A Novel Knowledge Distillation-Based Feature Selection for the Classification of ADHD
Abstract
:1. Introduction
2. Related Research
2.1. Structural Information Based Approaches
2.2. Functional Connectivity Based Approaches
2.3. Deep Learning-Based Approaches
3. Materials and Methods
3.1. Dataset
3.2. Preprocessing of the Dataset
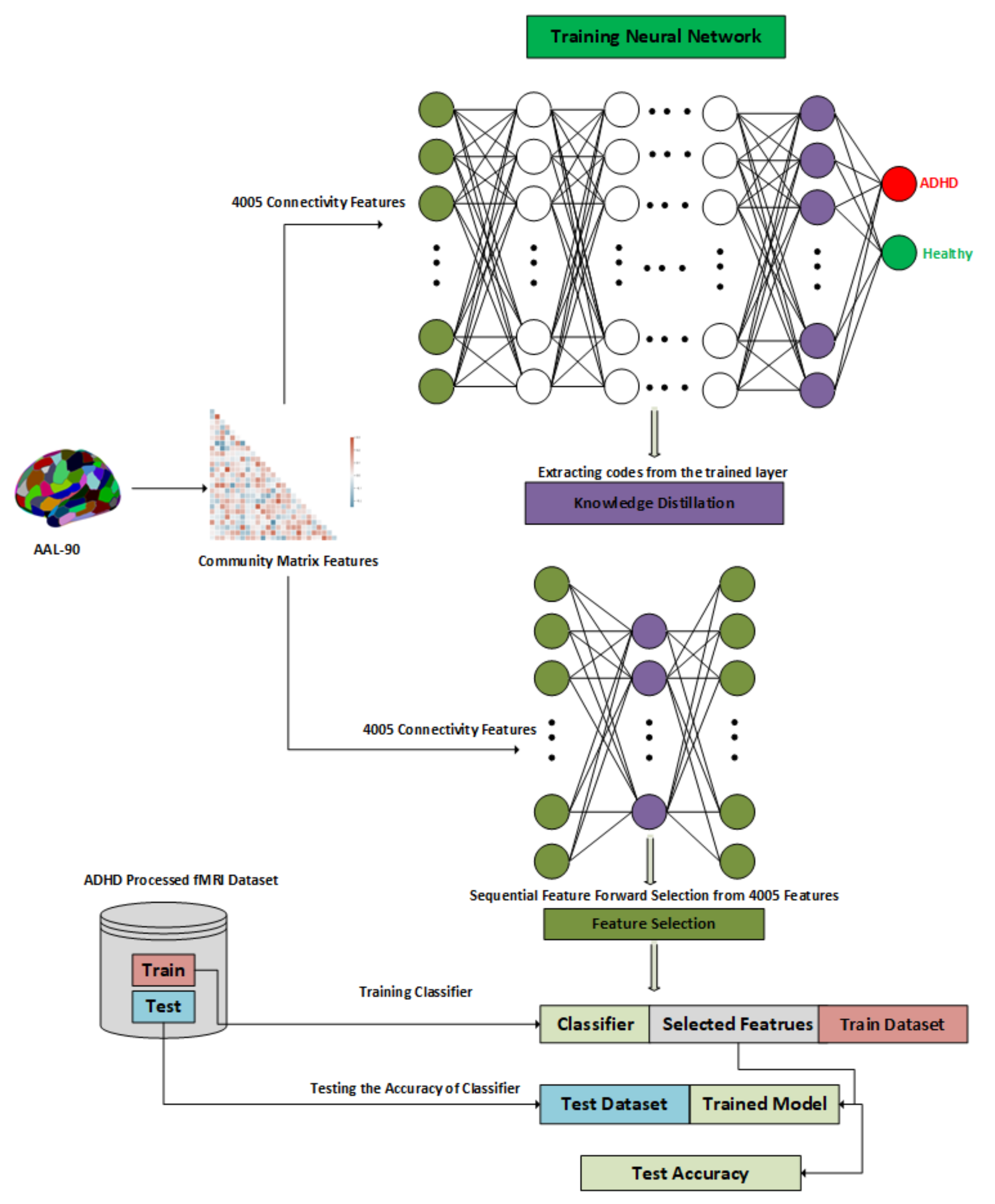
3.3. Methodology
3.3.1. Affinity Propagation
3.3.2. Architecture of the Proposed Approach
3.3.3. Latent Representation of Dataset
3.3.4. Knowledge Distillation
3.3.5. Sequential Forward Feature Selection
3.4. Algorithms
3.4.1. Connectivity Matrix
| Algorithm 1 Connectivity Matrix |
Input: , P Output:
|
3.4.2. Community Matrix
| Algorithm 2 Community Matrix Features |
Input: , Output:
|
3.4.3. Ranking of Features
| Algorithm 3 Features Ranking Algorithm |
Input: Output:
|
3.4.4. Features Subset Selection
| Algorithm 4 Sequential Feature Selection |
Input: , , Output: F
|
3.5. Experimental Settings
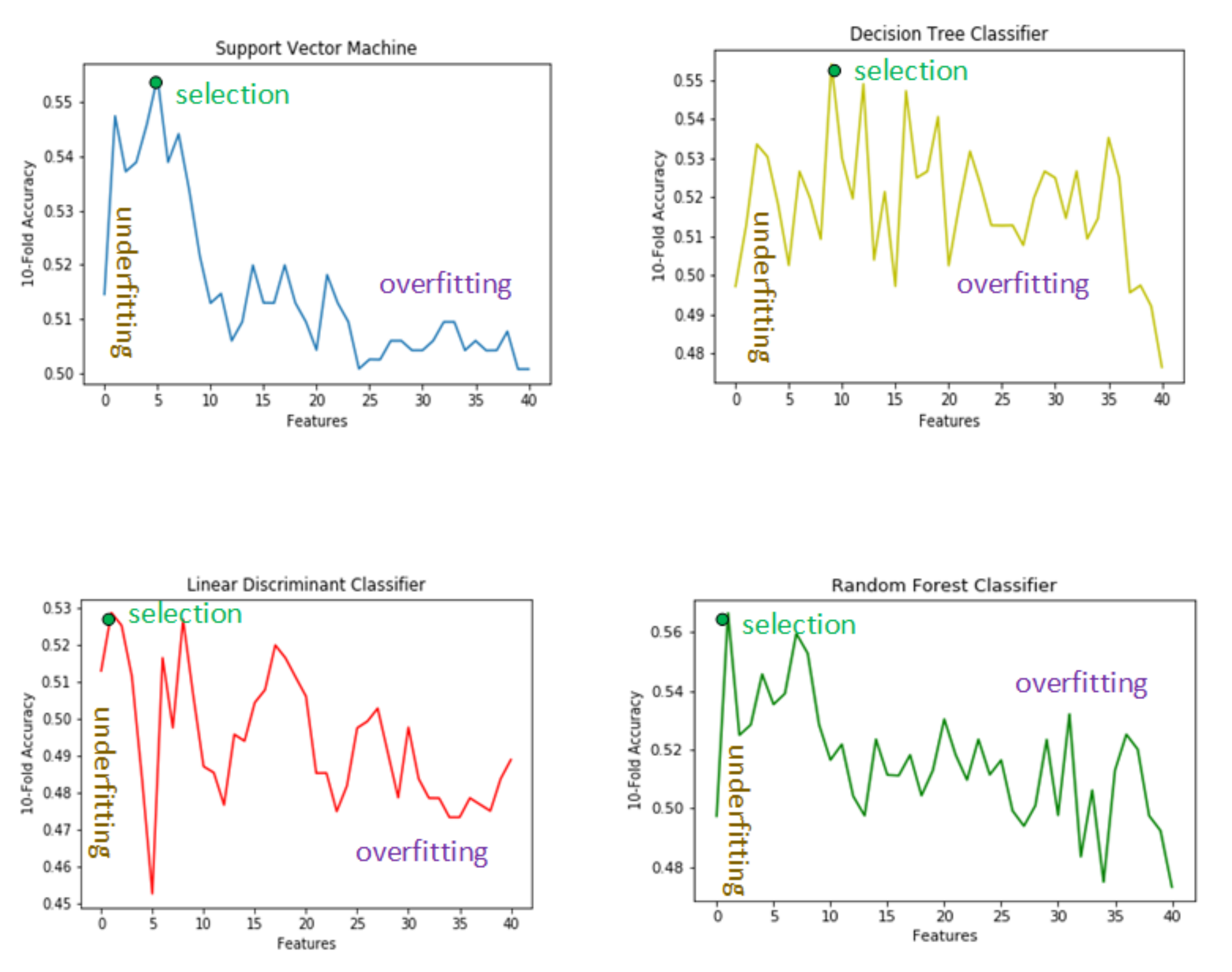
4. Features Selection
4.1. Low Number of Features
4.2. Large Number of Features
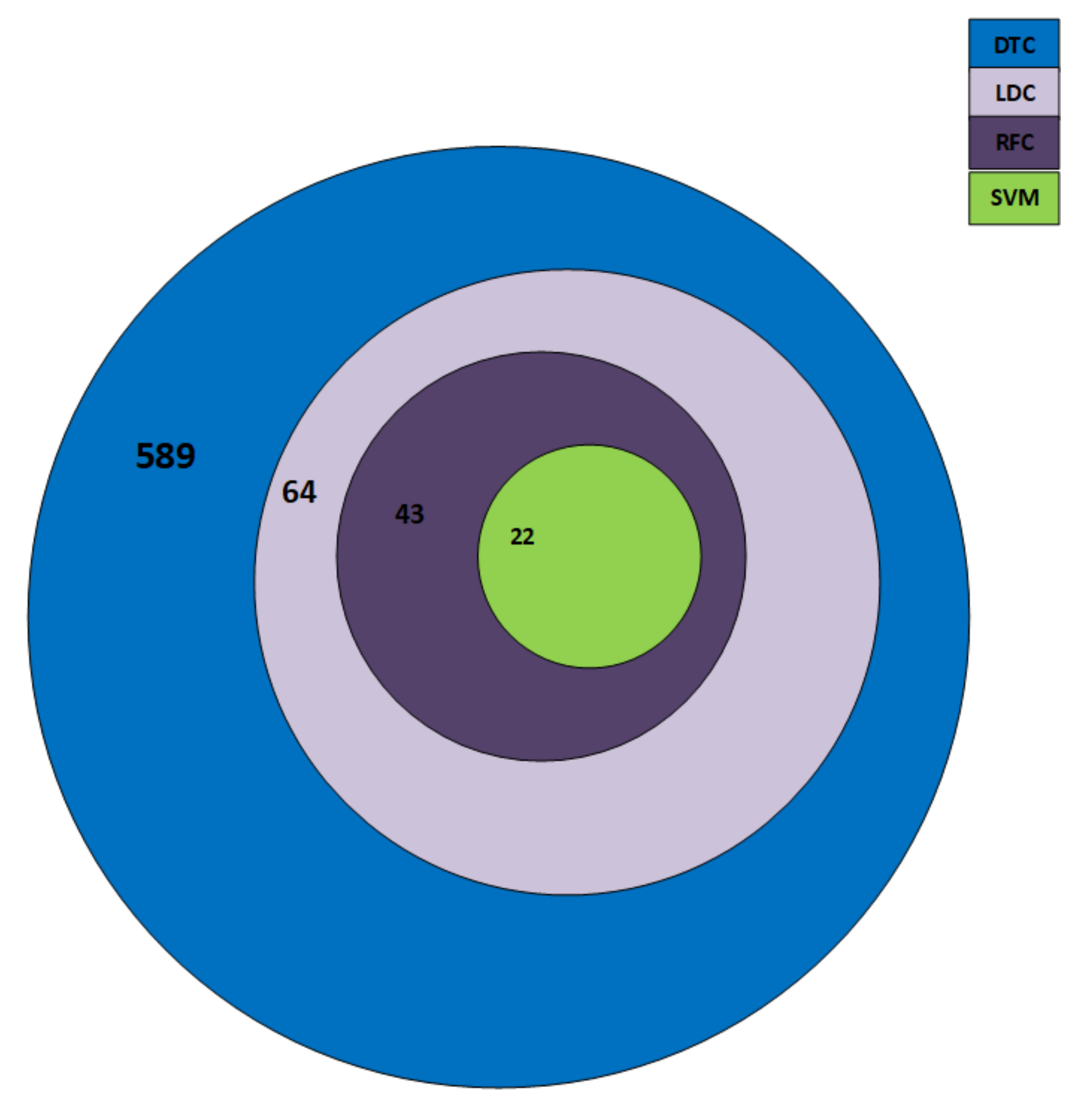
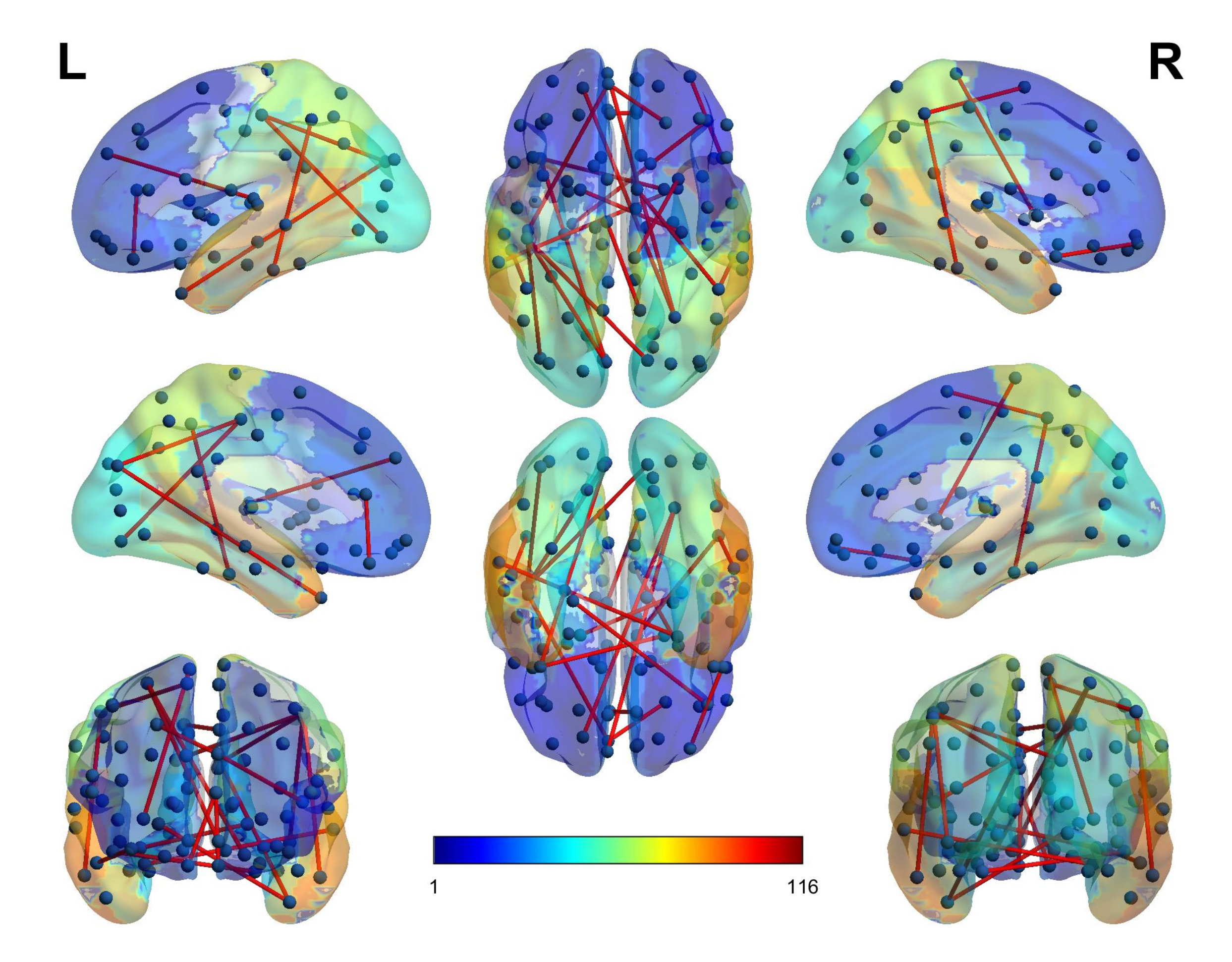
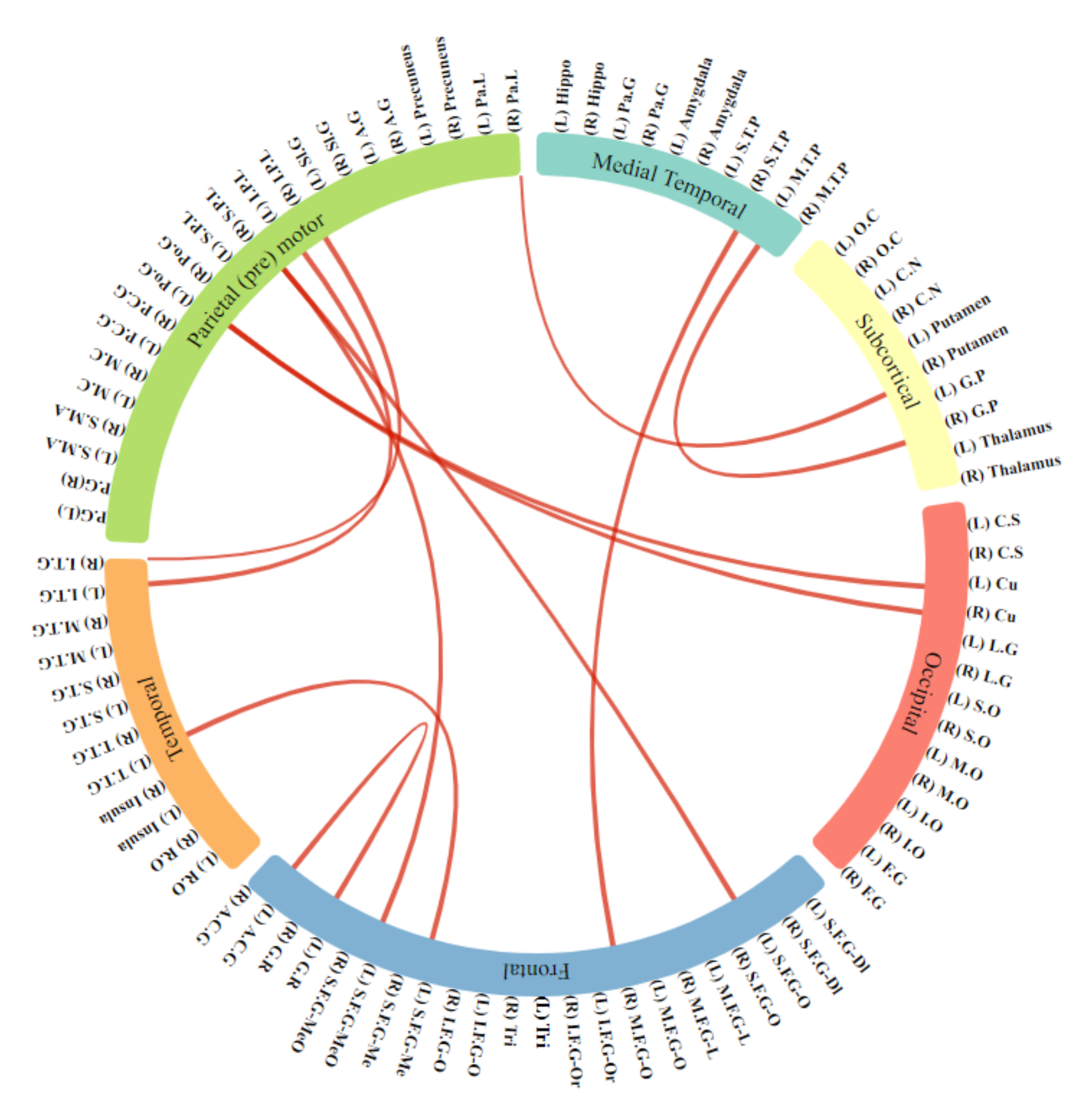
4.3. Selected Features
4.4. Support Vector Machine Features
5. Results
5.1. Training Using Combining Sites
5.2. Testing on Benchmark Datasets
6. Discussion
7. Conclusions
7.1. Applications
7.2. Future Directions
7.3. Limitations
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADHD | Attention Deficit Hyperactivity Disorder |
| DTC | Decision Tree Classifier |
| LDC | Linear Discriminant Classifier |
| RFC | Random Forest Classifier |
| SVM | Support Vector Machine |
| rs-fMRI | Resting State Functional Magnetic Resonance Imaging |
References
- Bassett, D.S.; Gazzaniga, M.S. Understanding complexity in the human brain. Trends Cogn. Sci. 2011, 15, 200–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bossaerts, P.; Murawski, C. Computational complexity and human decision-making. Trends Cogn. Sci. 2017, 21, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Anokhin, A.P.; Birbaumer, N.; Lutzenberger, W.; Nikolaev, A.; Vogel, F. Age increases brain complexity. Electroencephalogr. Clin. Neurophysiol. 1996, 99, 63–68. [Google Scholar] [CrossRef]
- Abbott, A. Solving the brain. Nature 2013, 499, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wender, P.H. Attention-deficit hyperactivity disorder in adults. Psychiatr. Clin. N. Am. 1998, 21, 761–774. [Google Scholar] [CrossRef]
- Swanson, J.M.; Volkow, N.D.; Newcorn, J.; Casey, B.; Moyzis, R.; Grandy, D.; Posner, M. Attention deficit hyperactivity disorder. Encycl. Cogn. Sci. 2006, 45, 642–657. [Google Scholar]
- Faraone, S.; Asherson, P.; Banaschewski, T.; Biederman, J.; Buitelaar, J.; Ramos-Quiroga, J.; Franke, B. Attention-deficit/hyperactivity disorder. Nat. Rev. Dis. Prim. 2015, 1, 15020. [Google Scholar] [CrossRef]
- Doyle, A.E.; Willcutt, E.G.; Seidman, L.J.; Biederman, J.; Chouinard, V.A.; Silva, J.; Faraone, S.V. Attention-deficit/hyperactivity disorder endophenotypes. Biol. Psychiatry 2005, 57, 1324–1335. [Google Scholar] [CrossRef]
- Bush, G. Attention-deficit/hyperactivity disorder and attention networks. Neuropsychopharmacology 2010, 35, 278–300. [Google Scholar] [CrossRef]
- Thomas, R.; Sanders, S.; Doust, J.; Beller, E.; Glasziou, P. Prevalence of attention-deficit/hyperactivity disorder: A systematic review and meta-analysis. Pediatrics 2015, 135, e994–e1001. [Google Scholar] [CrossRef] [Green Version]
- Franke, B.; Faraone, S.; Asherson, P.; Buitelaar, J.; Bau, C.; Ramos-Quiroga, J.A.; Mick, E.; Grevet, E.; Johansson, S.; Haavik, J.; et al. The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol. Psychiatry 2012, 17, 960–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, H.L.; Yung-Chin, H.; Chi-Yuan, S.; Tseng, W.Y.I.; Gau, S.S.F. White matter endophenotype candidates for ADHD: A diffusion imaging tractography study with sibling design. Psychol. Med. 2020, 50, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.G.; Ye, Z.; Li, Q.Y.; Liu, G.J.; Xie, B.; Wang, J. Changes of brain structure and function in ADHD children. Brain Topogr. 2011, 24, 243–252. [Google Scholar] [CrossRef] [PubMed]
- de Luis-García, R.; Cabús-Piñol, G.; Imaz-Roncero, C.; Argibay-Quiñones, D.; Barrio-Arranz, G.; Aja-Fernández, S.; Alberola-López, C. Attention deficit/hyperactivity disorder and medication with stimulants in young children: A DTI study. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2015, 57, 176–184. [Google Scholar] [CrossRef]
- Boutros, N.; Fraenkel, L.; Feingold, A. A four-step approach for developing diagnostic tests in psychiatry: EEG in ADHD as a test case. J. Neuropsychiatry Clin. Neurosci. 2005, 17, 455–464. [Google Scholar] [CrossRef]
- Micoulaud-Franchi, J.A.; Geoffroy, P.A.; Fond, G.; Lopez, R.; Bioulac, S.; Philip, P. EEG neurofeedback treatments in children with ADHD: An updated meta-analysis of randomized controlled trials. Front. Hum. Neurosci. 2014, 8, 906. [Google Scholar] [CrossRef] [Green Version]
- Heinrich, H.; Busch, K.; Studer, P.; Erbe, K.; Moll, G.H.; Kratz, O. EEG spectral analysis of attention in ADHD: Implications for neurofeedback training? Front. Hum. Neurosci. 2014, 8, 611. [Google Scholar] [CrossRef]
- Snyder, S.M.; Rugino, T.A.; Hornig, M.; Stein, M.A. Integration of an EEG biomarker with a clinician’s ADHD evaluation. Brain Behav. 2015, 5, e00330. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Song, Y.; Li, X. A deep learning framework for identifying children with ADHD using an EEG-based brain network. Neurocomputing 2019, 356, 83–96. [Google Scholar] [CrossRef]
- Dubreuil-Vall, L.; Ruffini, G.; Camprodon, J.A. Deep learning convolutional neural networks discriminate adult ADHD from healthy individuals on the basis of event-related spectral EEG. Front. Neurosci. 2020, 14, 251. [Google Scholar] [CrossRef] [PubMed]
- Deiber, M.P.; Hasler, R.; Colin, J.; Dayer, A.; Aubry, J.M.; Baggio, S.; Perroud, N.; Ros, T. Linking alpha oscillations, attention and inhibitory control in adult ADHD with EEG neurofeedback. NeuroImage Clin. 2020, 25, 102145. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.S.; Potenza, M.N.; Wang, Z.; Zhu, H.; Martin, A.; Marsh, R.; Plessen, K.J.; Yu, S. An FMRI study of the effects of psychostimulants on default-mode processing during Stroop task performance in youths with ADHD. Am. J. Psychiatry 2009, 166, 1286–1294. [Google Scholar] [CrossRef]
- Zuo, X.N. Modeling Resting-State Brain Functional Integration. Nat. Preced. 2010. [Google Scholar] [CrossRef]
- Cortese, S.; Kelly, C.; Chabernaud, C.; Proal, E.; Di Martino, A.; Milham, M.P.; Castellanos, F.X. Toward systems neuroscience of ADHD: A meta-analysis of 55 fMRI studies. Am. J. Psychiatry 2012, 169, 1038–1055. [Google Scholar] [CrossRef]
- Kaboodvand, N.; Iravani, B.; Fransson, P. Dynamic synergetic configurations of resting-state networks in ADHD. NeuroImage 2020, 207, 116347. [Google Scholar] [CrossRef] [PubMed]
- Sweitzer, M.M.; Kollins, S.H.; Kozink, R.V.; Hallyburton, M.; English, J.; Addicott, M.A.; Oliver, J.A.; McClernon, F.J. ADHD, smoking withdrawal, and inhibitory control: Results of a Neuroimaging Study with methylphenidate challenge. Neuropsychopharmacology 2018, 43, 851–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahathuduwa, C.N.; Wakefield, S.; West, B.D.; Blume, J.; Dassanayake, T.L.; Weerasinghe, V.S.; Mastergeorge, A. Effects of l-theanine–caffeine combination on sustained attention and inhibitory control among children with ADHD: A proof-of-concept neuroimaging RCT. Sci. Rep. 2020, 10, 13072. [Google Scholar] [CrossRef]
- Zimmer, L. Positron emission tomography neuroimaging for a better understanding of the biology of ADHD. Neuropharmacology 2009, 57, 601–607. [Google Scholar] [CrossRef]
- Spencer, T.J.; Biederman, J.; Madras, B.K.; Dougherty, D.D.; Bonab, A.A.; Livni, E.; Meltzer, P.C.; Martin, J.; Rauch, S.; Fischman, A.J. Further evidence of dopamine transporter dysregulation in ADHD: A controlled PET imaging study using altropane. Biol. Psychiatry 2007, 62, 1059–1061. [Google Scholar] [CrossRef] [Green Version]
- Cherkasova, M.V.; Faridi, N.; Casey, K.F.; Larcher, K.; O’Driscoll, G.A.; Hechtman, L.; Joober, R.; Baker, G.B.; Palmer, J.; Evans, A.C.; et al. Differential associations between cortical thickness and striatal dopamine in treatment-naïve adults with ADHD vs. healthy controls. Front. Hum. Neurosci. 2017, 11, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigurdardottir, H.L.; Kranz, G.S.; Rami-Mark, C.; James, G.M.; Vanicek, T.; Gryglewski, G.; Kautzky, A.; Hienert, M.; Traub-Weidinger, T.; Mitterhauser, M.; et al. Effects of norepinephrine transporter gene variants on NET binding in ADHD and healthy controls investigated by PET. Hum. Brain Mapp. 2016, 37, 884–895. [Google Scholar] [CrossRef]
- Kautzky, A.; Vanicek, T.; Philippe, C.; Kranz, G.; Wadsak, W.; Mitterhauser, M.; Hartmann, A.; Hahn, A.; Hacker, M.; Rujescu, D.; et al. Machine learning classification of ADHD and HC by multimodal serotonergic data. Transl. Psychiatry 2020, 10, 104. [Google Scholar] [CrossRef] [Green Version]
- Grodner, K.; Harcourt, S.; Sattuar, Z.; STrong, A.; Golden, C.; Amen, D.; Willeumier, K.; Taylor, D. B-01 Differentiating Attention Deficit/Hyperactivity Disorder (ADHD) Combined Type and ADHD Inattentive Type Using SPECT Imaging. Arch. Clin. Neuropsychol. 2016, 31, 613. [Google Scholar] [CrossRef]
- Klein, M.; Onnink, M.; van Donkelaar, M.; Wolfers, T.; Harich, B.; Shi, Y.; Dammers, J.; Arias-Vásquez, A.; Hoogman, M.; Franke, B. Brain imaging genetics in ADHD and beyond–mapping pathways from gene to disorder at different levels of complexity. Neurosci. Biobehav. Rev. 2017, 80, 115–155. [Google Scholar] [CrossRef]
- Karande, S.; Deshmukh, N.; Rangarajan, V.; Agrawal, A.; Sholapurwala, R. Brain SPECT scans in students with specific learning disability: Preliminary results. J. Postgrad. Med. 2019, 65, 33. [Google Scholar] [CrossRef] [PubMed]
- Akay, A.P.; Kaya, G.C.; Kose, S.; Yazıcıoğlu, Ç.E.; Erkuran, H.Ö.; Güney, S.A.; Oğuz, K.; Keskin, D.; Baykara, B.; Emiroğlu, N.İ.; et al. Genetic imaging study with [Tc-99m] TRODAT-1 SPECT in adolescents with ADHD using OROS-methylphenidate. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 86, 294–300. [Google Scholar] [CrossRef]
- Henderson, T.A.; van Lierop, M.J.; McLean, M.; Uszler, J.M.; Thornton, J.F.; Siow, Y.H.; Pavel, D.G.; Cardaci, J.; Cohen, P. Functional Neuroimaging in Psychiatry—Aiding in Diagnosis and Guiding Treatment. What the American Psychiatric Association Does Not Know. Front. Psychiatry 2020, 11, 276. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.R.; Sidhu, G.S.; Greiner, R.; Asgarian, N.; Bastani, M.; Silverstone, P.H.; Greenshaw, A.J.; Dursun, S.M. ADHD-200 Global Competition: Diagnosing ADHD using personal characteristic data can outperform resting state fMRI measurements. Front. Syst. Neurosci. 2012, 6, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoekzema, E.; Carmona, S.; Ramos-Quiroga, J.A.; Richarte Fernandez, V.; Bosch, R.; Soliva, J.C.; Rovira, M.; Bulbena, A.; Tobeña, A.; Casas, M.; et al. An independent components and functional connectivity analysis of resting state fMRI data points to neural network dysregulation in adult ADHD. Hum. Brain Mapp. 2014, 35, 1261–1272. [Google Scholar] [CrossRef]
- Riaz, A.; Asad, M.; Alonso, E.; Slabaugh, G. Fusion of fMRI and non-imaging data for ADHD classification. Comput. Med. Imaging Graph. 2018, 65, 115–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubia, K.; Criaud, M.; Wulff, M.; Alegria, A.; Brinson, H.; Barker, G.; Stahl, D.; Giampietro, V. Functional connectivity changes associated with fMRI neurofeedback of right inferior frontal cortex in adolescents with ADHD. NeuroImage 2019, 188, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, C.; Li, P.; Peng, Y.; Kang, X.; Jiang, C.; Li, F.; Zhu, X.; Yao, D.; Biswal, B.; et al. Separated Channel Attention Convolutional Neural Network (SC-CNN-Attention) to Identify ADHD in Multi-Site Rs-fMRI Dataset. Entropy 2020, 22, 893. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, A.; Pourahmadi, V.; Soltani, M.; Sheikhzadeh, H. Deep feature selection using a teacher-student network. Neurocomputing 2020, 383, 396–408. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.W.; Ho, C.C.; Chen, J.H. ADHD classification by a texture analysis of anatomical brain MRI data. Front. Syst. Neurosci. 2012, 6, 66. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Lin, P.; Zhang, T.; Wang, J. Extreme learning machine-based classification of ADHD using brain structural MRI data. PLoS ONE 2013, 8, e79476. [Google Scholar] [CrossRef]
- Fischl, B. FreeSurfer. NeuroImage 2012, 62, 774–781. [Google Scholar] [CrossRef] [Green Version]
- Lim, L.; Marquand, A.; Cubillo, A.A.; Smith, A.B.; Chantiluke, K.; Simmons, A.; Mehta, M.; Rubia, K. Disorder-specific predictive classification of adolescents with attention deficit hyperactivity disorder (ADHD) relative to autism using structural magnetic resonance imaging. PLoS ONE 2013, 8, e63660. [Google Scholar] [CrossRef]
- Qureshi, M.N.I.; Oh, J.; Min, B.; Jo, H.J.; Lee, B. Multi-modal, multi-measure, and multi-class discrimination of ADHD with hierarchical feature extraction and extreme learning machine using structural and functional brain MRI. Front. Hum. Neurosci. 2017, 11, 157. [Google Scholar] [PubMed] [Green Version]
- Sen, B.; Borle, N.C.; Greiner, R.; Brown, M.R. A general prediction model for the detection of ADHD and Autism using structural and functional MRI. PLoS ONE 2018, 13, e0194856. [Google Scholar] [CrossRef]
- Jiang, W.; Duan, K.; Rootes-Murdy, K.; Hoekstra, P.J.; Hartman, C.A.; Oosterlaan, J.; Heslenfeld, D.; Franke, B.; Buitelaar, J.; Arias-Vasquez, A.; et al. Structural brain alterations and their association with cognitive function and symptoms in Attention-deficit/Hyperactivity Disorder families. NeuroImage Clin. 2020, 27, 102273. [Google Scholar] [CrossRef]
- Sato, J.R.; Hoexter, M.Q.; Castellanos, X.F.; Rohde, L.A. Abnormal brain connectivity patterns in adults with ADHD: A coherence study. PLoS ONE 2012, 7, e45671. [Google Scholar]
- Choi, J.; Jeong, B.; Lee, S.W.; Go, H.J. Aberrant development of functional connectivity among resting state-related functional networks in medication-naive ADHD children. PLoS ONE 2013, 8, e83516. [Google Scholar] [CrossRef] [PubMed]
- McLeod, K.R.; Langevin, L.M.; Goodyear, B.G.; Dewey, D. Functional connectivity of neural motor networks is disrupted in children with developmental coordination disorder and attention-deficit/hyperactivity disorder. NeuroImage Clin. 2014, 4, 566–575. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, G.; Wang, P.; Rangaprakash, D.; Wilamowski, B. Fully connected cascade artificial neural network architecture for attention deficit hyperactivity disorder classification from functional magnetic resonance imaging data. IEEE Trans. Cybern. 2015, 45, 2668–2679. [Google Scholar] [CrossRef] [PubMed]
- Mowinckel, A.M.; Alnæs, D.; Pedersen, M.L.; Ziegler, S.; Fredriksen, M.; Kaufmann, T.; Sonuga-Barke, E.; Endestad, T.; Westlye, L.T.; Biele, G. Increased default-mode variability is related to reduced task-performance and is evident in adults with ADHD. NeuroImage Clin. 2017, 16, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Damiani, S.; Tarchi, L.; Scalabrini, A.; Marini, S.; Provenzani, U.; Rocchetti, M.; Oliva, F.; Politi, P. Beneath the surface: Hyper-connectivity between caudate and salience regions in ADHD fMRI at rest. Eur. Child Adolesc. Psychiatry 2020, 4, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yao, D.; Cao, Q.; Liu, L.; Zhao, Q.; Li, H.; Huang, F.; Wang, Y.; Qian, Q.; Wang, Y.; et al. Shared and distinct resting functional connectivity in children and adults with attention-deficit/hyperactivity disorder. Transl. Psychiatry 2020, 10, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, L.; You, Y.; Du, H.; Fu, D. Classification of ADHD with fMRI data and multi-objective optimization. Comput. Methods Prog. Biomed. 2020, 196, 105676. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, Y.; Wang, C.; Liu, X.; Zhao, L.; Wang, Z. ADHD classification by dual subspace learning using resting-state functional connectivity. Artif. Intell. Med. 2020, 103, 101786. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Gheisari, M.; Najafabadi, H.E.; Alzubi, J.A.; Gao, J.; Wang, G.; Abbasi, A.A.; Castiglione, A. OBPP: An ontology-based framework for privacy-preserving in IoT-based smart city. Future Gener. Comput. Syst. 2021, 123, 1–13. [Google Scholar] [CrossRef]
- Gao, J.; Wang, H.; Shen, H. Machine learning based workload prediction in cloud computing. In Proceedings of the 2020 29th International Conference on Computer Communications and Networks (ICCCN), Honolulu, HI, USA, 3–6 August 2020; pp. 1–9. [Google Scholar]
- Gao, J.; Wang, H.; Shen, H. Smartly handling renewable energy instability in supporting a cloud datacenter. In Proceedings of the 2020 IEEE International Parallel and Distributed Processing Symposium (IPDPS), New Orleans, LA, USA, 18–22 May 2020; pp. 769–778. [Google Scholar]
- Gao, J.; Wang, H.; Shen, H. Task failure prediction in cloud data centers using deep learning. IEEE Trans. Serv. Comput. 2020. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Mao, Z.; Su, Y.; Xu, G.; Wang, X.; Huang, Y.; Yue, W.; Sun, L.; Xiong, N. Spatio-temporal deep learning method for ADHD fMRI classification. Inf. Sci. 2019, 499, 1–11. [Google Scholar] [CrossRef]
- Riaz, A.; Asad, M.; Alonso, E.; Slabaugh, G. DeepFMRI: End-to-end deep learning for functional connectivity and classification of ADHD using fMRI. J. Neurosci. Methods 2020, 335, 108506. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhao, L.; Wang, X.; Xin, Q.; Zhao, J.; Guttery, D.S.; Zhang, Y.D. Deep Spatio-Temporal Representation and Ensemble Classification for Attention deficit/Hyperactivity disorder. IEEE Trans. Neural Syst. Rehabil. Eng. 2020, 29, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bellec, P.; Chu, C.; Chouinard-Decorte, F.; Benhajali, Y.; Margulies, D.S.; Craddock, R.C. The neuro bureau ADHD-200 preprocessed repository. NeuroImage 2017, 144, 275–286. [Google Scholar] [CrossRef]
- Lavoie-Courchesne, S.; Rioux, P.; Chouinard-Decorte, F.; Sherif, T.; Rousseau, M.E.; Das, S.; Adalat, R.; Doyon, J.; Craddock, C.; Margulies, D.; et al. Integration of a neuroimaging processing pipeline into a pan-canadian computing grid. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, , 2012; Volume 341, p. 012032. [Google Scholar]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Bellec, P.; Perlbarg, V.; Jbabdi, S.; Pélégrini-Issac, M.; Anton, J.L.; Doyon, J.; Benali, H. Identification of large-scale networks in the brain using fMRI. NeuroImage 2006, 29, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Frey, B.J.; Dueck, D. Clustering by passing messages between data points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benesty, J.; Chen, J.; Huang, Y.; Cohen, I. Pearson correlation coefficient. In Noise Reduction in Speech Processing; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–4. [Google Scholar]
- Rajpoot, K.; Riaz, A.; Majeed, W.; Rajpoot, N. Functional connectivity alterations in epilepsy from resting-state functional MRI. PLoS ONE 2015, 10, e0134944. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Mu, C.; Sun, C.; Yang, W.; Yang, X.; Zuo, X. Support vector machine-based optimized decision threshold adjustment strategy for classifying imbalanced data. Knowl. Based Syst. 2015, 76, 67–78. [Google Scholar] [CrossRef]
- Zhang, Y.; Fu, P.; Liu, W.; Chen, G. Imbalanced data classification based on scaling kernel-based support vector machine. Neural Comput. Appl. 2014, 25, 927–935. [Google Scholar] [CrossRef]
- Dey, S.; Rao, A.R.; Shah, M. Attributed graph distance measure for automatic detection of attention deficit hyperactive disordered subjects. Front. Neural Circ. 2014, 8, 64. [Google Scholar] [CrossRef] [Green Version]
- Nunez-Garcia, M.; Simpraga, S.; Jurado, M.A.; Garolera, M.; Pueyo, R.; Igual, L. FADR: Functional-anatomical discriminative regions for rest fMRI characterization. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2015; pp. 61–68. [Google Scholar]
- Hao, A.J.; He, B.L.; Yin, C.H. Discrimination of ADHD children based on Deep Bayesian Network. In Proceedings of the 2015 IET International Conference on Biomedical Image and Signal Processing (ICBISP 2015), Beijing, China, 19 November 2015; pp. 1–6. [Google Scholar]
- Bondy, J.A.; Murty, U.S.R. Graph Theory with Applications; Macmillan: London, UK, 1976; Volume 290. [Google Scholar]
- Xia, M.; Wang, J.; He, Y. BrainNet Viewer: A network visualization tool for human brain connectomics. PLoS ONE 2013, 8, e68910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolls, E.T.; Cheng, W.; Feng, J. Brain dynamics: The temporal variability of connectivity, and differences in schizophrenia and ADHD. Transl. Psychiatry 2021, 11, 70. [Google Scholar] [CrossRef]
- Jie, B.; Wee, C.Y.; Shen, D.; Zhang, D. Hyper-connectivity of functional networks for brain disease diagnosis. Med. Image Anal. 2016, 32, 84–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyeong, S.; Park, S.; Cheon, K.A.; Kim, J.J.; Song, D.H.; Kim, E. A new approach to investigate the association between brain functional connectivity and disease characteristics of attention-deficit/hyperactivity disorder: Topological neuroimaging data analysis. PLoS ONE 2015, 10, e0137296. [Google Scholar] [CrossRef]
- Liston, C.; Cohen, M.M.; Teslovich, T.; Levenson, D.; Casey, B. Atypical prefrontal connectivity in attention-deficit/hyperactivity disorder: Pathway to disease or pathological end point? Biol. Psychiatry 2011, 69, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Kumar, U.; Arya, A.; Agarwal, V. Neural network connectivity in ADHD children: An independent component and functional connectivity analysis of resting state fMRI data. Brain Imaging Behav. 2021, 15, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Aradhya, A.M.; Subbaraju, V.; Sundaram, S.; Sundararajan, N. Discriminant Spatial Filtering Method (DSFM) for the identification and analysis of abnormal resting state brain activities. Expert Syst. Appl. 2021, 181, 115074. [Google Scholar] [CrossRef]
| Training | Testing | ||||||
|---|---|---|---|---|---|---|---|
| Sr | Imaging Site | ADHD | HC | Total | ADHD | HC | Total |
| 1 | KKI | 18 | 51 | 69 | 3 | 8 | 11 |
| 2 | NI | 25 | 23 | 48 | 11 | 14 | 25 |
| 3 | NYU | 118 | 98 | 216 | 29 | 12 | 41 |
| 4 | OHSU | 37 | 42 | 79 | 6 | 28 | 34 |
| 5 | Peking_1 | 24 | 61 | 85 | 24 | 27 | 51 |
| Total | 497 | 162 | |||||
| Top Features | SVM | LDC | DTC | DTC |
|---|---|---|---|---|
| 1 | 0.49 | 0.5 | 0.49 | 0.49 |
| 5 | 0.53 | 0.55 | 0.51 | 0.53 |
| 10 | 0.54 | 0.56 | 0.52 | 0.54 |
| 15 | 0.55 | 0.56 | 0.53 | 0.56 |
| 20 | 0.55 | 0.57 | 0.54 | 0.56 |
| Top Features | SVM | LDC | DTC | DTC |
|---|---|---|---|---|
| 100 | 0.55 | 0.53 | 0.54 | 0.53 |
| 500 | 0.55 | 0.52 | 0.55 | 0.53 |
| 1000 | 0.54 | 0.52 | 0.52 | 0.52 |
| 1500 | 0.52 | 0.51 | 0.48 | 0.45 |
| 3000 | 0.3 | 0.2 | 0.4 | 0.3 |
| Classifier | Selected Features |
|---|---|
| SVM | 22 |
| LDC | 43 |
| DTC | 589 |
| RFC | 64 |
| Trained on Each Site | Trained on Combined Sites | ||||
|---|---|---|---|---|---|
| Sr | Site | Proposed | DeepFMRI (2020) [69] | Proposed | DeepFMRI (2020) [69] |
| 1 | NYU | 73 | 73.1 | 56 | 65.8 |
| 2 | NI | 70 | 67.9 | 64 | 60 |
| 3 | Peking | 60 | 62.7 | 60 | 43.1 |
| 4 | OHSU | 71 | - | 56 | - |
| 5 | KKI | 72 | - | 81 | - |
| Method | NYU | NI | Peking | OHSU | KKI |
|---|---|---|---|---|---|
| Proposed | 73.3 | 70 | 73.3 | 71 | 60 |
| DeepFMRI (2020) [69] | 73.1 | 67.9 | 62.7 | - | - |
| SC-CNN-ATT (2020) [42] | 77.7 | 75.3 | 60.4 | - | 65.2 |
| FCNet (2018) [40] | 67.4 | 72.9 | 25.4 | - | 85.3 |
| Nunez et al. (2015) [81] | - | - | 56 | - | 58 |
| AJHao et al. (2015) [82] | 64.7 | - | 66.3 | - | 59 |
| Dey et al. (2014) [80] | 81 | - | 56 | - | 58 |
| ADHD-200 (2012) [38] | - | 56.9 | 35.1 | - | 51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, N.A.; Waheeb, S.A.; Riaz, A.; Shang, X. A Novel Knowledge Distillation-Based Feature Selection for the Classification of ADHD. Biomolecules 2021, 11, 1093. https://doi.org/10.3390/biom11081093
Khan NA, Waheeb SA, Riaz A, Shang X. A Novel Knowledge Distillation-Based Feature Selection for the Classification of ADHD. Biomolecules. 2021; 11(8):1093. https://doi.org/10.3390/biom11081093
Chicago/Turabian StyleKhan, Naseer Ahmed, Samer Abdulateef Waheeb, Atif Riaz, and Xuequn Shang. 2021. "A Novel Knowledge Distillation-Based Feature Selection for the Classification of ADHD" Biomolecules 11, no. 8: 1093. https://doi.org/10.3390/biom11081093
APA StyleKhan, N. A., Waheeb, S. A., Riaz, A., & Shang, X. (2021). A Novel Knowledge Distillation-Based Feature Selection for the Classification of ADHD. Biomolecules, 11(8), 1093. https://doi.org/10.3390/biom11081093






