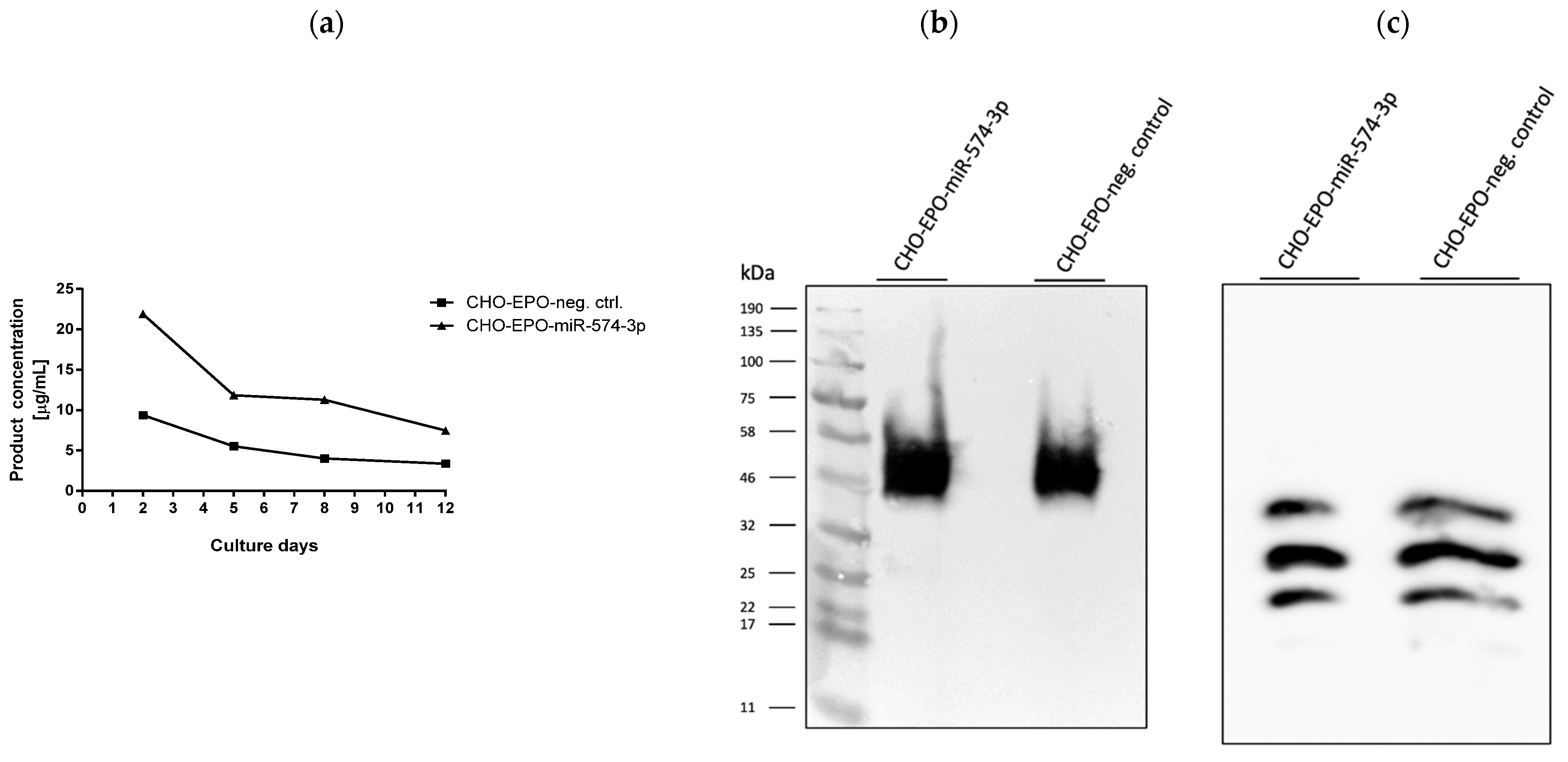
3.4. Stable Expression of miR-574-3p Increases the Level of Recombinant mRNA, Protein Production and the Anti-Apoptotic Effect in CHO Cells
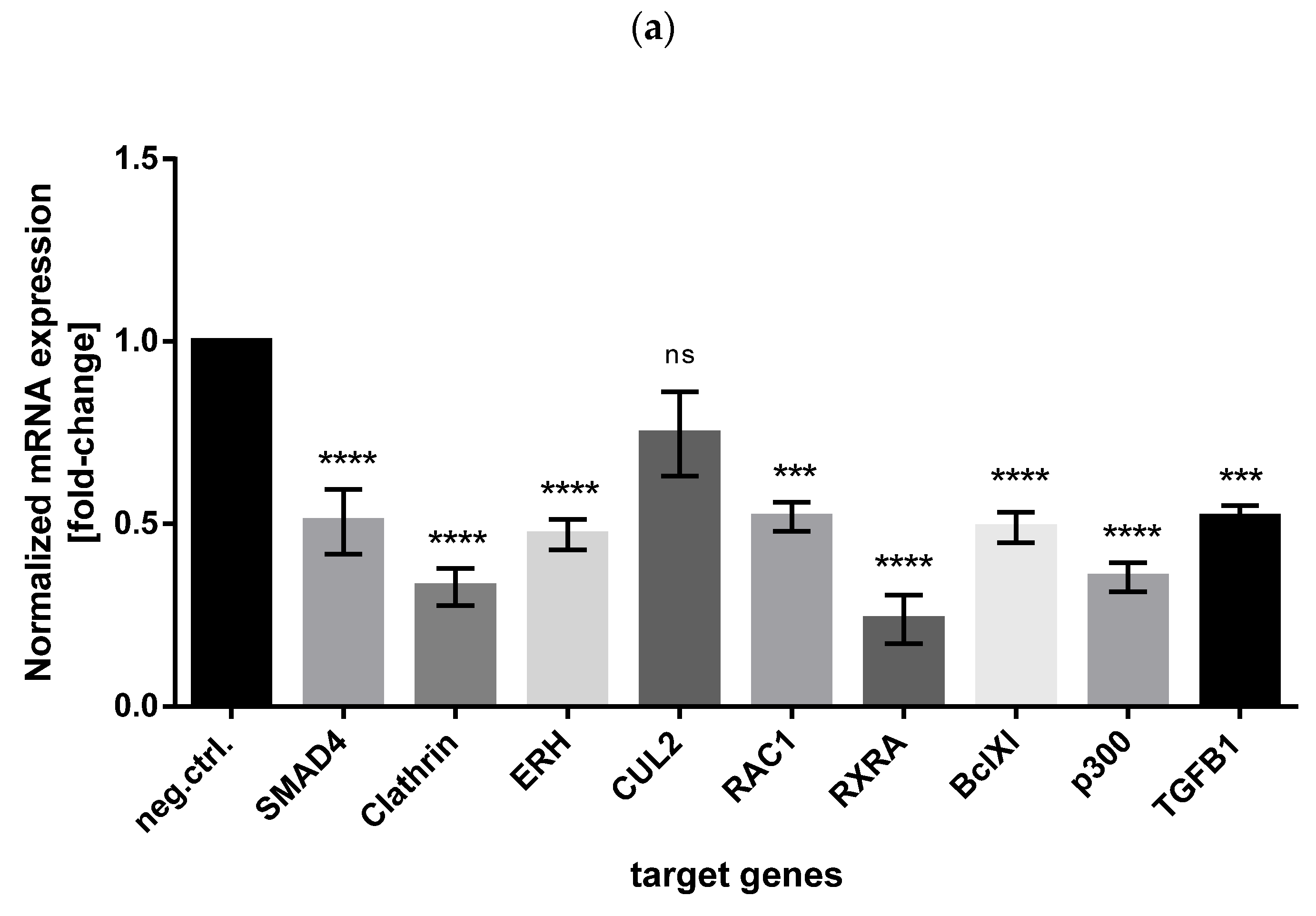
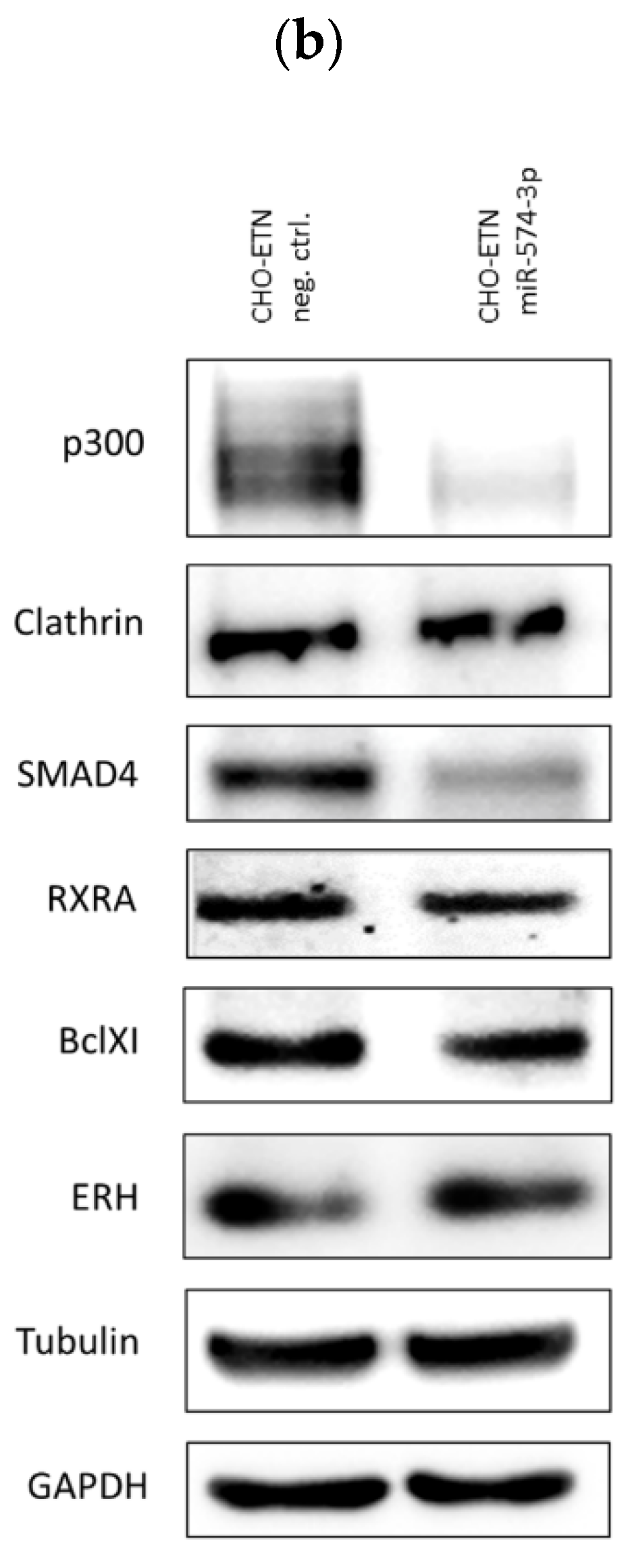
3.5. Validation of miR-574-3p Targets in CHO Cells
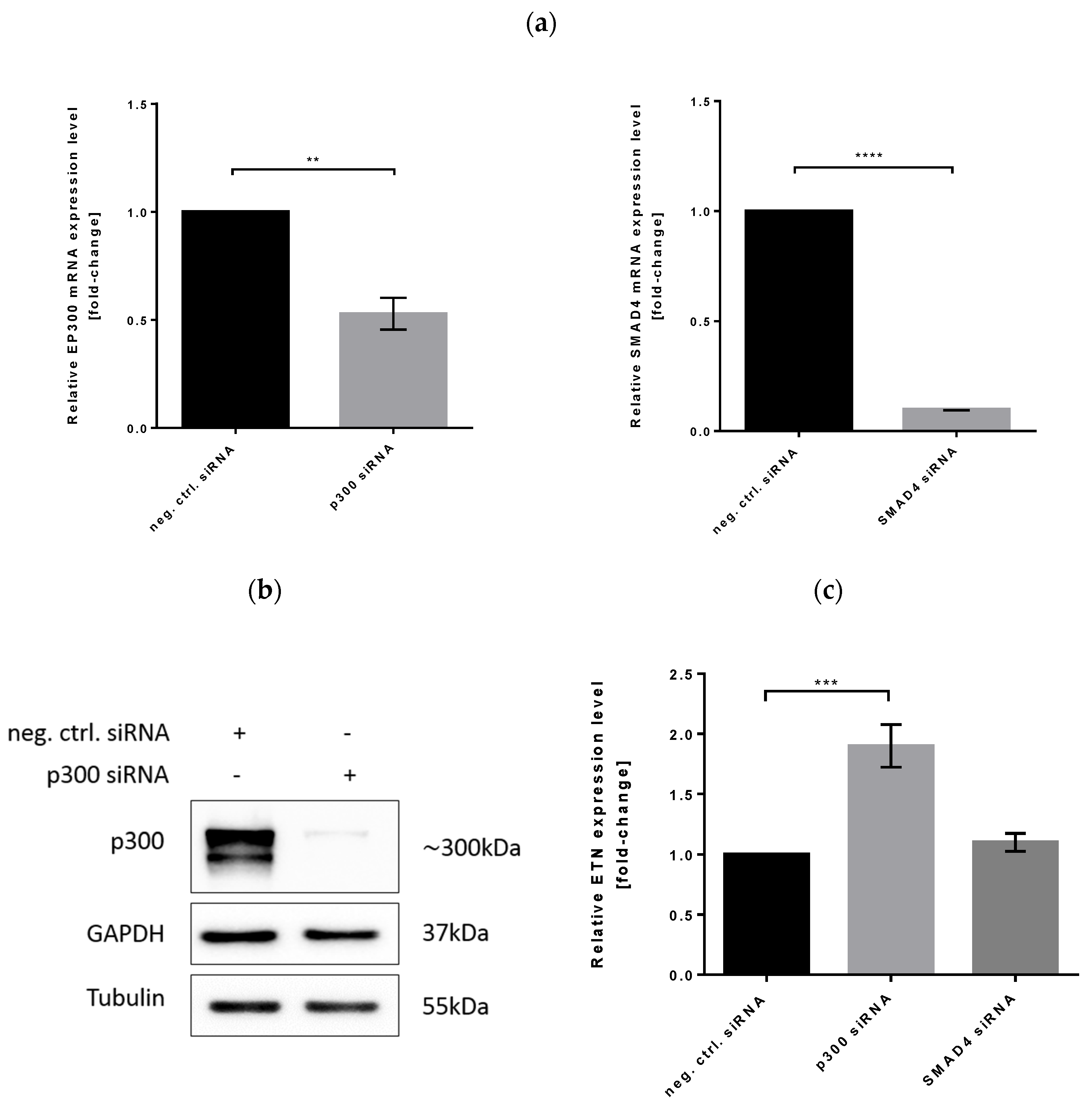
3.6. Effect of p300 and SMAD4 Knockdown on Heterologous Gene Expression
3.7. miR-574-3p Overexpression Induces p53 Downregulation in CHO Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Lalonde, M.E.; Durocher, Y. Therapeutic glycoprotein production in mammalian cells. J. Biotechnol. 2017, 251, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, Y.G.; Lee, G.M. CHO cells in biotechnology for production of recombinant proteins: Current state and further potential. Appl. Microbiol. Biotechnol. 2012, 93, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.; Yang, Y.; Ng, S. Advances in Mammalian Cell Line Development Technologies for Recombinant Protein Production. Pharmaceuticals 2013, 6, 579–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, L.M.; Dickson, A.J. Mammalian cell factories for efficient and stable protein expression. Curr. Opin. Biotechnol. 2006, 17, 381–386. [Google Scholar] [CrossRef]
- Butler, M.; Meneses-Acosta, A. Recent advances in technology supporting biopharmaceutical production from mammalian cells. Appl. Microbiol. Biotechnol. 2012, 96, 885–894. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the Root of miRNA-Mediated Gene Silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Bratkovič, T.; Glavan, G.; Štrukelj, B.; Živin, M.; Rogelj, B. Exploiting microRNAs for cell engineering and therapy. Biotechnol. Adv. 2012, 30, 753–765. [Google Scholar] [CrossRef]
- Xu, X.; Nagarajan, H.; Lewis, N.E.; Pan, S.; Cai, Z.; Liu, X.; Chen, W.; Xie, M.; Wang, W.; Hammond, S.; et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 2011, 29, 735–741. [Google Scholar] [CrossRef] [Green Version]
- Hammond, S.; Swanberg, J.C.; Kaplarevic, M.; Lee, K.H. Genomic sequencing and analysis of a Chinese hamster ovary cell line using Illumina sequencing technology. BMC Genom. 2011, 12, 1. [Google Scholar] [CrossRef] [Green Version]
- Hammond, S.; Swanberg, J.C.; Polson, S.W.; Lee, K.H. Profiling conserved microRNA expression in recombinant CHO cell lines using illumina sequencing. Biotechnol. Bioeng. 2012, 109, 1371–1375. [Google Scholar] [CrossRef] [Green Version]
- Hackl, M.; Jakobi, T.; Blom, J.; Doppmeier, D.; Brinkrolf, K.; Szczepanowski, R.; Bernhart, S.H.; zu Siederdissen, C.H.; Bort, J.A.H.; Wieser, M.; et al. Next-generation sequencing of the Chinese hamster ovary microRNA transcriptome: Identification, annotation and profiling of microRNAs as targets for cellular engineering. J. Biotechnol. 2011, 153, 62–75. [Google Scholar] [CrossRef] [Green Version]
- Klanert, G.; Jadhav, V.; Chanoumidou, K.; Grillari, J.; Borth, N.; Hackl, M. Endogenous microRNA clusters outperform chimeric sequence clusters in Chinese hamster ovary cells. Biotechnol. J. 2014, 9, 538–544. [Google Scholar] [CrossRef] [Green Version]
- Hackl, M.; Jadhav, V.; Jakobi, T.; Rupp, O.; Brinkrolf, K.; Goesmann, A.; Pühler, A.; Noll, T.; Borth, N.; Grillari, J. Computational identification of microRNA gene loci and precursor microRNA sequences in CHO cell lines. J. Biotechnol. 2012, 158, 151–155. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.C.; Jacob, N.M.; Nissom, P.M.; Hackl, M.; Lee, L.H.; Yap, M.; Hu, W.-S. Conserved microRNAs in Chinese hamster ovary cell lines. Biotechnol. Bioeng. 2011, 108, 475–480. [Google Scholar] [CrossRef]
- Barron, N.; Kumar, N.; Sanchez, N.; Doolan, P.; Clarke, C.; Meleady, P.; O’Sullivan, F.; Clynes, M. Engineering CHO cell growth and recombinant protein productivity by overexpression of miR-7. J. Biotechnol. 2011, 151, 204–211. [Google Scholar] [CrossRef]
- Druz, A.; Chu, C.; Majors, B.; Santuary, R.; Betenbaugh, M.; Shiloach, J. A novel microRNA mmu-miR-466h affects apoptosis regulation in mammalian cells. Biotechnol. Bioeng. 2011, 108, 1651–1661. [Google Scholar] [CrossRef] [Green Version]
- Druz, A.; Son, Y.; Betenbaugh, M.; Shiloach, J. Stable inhibition of mmu-miR-466h-5p improves apoptosis resistance and protein production in CHO cells. Metab. Eng. 2013, 16, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Loh, W.P.; Loo, B.; Zhou, L.; Zhang, P.; Lee, D.-Y.; Yang, Y.; Lam, K.P. Overexpression of microRNAs enhances recombinant protein production in Chinese hamster ovary cells. Biotechnol. J. 2014, 9, 1140–1151. [Google Scholar] [CrossRef]
- Strotbek, M.; Florin, L.; Koenitzer, J.; Tolstrup, A.; Kaufmann, H.; Hausser, A.; Olayioye, M.A. Stable microRNA expression enhances therapeutic antibody productivity of Chinese hamster ovary cells. Metab. Eng. 2013, 20, 157–166. [Google Scholar] [CrossRef]
- Jadhav, V.; Hackl, M.; Klanert, G.; Hernandez Bort, J.A.; Kunert, R.; Grillari, J.; Borth, N. Stable overexpression of miR-17 enhances recombinant protein production of CHO cells. J. Biotechnol. 2014, 175, 38–44. [Google Scholar] [CrossRef] [Green Version]
- Emmerling, V.V.; Fischer, S.; Stiefel, F.; Holzmann, K.; Handrick, R.; Hesse, F.; Hörer, M.; Kochanek, S.; Otte, K. Temperature-sensitive miR-483 is a conserved regulator of recombinant protein and viral vector production in mammalian cells. Biotechnol. Bioeng. 2015, 113, 830–841. [Google Scholar] [CrossRef]
- Fischer, S.; Buck, T.; Wagner, A.; Ehrhart, C.; Giancaterino, J.; Mang, S.; Schad, M.; Mathias, S.; Aschrafi, A.; Handrick, R.; et al. A functional high-content miRNA screen identifies miR-30 family to boost recombinant protein production in CHO cells. Biotechnol. J. 2014, 9, 1279–1292. [Google Scholar] [CrossRef]
- Fischer, S.; Paul, A.J.; Wagner, A.; Mathias, S.; Geiss, M.; Schandock, F.; Domnowski, M.; Zimmermann, J.; Handrick, R.; Hesse, F.; et al. miR-2861 as novel HDAC5 inhibitor in CHO cells enhances productivity while maintaining product quality. Biotechnol. Bioeng. 2015, 112, 2142–2153. [Google Scholar] [CrossRef]
- Schoellhorn, M.; Fischer, S.; Wagner, A.; Handrick, R.; Otte, K. miR-143 targets MAPK7 in CHO cells and induces a hyperproductive phenotype to enhance production of difficult-to-express proteins. Biotechnol. Prog. 2017, 33, 1046–1058. [Google Scholar] [CrossRef]
- Martinez-Lopez, J.E.; Coleman, O.; Meleady, P.; Clynes, M. Transfection of miR-31* boosts oxidative phosphorylation metabolism in the mitochondria and enhances recombinant protein production in Chinese hamster ovary cells. J. Biotechnol. 2021, 333, 86–96. [Google Scholar] [CrossRef]
- Bryan, L.; Henry, M.; Barron, N.; Gallagher, C.; Kelly, R.M.; Frye, C.C.; Osborne, M.D.; Clynes, M.; Meleady, P. Differential expression of miRNAs and functional role of mir-200a in high and low productivity CHO cells expressing an Fc fusion protein. Biotechnol. Lett. 2021, 43, 1551–1563. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.S.; Breen, L.; Gallagher, C.; Kelly, S.; Henry, M.; Lao, N.T.; Meleady, P.; O’Gorman, D.; Clynes, M.; Barron, N. Re-programming CHO cell metabolism using miR-23 tips the balance towards a highly productive phenotype. Biotechnol. J. 2015, 10, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Raab, N.; Mathias, S.; Alt, K.; Handrick, R.; Fischer, S.; Schmieder, V.; Jadhav, V.; Borth, N.; Otte, K. CRISPR/Cas9-Mediated Knockout of MicroRNA-744 Improves Antibody Titer of CHO Production Cell Lines. Biotechnol. J. 2019, 14, e1800477. [Google Scholar] [CrossRef] [PubMed]
- Peltier, H.J.; Latham, G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008, 14, 844–852. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.-G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.-S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Wu, Y.; Choi, J.-G.; Wu, H. Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site. Proc. Natl. Acad. Sci. USA 2013, 110, 20687–20692. [Google Scholar] [CrossRef] [Green Version]
- Roden, C.; Gaillard, J.; Kanoria, S.; Rennie, W.; Barish, S.; Cheng, J.; Pan, W.; Liu, J.; Cotsapas, C.; DIng, Y.; et al. Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res. 2017, 27, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Cullen, B.R. Sequence requirements for micro RNA processing and function in human cells. RNA 2003, 9, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Cullen, B.R. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J. Biol. Chem. 2005, 280, 27595–27603. [Google Scholar] [CrossRef] [Green Version]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Brown, A.J.; Gibson, S.; Hatton, D.; James, D.C. Transcriptome-Based Identification of the Optimal Reference CHO Genes for Normalisation of qPCR Data. Biotechnol. J. 2018, 13, 1700259. [Google Scholar] [CrossRef]
- Wasley, L.C.; Timony, G.; Murtha, P.; Stoudemire, J.; Dorner, A.J.; Caro, J.; Krieger, M.; Kaufman, R.J. The importance of N- and O-linked oligosaccharides for the biosynthesis and in vitro and in vivo biologic activities of erythropoietin. Blood 1991, 77, 2624–2632. [Google Scholar] [CrossRef] [Green Version]
- Dancy, B.M.; Cole, P.A. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015, 115, 2419–2452. [Google Scholar] [CrossRef]
- Janknecht, R.; Wells, N.J.; Hunter, T. TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 1998, 12, 2114–2119. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.X.; Tian, M.; Zhao, B.X.; Li, G.D.; Liu, B.; Zhan, Y.Y.; Chen, H.Z.; Wu, Q. Orphan receptor TR3 attenuates the p300-induced acetylation of retinoid X receptor-α. Mol. Endocrinol. 2007, 21, 2877–2889. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Ito, A.; Lai, C.H.; Zhao, X.; Saito, S.; Hamilton, M.H.; Appella, E.; Yao, T.P. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001, 20, 1331–1340. [Google Scholar] [CrossRef] [Green Version]
- Brooks, C.L.; Gu, W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2011, 2, 456–462. [Google Scholar] [CrossRef] [Green Version]
- Seto, E.; Usheva, A.; Zambetti, G.P.; Momand, J.; Horikoshi, N.; Weinmann, R.; Levine, A.J.; Shenk, T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 1992, 89, 12028–12032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mack, D.H.; Vartikar, J.; Pipas, J.M.; Laimins, L.A. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nat. Cell Biol. 1993, 363, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Ragimov, N.; Krauskopf, A.; Navot, N.; Rotter, V.; Oren, M.; Aloni, Y. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene 1993, 8, 1183–1193. [Google Scholar] [PubMed]
- Subbaramaiah, K.; Altorkil, N.; Chung, W.J.; Mestre, J.R.; Sampat, A.; Dannenberg, A.J. Inhibition of cyclooxygenase-2 gene expression by p53. J. Biol. Chem. 1999, 274, 10911–10915. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Mehew, J.W.; Heckman, C.A.; Arcinas, M.; Boxer, L.M. Negative regulation of bcl-2 expression by p53 in hematopoietic cells. Oncogene 2001, 20, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Farmer, G.; Colgan, J.; Nakatani, Y.; Manley, J.L.; Prives, C. Functional interaction between p53, the TATA-binding protein (TBP), andTBP-associated factors in vivo. Mol. Cell. Biol. 1996, 16, 4295–4304. [Google Scholar] [CrossRef] [Green Version]
- Crighton, D.; Woiwode, A.; Zhang, C.; Mandavia, N.; Morton, J.P.; Warnock, L.J.; Milner, J.; White, R.J.; Johnson, D.L. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 2003, 22, 2810–2820. [Google Scholar] [CrossRef] [Green Version]
- Rodova, M.; Jayini, R.; Singasani, R.; Chipps, E.; Islam, M.R. CMV promoter is repressed by p53 and activated by JNK pathway. Plasmid 2013, 69, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Jadhav, V.; Hackl, M.; Druz, A.; Shridhar, S.; Chung, C.Y.; Heffner, K.M.; Kreil, D.P.; Betenbaugh, M.; Shiloach, J.; Barron, N.; et al. CHO microRNA engineering is growing up: Recent successes and future challenges. Biotechnol. Adv. 2013, 31, 1501–1513. [Google Scholar] [CrossRef] [Green Version]
- Hernández Bort, J.A.; Hackl, M.; Höflmayer, H.; Jadhav, V.; Harreither, E.; Kumar, N.; Ernst, W.; Grillari, J.; Borth, N. Dynamic mRNA and miRNA profiling of CHO-K1 suspension cell cultures. Biotechnol. J. 2012, 7, 500–515. [Google Scholar] [CrossRef]
- Lin, N.; Davis, A.; Bahr, S.; Borgschulte, T.; Achtien, K.; Kayser, K. Profiling highly conserved microrna expression in recombinant IgG-producing and parental Chinese hamster ovary cells. Biotechnol. Prog. 2011, 27, 1163–1171. [Google Scholar] [CrossRef]
- Maccani, A.; Hackl, M.; Leitner, C.; Steinfellner, W.; Graf, A.B.; Tatto, N.E.; Karbiener, M.; Scheideler, M.; Grillari, J.; Mattanovich, D.; et al. Identification of microRNAs specific for high producer CHO cell lines using steady-state cultivation. Appl. Microbiol. Biotechnol. 2014, 98, 7535–7548. [Google Scholar] [CrossRef] [Green Version]
- Klanert, G.; Jadhav, V.; Shanmukam, V.; Diendorfer, A.; Karbiener, M.; Scheideler, M.; Bort, J.H.; Grillari, J.; Hackl, M.; Borth, N. A signature of 12 microRNAs is robustly associated with growth rate in a variety of CHO cell lines. J. Biotechnol. 2016, 235, 150–161. [Google Scholar] [CrossRef] [Green Version]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.P.; Keller, A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8, 107167–107175. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Wang, Y.; Sun, Z.; Wang, H.; Zhou, H.; Zhang, L.; Zhang, S.; Cao, X. MiRNA-132-3p inhibits osteoblast differentiation by targeting Ep300 in simulated microgravity. Sci. Rep. 2015, 5, 18655. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Z.M.; Huang, Y.; Ishiko, T.; Nakada, S.; Utsugisawa, T.; Shioya, H.; Utsugisawa, Y.; Shi, Y.; Weichselbaum, R.; Kufe, D. Function for p300 and not CBP in the apoptotic response to DNA damage. Oncogene 1999, 18, 5714–5717. [Google Scholar] [CrossRef] [Green Version]
- Perrem, K.; Rayner, J.; Voss, T.; Sturzbecher, H.; Jackson, P.; Braithwaite, A. p53 represses SV40 transcription by preventing formation of transcription complexes. Oncogene 1995, 11, 1299–1307. [Google Scholar]
- Li, M.; Luo, J.; Brooks, C.L.; Gu, W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002, 277, 50607–50611. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Poetz, F.; Bruer, M.; Ly-Hartig, T.B.N.; Schott, J.; Séraphin, B.; Stoecklin, G. Acetylation-Dependent Control of Global Poly(A) RNA Degradation by CBP/p300 and HDAC1/2. Mol. Cell 2016, 63, 927–938. [Google Scholar] [CrossRef] [Green Version]
- Wahle, E.; Winkler, G.S. RNA decay machines: Deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 561–570. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Švab, Ž.; Braga, L.; Guarnaccia, C.; Labik, I.; Herzog, J.; Baralle, M.; Giacca, M.; Skoko, N. High Throughput miRNA Screening Identifies miR-574-3p Hyperproductive Effect in CHO Cells. Biomolecules 2021, 11, 1125. https://doi.org/10.3390/biom11081125
Švab Ž, Braga L, Guarnaccia C, Labik I, Herzog J, Baralle M, Giacca M, Skoko N. High Throughput miRNA Screening Identifies miR-574-3p Hyperproductive Effect in CHO Cells. Biomolecules. 2021; 11(8):1125. https://doi.org/10.3390/biom11081125
Chicago/Turabian StyleŠvab, Živa, Luca Braga, Corrado Guarnaccia, Ivan Labik, Jeremias Herzog, Marco Baralle, Mauro Giacca, and Nataša Skoko. 2021. "High Throughput miRNA Screening Identifies miR-574-3p Hyperproductive Effect in CHO Cells" Biomolecules 11, no. 8: 1125. https://doi.org/10.3390/biom11081125
APA StyleŠvab, Ž., Braga, L., Guarnaccia, C., Labik, I., Herzog, J., Baralle, M., Giacca, M., & Skoko, N. (2021). High Throughput miRNA Screening Identifies miR-574-3p Hyperproductive Effect in CHO Cells. Biomolecules, 11(8), 1125. https://doi.org/10.3390/biom11081125






