Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition
Abstract
1. Introduction
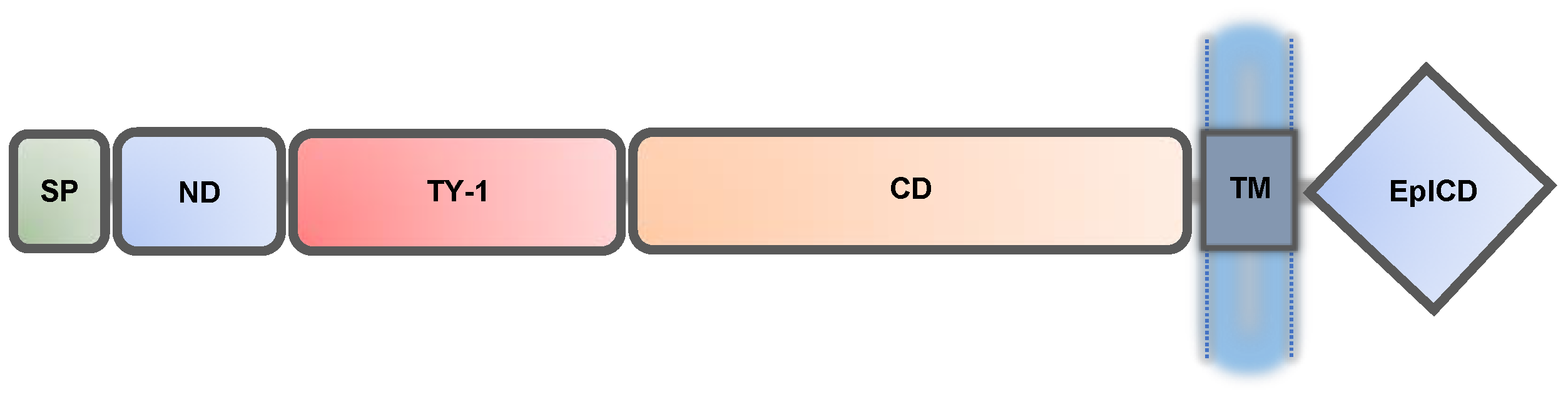
2. EpCAM Forms Intercellular Adhesions and Promotes Cellular Segregation and Aggregation
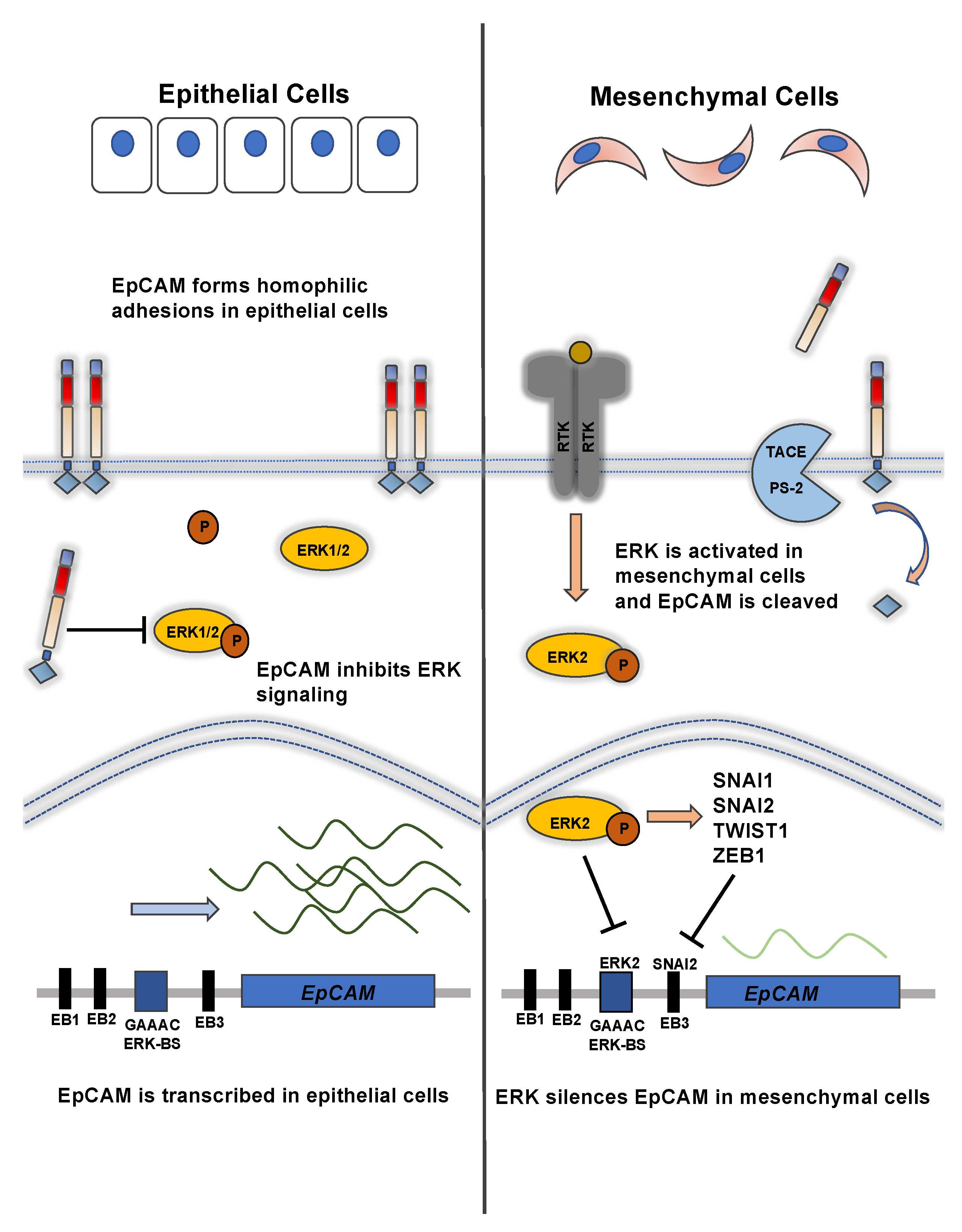
3. Transcriptional Control of EpCAM Expression and its Downregulation during EMT
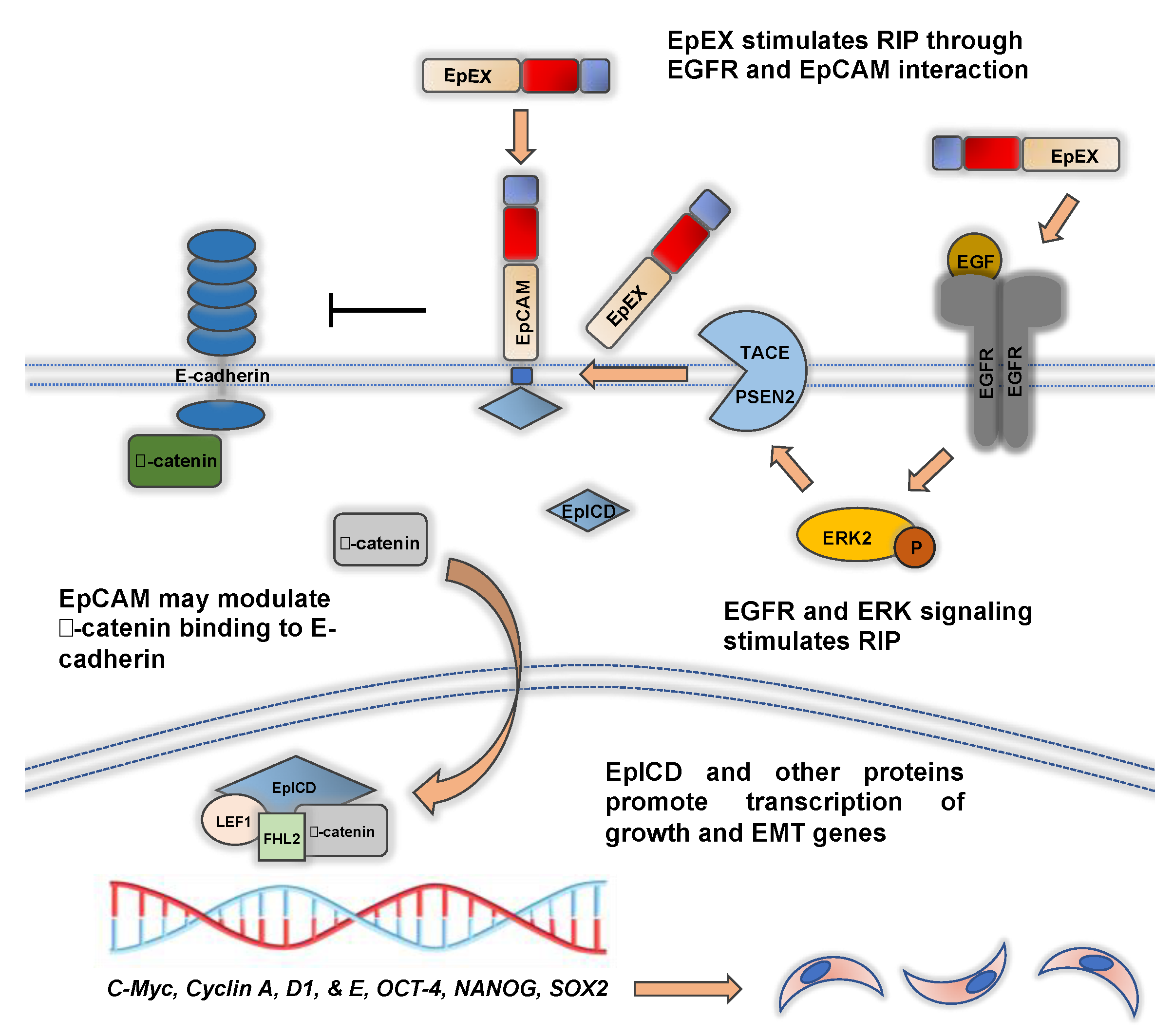
4. Regulated Intramembrane Proteolysis of EpCAM and EpICD Signaling during EMT
5. EpCAM Is Regulated by EGFR Signaling during EMT and EpEX Is an EGFR Ligand that Affects EMT
6. EpCAM Mediates Migration and Invasion through Various Signaling Pathways and Mechanisms
7. EpCAM Regulates the Formation of Epithelial Junctional Protein Complexes and May Affect EMT
8. EpCAM Expression in Epithelial and Mesenchymal Cancers
9. EpCAM, Circulating Tumor Cells, and EMT
10. EpCAM, Cancer Stem Cells, and EMT
11. EpCAM and Drug Resistance during EMT
12. Future Directions
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADAM17 | ADAM metallopeptidase 17 |
| AP-1 | activator protein 1 |
| ATC | anaplastic thyroid cancer |
| BAA | bead aggregation assays |
| CAM | cell adhesion molecule |
| CD | carboxyl-terminal domain |
| CSC | cancer stem cells |
| CTC | circulating tumor cells |
| CTE | congenital tufting enteropathy |
| CTSL | cathepsins L |
| DTCs | disseminated tumor cells |
| ECM | extracellular matrix |
| EGFR | epidermal growth factor receptor |
| ERK | extracellular signal-regulated kinases |
| EpCAM | epithelial cell adhesion molecular |
| EpEX | EpCAM extracellular domain |
| EpICD | EpCAM intracellular domain |
| FHL2 | four-and-a-half LIM domains 2 |
| FILM-FRET | fluorescence-lifetime imaging based Förster resonance energy transfer |
| EMT | epithelial-to-mesenchymal transition |
| JMJD2D | histone lysine demethylase 4D |
| kb | kilobases |
| kD | kilodalton |
| LC | Langerhans cells |
| LEF1 | lymphoid enhancer binder factor 1 |
| MET | mesenchymal-to-epithelial transition (MET) |
| MMPs | matrix metallopeptidases |
| ND | N-terminal domain |
| nPKC | novel protein kinase C |
| OCT4 | octamer-binding transcription factor 4 |
| PDTC | poorly differentiated thyroid cancer |
| PSEN2 | presenilin 2 |
| RIP | regulated intramembrane proteolysis |
| shRNA | short-hairpin RNA |
| SAXS | small angle X-ray scattering |
| SP | signal peptide |
| TACE | tumor necrosis factor-α-converting enzyme |
| TCF4 | transcription factor 4 |
| TGF-β1 | transforming growth factor- β1 |
| TNBC | triple negative breast cancer |
| TM | transmembrane |
| TY-1 | type-1 thyroglobulin |
| ZO-1 | zona occludens-1 |
References
- Schnell, U.; Cirulli, V.; Giepmans, B.N. EpCAM: Structure and function in health and disease. Biochim. Biophys. Acta (BBA) 2013, 1828, 1989–2001. [Google Scholar] [CrossRef]
- Momburg, F.; Moldenhauer, G.; Hämmerling, G.J.; Möller, P. Immunohistochemical study of the expression of a Mr 34,000 human epithelium-specific surface glycoprotein in normal and malignant tissues. Cancer Res. 1987, 47, 2883–2891. [Google Scholar] [PubMed]
- Moldenhauer, G.; Momburg, F.; Möller, P.; Schwartz, R.; Hämmerling, G.J. Epithelium-specific surface glycoprotein of Mr 34,000 is a widely distributed human carcinoma marker. Br. J. Cancer 1987, 56, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Nagao, K.; Zhu, J.; Heneghan, M.B.; Hanson, J.C.; Morasso, M.I.; Tessarollo, L.; Mackem, S.; Udey, M.C. Abnormal Placental Development and Early Embryonic Lethality in EpCAM-Null Mice. PLoS ONE 2009, 4, e8543. [Google Scholar] [CrossRef]
- Das, B.; Sivagnanam, M. Congenital Tufting Enteropathy: Biology, Pathogenesis and Mechanisms. J. Clin. Med. 2020, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Schnell, U.; Kuipers, J.; Mueller, J.L.; Veenstra-Algra, A.; Sivagnanam, M.; Giepmans, B.N. Absence of cell-surface EpCAM in congenital tufting enteropathy. Hum. Mol. Genet. 2013, 22, 2566–2571. [Google Scholar] [CrossRef]
- Sivagnanam, M.; Mueller, J.L.; Lee, H.; Chen, Z.; Nelson, S.F.; Turner, D.; Zlotkin, S.H.; Pencharz, P.B.; Ngan, B.; Libiger, O.; et al. Identification of EpCAM as the Gene for Congenital Tufting Enteropathy. Gastroenterology 2008, 135, 429–437. [Google Scholar] [CrossRef]
- Patey, N.; Scoazec, J.Y.; Cuenod-Jabri, B.; Canioni, D.; Kedinger, M.; Goulet, O.; Brousse, N. Distribution of cell adhesion molecules in infants with intestinal epithelial dysplasia (tufting enteropathy). Gastroenterology 1997, 113, 833–843. [Google Scholar] [CrossRef]
- Guerra, E.; Lattanzio, R.; La Sorda, R.; Dini, F.; Tiboni, G.M.; Piantelli, M.; Alberti, S. mTrop1/Epcam Knockout Mice Develop Congenital Tufting Enteropathy through Dysregulation of Intestinal E-cadherin/β-catenin. PLoS ONE 2012, 7, e49302. [Google Scholar] [CrossRef]
- Mueller, J.L.; McGeough, M.D.; Peña, C.A.; Sivagnanam, M. Functional consequences of EpCam mutation in mice and men. Am. J. Physiol. Liver Physiol. 2014, 306, G278–G288. [Google Scholar] [CrossRef]
- Kozan, P.A.; McGeough, M.D.; Pena, C.A.; Mueller, J.L.; Barrett, K.E.; Marchelletta, R.R.; Sivagnanam, M. Mutation of EpCAM leads to intestinal barrier and ion transport dysfunction. J. Mol. Med. 2015, 93, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Maetzel, D.; Denzel, S.; Mack, B.; Canis, M.; Went, P.T.; Benk, M.; Kieu, C.; Papior, P.; Baeuerle, P.A.; Munz, M.; et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 2009, 11, 162–171. [Google Scholar] [CrossRef]
- Pan, M.; Schinke, H.; Luxenburger, E.; Kranz, G.; Shakhtour, J.; Libl, D.; Huang, Y.; Gaber, A.; Pavšič, M.; Lenarčič, B.; et al. EpCAM ectodomain EpEX is a ligand of EGFR that counteracts EGF-mediated epithelial-mesenchymal transition through modulation of phospho-ERK1/2 in head and neck cancers. PLoS Biol. 2018, 16, e2006624. [Google Scholar] [CrossRef] [PubMed]
- Herlyn, M.; Steplewski, Z.; Herlyn, D.; Koprowski, H. Colorectal carcinoma-specific antigen: Detection by means of monoclonal antibodies. Proc. Natl. Acad. Sci. USA 1979, 76, 1438–1442. [Google Scholar] [CrossRef] [PubMed]
- Went, P.T.; Lugli, A.; Meier, S.; Bundi, M.; Mirlacher, M.; Sauter, G.; Dirnhofer, S. Frequent EpCam protein expression in human carcinomas. Hum. Pathol. 2004, 35, 122–128. [Google Scholar] [CrossRef]
- Sears, H.; Mattis, J.; Herlyn, D.; Häyry, P.; Atkinson, B.; Ernst, C.; Steplewski, Z.; Koprowski, H. Phase-I clinical trial of monoclonal antibody in treatment of gastrointestinal tumours. Lancet 1982, 319, 762–765. [Google Scholar] [CrossRef]
- Riethmüller, G.; Holz, E.; Schlimok, G.; Schmiegel, W.; Raab, R.; Höffken, K.; Gruber, R.; Funke, I.; Pichlmaier, H.; Hirche, H.; et al. Monoclonal antibody therapy for resected Dukes’ C colorectal cancer: Seven-year outcome of a multicenter randomized trial. J. Clin. Oncol. 1998, 16, 1788–1794. [Google Scholar] [CrossRef] [PubMed]
- Linke, R.; Klein, A.; Seimetz, D. Catumaxomab. mAbs 2010, 2, 129–136. [Google Scholar] [CrossRef]
- Ruf, P.; Kluge, M.; Jäger, M.; Burges, A.; Volovat, C.; Heiss, M.M.; Hess, J.; Wimberger, P.; Brandt, B.; Lindhofer, H. Pharmacokinetics, immunogenicity and bioactivity of the therapeutic antibody catumaxomab intraperitoneally administered to cancer patients. Br. J. Clin. Pharmacol. 2010, 69, 617–625. [Google Scholar] [CrossRef]
- Seeber, A.; Martowicz, A.; Spizzo, G.; Buratti, T.; Obrist, P.; Fong, D.; Gastl, G.; Untergasser, G. Soluble EpCAM levels in ascites correlate with positive cytology and neutralize catumaxomab activity in vitro. BMC Cancer 2015, 15, 1–12. [Google Scholar] [CrossRef][Green Version]
- Knödler, M.; Körfer, J.; Kunzmann, V.; Trojan, J.; Daum, S.; Schenk, M.; Kullmann, F.; Schroll, S.; Behringer, D.; Stahl, M.; et al. Randomised phase II trial to investigate catumaxomab (anti-EpCAM × anti-CD3) for treatment of peritoneal carcinomatosis in patients with gastric cancer. Br. J. Cancer 2018, 119, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Hay, E.D. An Overview of Epithelio-Mesenchymal Transformation. Cells Tissues Organs 1995, 154, 8–20. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Pavšič, M.; Gunčar, G.; Djinović-Carugo, K.; Lenarčič, B. Crystal structure and its bearing towards an understanding of key biological functions of EpCAM. Nat. Commun. 2014, 5, 4764. [Google Scholar] [CrossRef] [PubMed]
- Gaber, A.; Kim, S.J.; Kaake, R.M.; Benčina, M.; Krogan, N.; Šali, A.; Pavšič, M.; Lenarčič, B. EpCAM homo-oligomerization is not the basis for its role in cell-cell adhesion. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, S.V.; Velders, M.P.; Bakker, H.A.; Fleuren, G.J.; Warnaar, S.O. Ep-CAM: A human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 1994, 125, 437–446. [Google Scholar] [CrossRef]
- Litvinov, S.V.; Bakker, H.A.M.; Gourevitch, M.M.; Velders, M.P.; Warnaar, S.O. Evidence for a Role of the Epithelial Glycoprotein 40 (Ep-CAM) in Epithelial Cell-Cell Adhesion. Cell Adhes. Commun. 1994, 2, 417–428. [Google Scholar] [CrossRef]
- Balzar, M.; Prins, F.A.; Bakker, H.A.; Fleuren, G.J.; Warnaar, S.O.; Litvinov, S.V. The Structural Analysis of Adhesions Mediated by Ep-CAM. Exp. Cell Res. 1999, 246, 108–121. [Google Scholar] [CrossRef]
- Balzar, M.; Bakker, H.A.M.; Briaire-De-Bruijn, I.H.; Fleuren, G.J.; Warnaar, S.O.; Litvinov, S.V. Cytoplasmic Tail Regulates the Intercellular Adhesion Function of the Epithelial Cell Adhesion Molecule. Mol. Cell. Biol. 1998, 18, 4833–4843. [Google Scholar] [CrossRef]
- Trebak, M.; Begg, G.E.; Chong, J.M.; Kanazireva, E.V.; Herlyn, D.; Speicher, D.W. Oligomeric State of the Colon Carcinoma-associated Glycoprotein GA733-2 (Ep-CAM/EGP40) and Its Role in GA733-mediated Homotypic Cell-Cell Adhesion. J. Biol. Chem. 2001, 276, 2299–2309. [Google Scholar] [CrossRef] [PubMed]
- Balzar, M.; Bruijn, I.H.B.-D.; Rees-Bakker, H.A.M.; Prins, F.A.; Helfrich, W.; de Leij, L.; Riethmüller, G.; Alberti, S.; Warnaar, S.O.; Fleuren, G.J.; et al. Epidermal Growth Factor-Like Repeats Mediate Lateral and Reciprocal Interactions of Ep-CAM Molecules in Homophilic Adhesions. Mol. Cell. Biol. 2001, 21, 2570–2580. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fagotto, F.; Aslemarz, A. EpCAM cellular functions in adhesion and migration, and potential impact on invasion: A critical review. Biochim. Biophys. Acta 2020, 1874, 188436. [Google Scholar] [CrossRef]
- Van Der Gun, B.T.; Melchers, L.J.; Ruiters, M.H.; De Leij, L.F.M.H.; McLaughlin, P.M.; Rots, M.G. EpCAM in carcinogenesis: The good, the bad or the ugly. Carcinogenesis 2010, 31, 1913–1921. [Google Scholar] [CrossRef] [PubMed]
- Byers, L.A.; Diao, L.; Wang, J.; Saintigny, P.; Girard, L.; Peyton, M.; Shen, L.; Fan, Y.; Giri, U.; Tumula, P.K.; et al. An Epithelial–Mesenchymal Transition Gene Signature Predicts Resistance to EGFR and PI3K Inhibitors and Identifies Axl as a Therapeutic Target for Overcoming EGFR Inhibitor Resistance. Clin. Cancer Res. 2013, 19, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Ferrell, J.E. Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 2002, 14, 140–148. [Google Scholar] [CrossRef]
- Sankpal, N.V.; Fleming, T.P.; Sharma, P.K.; Wiedner, H.J.; Gillanders, W.E. A double-negative feedback loop between EpCAM and ERK contributes to the regulation of epithelial–mesenchymal transition in cancer. Oncogene 2017, 36, 3706–3717. [Google Scholar] [CrossRef]
- Ahodantin, J.; Lekbaby, B.; Nader, M.B.; Soussan, P.; Kremsdorf, D. Hepatitis B virus X protein enhances the development of liver fibrosis and the expression of genes associated with epithelial-mesenchymal transitions and tumor progenitor cells. Carcinogenesis 2019, 41, 358–367. [Google Scholar] [CrossRef]
- Gemmill, R.M.; Roche, J.; Potiron, V.A.; Nasarre, P.; Mitas, M.; Coldren, C.D.; Helfrich, B.A.; Garrett-Mayer, E.; Bunn, P.A.; Drabkin, H.A. ZEB1-responsive genes in non-small cell lung cancer. Cancer Lett. 2011, 300, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Nasarre, P.; Gemmill, R.; Baldys, A.; Pontis, J.; Korch, C.; Guilhot, J.; Ait-Si-Ali, S.; Drabkin, H. Global Decrease of Histone H3K27 Acetylation in ZEB1-Induced Epithelial to Mesenchymal Transition in Lung Cancer Cells. Cancers 2013, 5, 334–356. [Google Scholar] [CrossRef]
- Vannier, C.; Mock, K.; Brabletz, T.; Driever, W. Zeb1 Regulates E-cadherin and Epcam (Epithelial Cell Adhesion Molecule) Expression to Control Cell Behavior in Early Zebrafish Development. J. Biol. Chem. 2013, 288, 18643–18659. [Google Scholar] [CrossRef]
- Van Der Gun, B.T.F.; De Groote, M.L.; Kazemier, H.G.; Arendzen, A.J.; Terpstra, P.; Ruiters, M.H.J.; McLaughlin, P.M.J.; Rots, M.G. Transcription factors and molecular epigenetic marks underlying EpCAM overexpression in ovarian cancer. Br. J. Cancer 2011, 105, 312–319. [Google Scholar] [CrossRef]
- Huber, M.A.; Azoitei, N.; Baumann, B.; Grunert, S.; Sommer, A.; Pehamberger, H.; Kraut, N.; Beug, H.; Wirth, T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Investig. 2004, 114, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-T.; Osmulski, P.; Wang, Y.; Huang, Y.-W.; Liu, L.; Ruan, J.; Jin, V.X.; Kirma, N.B.; Gaczynska, M.E.; Huang, T.H.-M. EpCAM-Regulated Transcription Exerts Influences on Nanomechanical Properties of Endometrial Cancer Cells That Promote Epithelial-to-Mesenchymal Transition. Cancer Res. 2016, 76, 6171–6182. [Google Scholar] [CrossRef] [PubMed]
- Rokavec, M.; Öner, M.G.; Li, H.; Jackstadt, R.; Jiang, L.; Lodygin, D.; Kaller, M.; Horst, D.; Ziegler, P.K.; Schwitalla, S.; et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 2014, 124, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Yao, W.; Yang, T.; Yang, Y.; Liu, Y.; Shen, Q.; Zhang, J.; Qi, W.; Wang, J. aPKC-ι/P-Sp1/Snail signaling induces epithelial-mesenchymal transition and immunosuppression in cholangiocarcinoma. Hepatology 2017, 66, 1165–1182. [Google Scholar] [CrossRef]
- Dittmer, J. The role of the transcription factor Ets1 in carcinoma. Semin. Cancer Biol. 2015, 35, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Tillo, E.; de Barrios, O.; Siles, L.; Cuatrecasas, M.; Castells, A.; Postigo, A. β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc. Natl. Acad. Sci. USA 2011, 108, 19204–19209. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Budhu, A.; Forgues, M.; Wang, X.W. Activation of Hepatic Stem Cell Marker EpCAM by Wnt–β-Catenin Signaling in Hepatocellular Carcinoma. Cancer Res. 2007, 67, 10831–10839. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Zhang, X.; Wang, H.; Rui, D.; Yin, A.; Qiu, G.; He, Y. CpG island methylation status in the EpCAM promoter region and gene expression. Oncol. Rep. 1994, 20, 1061–1067. [Google Scholar] [CrossRef]
- Shiah, S.-G.; Chang, L.-C.; Tai, K.-Y.; Lee, G.-H.; Wu, C.-W.; Shieh, Y.-S. The involvement of promoter methylation and DNA methyltransferase-1 in the regulation of EpCAM expression in oral squamous cell carcinoma. Oral Oncol. 2009, 45, e1–e8. [Google Scholar] [CrossRef]
- Sankpal, N.V.; Willman, M.W.; Fleming, T.P.; Mayfield, J.D.; Gillanders, W.E. Transcriptional Repression of Epithelial Cell Adhesion Molecule Contributes to p53 Control of Breast Cancer Invasion. Cancer Res. 2009, 69, 753–757. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nasr, A.F.; Nutini, M.; Palombo, B.; Guerra, E.; Alberti, S. Mutations of TP53 induce loss of DNA methylation and amplification of the TROP1 gene. Oncogene 2003, 22, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Li, M.; Zhuo, M.; Guo, P.; Chen, Q.; Mo, P.; Li, W.; Yu, C. Histone demethylase JMJD2D promotes the self-renewal of liver cancer stem-like cells by enhancing EpCAM and Sox9 expression. J. Biol. Chem. 2021, 296, 100121. [Google Scholar] [CrossRef] [PubMed]
- Denzel, S.; Maetzel, D.; Mack, B.; Eggert, C.; Bärr, G.; Gires, O. Initial activation of EpCAM cleavage via cell-to-cell contact. BMC Cancer 2009, 9, 402. [Google Scholar] [CrossRef]
- Liang, K.-H.; Tso, H.-C.; Hung, S.-H.; Kuan, I.-I.; Lai, J.-K.; Ke, F.-Y.; Chuang, Y.-T.; Liu, I.-J.; Wang, Y.-P.; Chen, R.-H.; et al. Extracellular domain of EpCAM enhances tumor progression through EGFR signaling in colon cancer cells. Cancer Lett. 2018, 433, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Pérez, A.; Mack, B.; Maetzel, D.; Kremling, H.; Eggert, C.H.; Harreus, U.; Gires, O. EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene 2013, 32, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-W.; Liao, M.-Y.; Lin, W.-W.; Wang, Y.-P.; Lu, T.-Y.; Wu, H.-C. Epithelial Cell Adhesion Molecule Regulates Tumor Initiation and Tumorigenesis via Activating Reprogramming Factors and Epithelial-Mesenchymal Transition Gene Expression in Colon Cancer. J. Biol. Chem. 2012, 287, 39449–39459. [Google Scholar] [CrossRef]
- Huang, Y.; Chanou, A.; Kranz, G.; Pan, M.; Kohlbauer, V.; Ettinger, A.; Gires, O. Membrane-associated epithelial cell adhesion molecule is slowly cleaved by γ-secretase prior to efficient proteasomal degradation of its intracellular domain. J. Biol. Chem. 2019, 294, 3051–3064. [Google Scholar] [CrossRef]
- Nelson, W.J. Convergence of Wnt, -Catenin, and Cadherin Pathways. Science 2004, 303, 1483–1487. [Google Scholar] [CrossRef]
- Osta, W.A.; Chen, Y.; Mikhitarian, K.; Mitas, M.; Salem, M.; Hannun, Y.A.; Cole, D.J.; Gillanders, W.E. EpCAM Is Overexpressed in Breast Cancer and Is a Potential Target for Breast Cancer Gene Therapy. Cancer Res. 2004, 64, 5818–5824. [Google Scholar] [CrossRef]
- Shi, R.; Liu, L.; Wang, F.; He, Y.; Niu, Y.; Wang, C.; Zhang, X.; Zhang, X.; Zhang, H.; Chen, M.; et al. Downregulation of cytokeratin 18 induces cellular partial EMT and stemness through increasing EpCAM expression in breast cancer. Cell. Signal. 2020, 76, 109810. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Hopfinger, N.R.; Nguyen, T.D.; Pogash, T.J.; Santucci-Pereira, J.; Russo, J. Epigenetic reprogramming of epithelial mesenchymal transition in triple negative breast cancer cells with DNA methyltransferase and histone deacetylase inhibitors. J. Exp. Clin. Cancer Res. 2018, 37, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, R.T.; Kaur, J.; Leong, I.; Macmillan, C.; Witterick, I.J.; Walfish, P.G.; Ralhan, R. Subcellular differential expression of Ep-ICD in oral dysplasia and cancer is associated with disease progression and prognosis. BMC Cancer 2016, 16, 486. [Google Scholar] [CrossRef]
- Kunavisarut, T.; Kak, I.; Macmillan, C.; Ralhan, R.; Walfish, P.G. Immunohistochemical analysis based Ep-ICD subcellular localization index (ESLI) is a novel marker for metastatic papillary thyroid microcarcinoma. BMC Cancer 2012, 12, 523. [Google Scholar] [CrossRef]
- Srivastava, G.; Assi, J.; Kashat, L.; Matta, A.; Chang, M.; Walfish, P.G.; Ralhan, R. Nuclear Ep-ICD accumulation predicts aggressive clinical course in early stage breast cancer patients. BMC Cancer 2014, 14, 726. [Google Scholar] [CrossRef]
- Jachin, S.; Bae, J.S.; Sung, J.J.; Park, H.S.; Jang, K.Y.; Chung, M.J.; Kim, D.G.; Moon, W.S. The role of nuclear EpICD in extrahepatic cholangiocarcinoma: Association with β-catenin. Int. J. Oncol. 2014, 45, 691–698. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fong, D.; Moser, P.; Kasal, A.; Seeber, A.; Gastl, G.; Martowicz, A.; Wurm, M.; Mian, C.; Obrist, P.; Mazzoleni, G.; et al. Loss of membranous expression of the intracellular domain of EpCAM is a frequent event and predicts poor survival in patients with pancreatic cancer. Histopathology 2014, 64, 683–692. [Google Scholar] [CrossRef]
- Ralhan, R.; He, H.C.-H.; So, A.K.-C.; Tripathi, S.C.; Kumar, M.; Hasan, R.; Kaur, J.; Kashat, L.; Macmillan, C.; Chauhan, S.S.; et al. Nuclear and Cytoplasmic Accumulation of Ep-ICD Is Frequently Detected in Human Epithelial Cancers. PLoS ONE 2010, 5, e14130. [Google Scholar] [CrossRef]
- Assi, J.; Srivastava, G.; Matta, A.; Macmillan, C.; Ralhan, R.; Walfish, P.G. Nuclear Ep-ICD Expression Is a Predictor of Poor Prognosis in “Low Risk” Prostate Adenocarcinomas. PLoS ONE 2015, 10, e0107586. [Google Scholar] [CrossRef]
- Hsu, Y.-T.; Gu, F.; Huang, Y.-W.; Liu, J.; Ruan, J.; Huang, R.-L.; Wang, C.-M.; Chen, C.-L.; Jadhav, R.; Lai, H.-C.; et al. Promoter Hypomethylation of EpCAM-Regulated Bone Morphogenetic Protein Gene Family in Recurrent Endometrial Cancer. Clin. Cancer Res. 2013, 19, 6272–6285. [Google Scholar] [CrossRef]
- Chen, H.-N.; Liang, K.-H.; Lai, J.-K.; Lan, C.-H.; Liao, M.-Y.; Hung, S.-H.; Chuang, Y.-T.; Chen, K.-C.; Tsuei, W.W.-F.; Wu, H.-C. EpCAM Signaling Promotes Tumor Progression and Protein Stability of PD-L1 through the EGFR Pathway. Cancer Res. 2020, 80, 5035–5050. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhan, H. Communication between EMT and PD-L1 signaling: New insights into tumor immune evasion. Cancer Lett. 2020, 468, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Kuan, I.-I.; Lee, C.-C.; Chen, C.-H.; Lu, J.; Kuo, Y.-S.; Wu, H.-C. The extracellular domain of epithelial cell adhesion molecule (EpCAM) enhances multipotency of mesenchymal stem cells through EGFR–LIN28–LET7 signaling. J. Biol. Chem. 2019, 294, 7769–7786. [Google Scholar] [CrossRef]
- Wang, M.-H.; Sun, R.; Zhou, X.-M.; Zhang, M.-Y.; Lu, J.-B.; Yang, Y.; Zeng, L.-S.; Yang, X.-Z.; Shi, L.; Xiao, R.-W.; et al. Epithelial cell adhesion molecule overexpression regulates epithelial-mesenchymal transition, stemness and metastasis of nasopharyngeal carcinoma cells via the PTEN/AKT/mTOR pathway. Cell Death Dis. 2018, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Driemel, C.; Kremling, H.; Schumacher, S.; Will, D.; Wolters, J.; Lindenlauf, N.; Mack, B.; Baldus, S.A.; Hoya, V.; Pietsch, J.M.; et al. Context-dependent adaption of EpCAM expression in early systemic esophageal cancer. Oncogene 2013, 33, 4904–4915. [Google Scholar] [CrossRef]
- Zhou, N.; Wang, H.; Liu, H.; Xue, H.; Lin, F.; Meng, X.; Liang, A.; Zhao, Z.; Liu, Y.; Qian, H. MTA1-upregulated EpCAM is associated with metastatic behaviors and poor prognosis in lung cancer. J. Exp. Clin. Cancer Res. 2015, 34, 1–10. [Google Scholar] [CrossRef]
- Martowicz, A.; Rainer, J.; Lelong, J.; Spizzo, G.; Gastl, G.; Untergasser, G. EpCAM overexpression prolongs proliferative capacity of primary human breast epithelial cells and supports hyperplastic growth. Mol. Cancer 2013, 12, 56. [Google Scholar] [CrossRef]
- Sankpal, N.V.; Mayfield, J.D.; Willman, M.W.; Fleming, T.P.; Gillanders, W.E. Activator protein 1 (AP-1) contributes to EpCAM-dependent breast cancer invasion. Breast Cancer Res. 2011, 13, R124. [Google Scholar] [CrossRef]
- Gao, J.; Yan, Q.; Wang, J.; Liu, S.; Yang, X. Epithelial-to-Mesenchymal Transition Induced by TGF-β1 Is Mediated by AP1-Dependent EpCAM Expression in MCF-7 Cells. J. Cell. Physiol. 2015, 230, 775–782. [Google Scholar] [CrossRef]
- Sankpal, N.V.; Fleming, T.P.; Gillanders, W.E. EpCAM modulates NF-κB signaling and interleukin-8 expression in breast cancer. Mol. Cancer Res. 2013, 11, 418–426. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, L.; Liu, X.; Gao, J.; Liu, T.; Yan, Q.; Yang, X. Hypoxia modulates stem cell properties and induces EMT through N -glycosylation of EpCAM in breast cancer cells. J. Cell. Physiol. 2020, 235, 3626–3633. [Google Scholar] [CrossRef]
- Martowicz, A.; Spizzo, G.; Gastl, G.; Untergasser, G. Phenotype-dependent effects of EpCAM expression on growth and invasion of human breast cancer cell lines. BMC Cancer 2012, 12, 501. [Google Scholar] [CrossRef]
- Barth, A.I.M.; Kim, H.; Riedel-Kruse, I.H. Regulation of epithelial migration by epithelial cell adhesion molecule requires its Claudin-7 interaction domain. PLoS ONE 2018, 13, e0204957. [Google Scholar] [CrossRef]
- Fan, Q.; Cheng, J.-C.; Qiu, X.; Chang, H.-M.; Leung, P.C. EpCAM is up-regulated by EGF via ERK1/2 signaling and suppresses human epithelial ovarian cancer cell migration. Biochem. Biophys. Res. Commun. 2015, 457, 256–261. [Google Scholar] [CrossRef]
- Weng, C.-C.; Ding, P.-Y.; Liu, Y.-H.; Hawse, J.R.; Subramaniam, M.; Wu, C.-C.; Lin, Y.-C.; Chen, C.-Y.; Hung, W.-C.; Cheng, K.-H. Mutant Kras-induced upregulation of CD24 enhances prostate cancer stemness and bone metastasis. Oncogene 2019, 38, 2005–2019. [Google Scholar] [CrossRef]
- Gaiser, M.R.; Lammermann, T.; Feng, X.; Igyarto, B.Z.; Kaplan, D.H.; Tessarollo, L.; Germain, R.N.; Udey, M.C. Cancer-associated epithelial cell adhesion molecule (EpCAM.; CD326) enables epidermal Langerhans cell motility and migration in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, E889–E897. [Google Scholar] [CrossRef] [PubMed]
- Maghzal, N.; Vogt, E.; Reintsch, W.; Fraser, J.S.; Fagotto, F. The tumor-associated EpCAM regulates morphogenetic movements through intracellular signaling. J. Cell Biol. 2010, 191, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Maghzal, N.; Kayali, H.A.; Rohani, N.; Kajava, A.V.; Fagotto, F. EpCAM Controls Actomyosin Contractility and Cell Adhesion by Direct Inhibition of PKC. Dev. Cell 2013, 27, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Ozanne, B.W.; Spence, H.J.; McGarry, L.C.; Hennigan, R.F. Transcription factors control invasion: AP-1 the first among equals. Oncogene 2006, 26, 1–10. [Google Scholar] [CrossRef]
- Theveneau, E.; Mayor, R. Cadherins in collective cell migration of mesenchymal cells. Curr. Opin. Cell Biol. 2012, 24, 677–684. [Google Scholar] [CrossRef]
- Loh, C.-Y.; Chai, J.; Tang, T.; Wong, W.; Sethi, G.; Shanmugam, M.; Chong, P.; Looi, C. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Litvinov, S.V.; Balzar, M.; Winter, M.J.; Bakker, H.A.; Bruijn, I.H.B.-D.; Prins, F.; Fleuren, G.J.; Warnaar, S.O. Epithelial Cell Adhesion Molecule (Ep-CAM) Modulates Cell–Cell Interactions Mediated by Classic Cadherins. J. Cell Biol. 1997, 139, 1337–1348. [Google Scholar] [CrossRef]
- Winter, M.J.; Nagelkerken, B.; Mertens, A.E.; Rees-Bakker, H.A.; Bruijn, I.H.B.-D.; Litvinov, S.V. Expression of Ep-CAM shifts the state of cadherin-mediated adhesions from strong to weak. Exp. Cell Res. 2003, 285, 50–58. [Google Scholar] [CrossRef]
- Slanchev, K.; Carney, T.J.; Stemmler, M.P.; Koschorz, B.; Amsterdam, A.; Schwarz, H.; Hammerschmidt, M. The Epithelial Cell Adhesion Molecule EpCAM Is Required for Epithelial Morphogenesis and Integrity during Zebrafish Epiboly and Skin Development. PLoS Genet. 2009, 5, e1000563. [Google Scholar] [CrossRef] [PubMed]
- Wolburg, H.; Wolburg-Buchholz, K.; Kraus, J.; Rascher-Eggstein, G.; Liebner, S.; Hamm, S.; Duffner, F.; Grote, E.-H.; Risau, W.; Engelhardt, B. Localization of claudin-3 in tight junctions of the blood-brain barrier is selectively lost during experimental autoimmune encephalomyelitis and human glioblastoma multiforme. Acta Neuropathol. 2003, 105, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Kominsky, S.L.; Argani, P.; Korz, D.; Evron, E.; Raman, V.; Garrett, E.; Rein, A.; Sauter, G.; Kallioniemi, O.-P.; Sukumar, S. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene 2003, 22, 2021–2033. [Google Scholar] [CrossRef] [PubMed]
- Michl, P.; Barth, C.; Buchholz, M.; Lerch, M.M.; Rolke, M.; Holzmann, K.-H.; Menke, A.; Fensterer, H.; Giehl, K.; Löhr, M.; et al. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003, 63, 6265–6271. [Google Scholar]
- Ladwein, M.; Pape, U.-F.; Schmidt, D.-S.; Schnölzer, M.; Fiedler, S.; Langbein, L.; Franke, W.W.; Moldenhauer, G.; Zöller, M. The cell–cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp. Cell Res. 2005, 309, 345–357. [Google Scholar] [CrossRef]
- Wu, C.-J.; Mannan, P.; Lu, M.; Udey, M.C. Epithelial Cell Adhesion Molecule (EpCAM) Regulates Claudin Dynamics and Tight Junctions. J. Biol. Chem. 2013, 288, 12253–12268. [Google Scholar] [CrossRef]
- Kuhn, S.; Koch, M.; Nübel, T.; Ladwein, M.; Antolovic, D.; Klingbeil, P.; Hildebrand, D.; Moldenhauer, G.; Langbein, L.; Franke, W.W.; et al. A Complex of EpCAM, Claudin-7, CD44 Variant Isoforms, and Tetraspanins Promotes Colorectal Cancer Progression. Mol. Cancer Res. 2007, 5, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Nübel, T.; Preobraschenski, J.; Tuncay, H.; Weiss, T.; Kuhn, S.; Ladwein, M.; Langbein, L.; Zöller, M. Claudin-7 Regulates EpCAM-Mediated Functions in Tumor Progression. Mol. Cancer Res. 2009, 7, 285–299. [Google Scholar] [CrossRef]
- Philip, R.; Heiler, S.; Mu, W.; Büchler, M.W.; Zöller, M.; Thuma, F. Claudin-7 promotes the epithelial–mesenchymal transition in human colorectal cancer. Oncotarget 2014, 6, 2046–2063. [Google Scholar] [CrossRef]
- Craig, S.E.; Brady-Kalnay, S.M. Cancer Cells Cut Homophilic Cell Adhesion Molecules and Run. Cancer Res. 2010, 71, 303–309. [Google Scholar] [CrossRef]
- Varga, M.; Obrist, P.; Schneeberger, S.; Mühlmann, G.; Felgel-Farnholz, C.; Fong, D.; Zitt, M.; Brunhuber, T.; Schäfer, G.; Gastl, G.; et al. Overexpression of Epithelial Cell Adhesion Molecule Antigen in Gallbladder Carcinoma Is an Independent Marker for Poor Survival. Clin. Cancer Res. 2004, 10, 3131–3136. [Google Scholar] [CrossRef]
- Kroepil, F.; Dulian, A.; Vallböhmer, D.; Geddert, H.; Krieg, A.; Vay, C.; Topp, S.A.; Esch, J.S.A.; Baldus, S.E.; Gires, O.; et al. High EpCAM expression is linked to proliferation and lauren classification in gastric cancer. BMC Res. Notes 2013, 6, 253. [Google Scholar] [CrossRef]
- Fong, D.; Steurer, M.; Obrist, P.; Barbieri, V.; Margreiter, R.; Amberger, A.; Laimer, K.; Gastl, G.; Tzankov, A.; Spizzo, G. Ep-CAM expression in pancreatic and ampullary carcinomas: Frequency and prognostic relevance. J. Clin. Pathol. 2007, 61, 31–35. [Google Scholar] [CrossRef]
- Mukherjee, S.; Richardson, A.M.; Rodriguez-Canales, J.; Ylaya, K.; Erickson, H.S.; Player, A.; Kawasaki, E.S.; Pinto, P.A.; Choyke, P.L.; Merino, M.J.; et al. Identification of EpCAM as a Molecular Target of Prostate Cancer Stroma. Am. J. Pathol. 2009, 175, 2277–2287. [Google Scholar] [CrossRef]
- Massoner, P.; Thomm, T.; Mack, B.; Untergasser, G.; Martowicz, A.; Bobowski, K.; Klocker, H.; Gires, O.; Puhr, M. EpCAM is overexpressed in local and metastatic prostate cancer, suppressed by chemotherapy and modulated by MET-associated miRNA-200c/205. Br. J. Cancer 2014, 111, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Herlyn, R.; Herlyn, M.; Steplewski, Z.; Koprowski, H. Monoclonal antibodies in cell-mediated cytotoxicity against human melanoma and colorectal carcinoma. Eur. J. Immunol. 1979, 9, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, G.; Fong, D.; Wurm, M.; Ensinger, C.; Obrist, P.; Hofer, C.; Mazzoleni, G.; Gastl, G.; Went, P. EpCAM expression in primary tumour tissues and metastases: An immunohistochemical analysis. J. Clin. Pathol. 2011, 64, 415–420. [Google Scholar] [CrossRef]
- Tombolan, L.; Rossi, E.; Zin, A.; Santoro, L.; Bonvini, P.; Zamarchi, R.; Bisogno, G. Pediatric sarcomas display a variable EpCAM expression in a histology-dependent manner. Transl. Oncol. 2020, 13, 100846. [Google Scholar] [CrossRef]
- Cimino, A.; Halushka, M.; Illei, P.; Wu, X.; Sukumar, S.; Argani, P. Epithelial cell adhesion molecule (EpCAM) is overexpressed in breast cancer metastases. Breast Cancer Res. Treat. 2009, 123, 701–708. [Google Scholar] [CrossRef]
- Spizzo, G.; Gastl, G.; Obrist, P.; Went, P.; Dirnhofer, S.; Bischoff, S.; Mirlacher, M.; Sauter, G.; Simon, R.; Stopatschinskaya, S.; et al. High Ep-CAM Expression is Associated with Poor Prognosis in Node-positive Breast Cancer. Breast Cancer Res. Treat. 2004, 86, 207–213. [Google Scholar] [CrossRef]
- Gastl, G.; Spizzo, G.; Obrist, P.; Dünser, M.; Mikuz, G. Ep-CAM overexpression in breast cancer as a predictor of survival. Lancet 2000, 356, 1981–1982. [Google Scholar] [CrossRef]
- Soysal, S.D.; Muenst, S.; Barbie, T.; Fleming, T.; Gao, F.; Spizzo, G.; Oertli, D.; Viehl, C.T.; Obermann, E.C.; Gillanders, W.E. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2+, basal-like, and HER2 intrinsic subtypes of breast cancer. Br. J. Cancer 2013, 108, 1480–1487. [Google Scholar] [CrossRef] [PubMed]
- Reed, A.E.M.; Kutasovic, J.R.; Vargas, A.C.; Jayanthan, J.; Al-Murrani, A.; Reid, L.E.; Chambers, R.; Da Silva, L.; Melville, L.; Evans, E.; et al. An epithelial to mesenchymal transition programme does not usually drive the phenotype of invasive lobular carcinomas. J. Pathol. 2016, 238, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating Breast Tumor Cells Exhibit Dynamic Changes in Epithelial and Mesenchymal Composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, J.; Liu, Y.; Gong, X. Epithelial cell adhesion molecule and epithelial-mesenchymal transition are associated with vasculogenic mimicry, poor prognosis, and metastasis of triple negative breast cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 1678–1689. [Google Scholar]
- Ensinger, C.; Kremser, R.; Prommegger, R.; Spizzo, G.; Schmid, K.W. EpCAM Overexpression in Thyroid Carcinomas. J. Immunother. 2006, 29, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Hardin, H.; Montemayor-Garcia, C.; Lloyd, R.V. Thyroid cancer stem-like cells and epithelial-mesenchymal transition in thyroid cancers. Hum. Pathol. 2013, 44, 1707–1713. [Google Scholar] [CrossRef]
- Montemayor-Garcia, C.; Hardin, H.; Guo, Z.; Larrain, C.; Buehler, D.; Asioli, S.; Chen, H.; Lloyd, R.V. The role of epithelial mesenchymal transition markers in thyroid carcinoma progression. Endocr. Pathol. 2013, 24, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Stoecklein, N.H.; Lin, P.P.; Gires, O. Circulating and disseminated tumor cells: Diagnostic tools and therapeutic targets in motion. Oncotarget 2017, 8, 1884–1912. [Google Scholar] [CrossRef] [PubMed]
- Bulfoni, M.; Turetta, M.; Del Ben, F.; Di Loreto, C.; Beltrami, A.P.; Cesselli, D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. Int. J. Mol. Sci. 2016, 17, 1775. [Google Scholar] [CrossRef]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells 2020, 9, 1836. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Schulze, K.; Gasch, C.; Staufer, K.; Nashan, B.; Lohse, A.W.; Pantel, K.; Riethdorf, S.; Wege, H. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int. J. Cancer 2013, 133, 2165–2171. [Google Scholar] [CrossRef] [PubMed]
- Danila, D.C.; Heller, G.; Gignac, G.A.; Gonzalez-Espinoza, R.; Anand, A.; Tanaka, E.; Lilja, H.; Schwartz, L.; Larson, S.; Fleisher, M.; et al. Circulating Tumor Cell Number and Prognosis in Progressive Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2007, 13, 7053–7058. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Khan, M.S.; Tsigani, T.; Rashid, M.; Rabouhans, J.S.; Yu, D.; Luong, T.V.; Caplin, M.; Meyer, T. Circulating Tumor Cells and EpCAM Expression in Neuroendocrine Tumors. Clin. Cancer Res. 2011, 17, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Mani, S.A.; Levine, H. Hybrid epithelial/mesenchymal phenotype(s): The ‘fittest’ for metastasis? Biochim. Biophys. Acta 2018, 1870, 151–157. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Cadilha, B.L.; Markota, A.; Voigt, C.; Huang, Z.; Lin, P.P.; Wang, D.D.; Dai, J.; Kranz, G.; et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci. Adv. 2019, 5, eaav4275. [Google Scholar] [CrossRef] [PubMed]
- Ruscetti, M.; Quach, B.; Dadashian, E.L.; Mulholland, D.J.; Wu, H. Tracking and Functional Characterization of Epithelial–Mesenchymal Transition and Mesenchymal Tumor Cells during Prostate Cancer Metastasis. Cancer Res. 2015, 75, 2749–2759. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Marengo, M.S.; Oltean, S.; Kemeny, G.; Bitting, R.L.; Turnbull, J.D.; Herold, C.I.; Marcom, P.K.; George, D.J.; Garcia-Blanco, M.A. Circulating Tumor Cells from Patients with Advanced Prostate and Breast Cancer Display Both Epithelial and Mesenchymal Markers. Mol. Cancer Res. 2011, 9, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Strati, A.; Nikolaou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic Significance of TWIST1, CD24, CD44, and ALDH1 Transcript Quantification in EpCAM-Positive Circulating Tumor Cells from Early Stage Breast Cancer Patients. Cells 2019, 8, 652. [Google Scholar] [CrossRef]
- Alonso-Alconada, L.; Muinelo-Romay, L.; Madissoo, K.; Díaz-López, A.; Krakstad, C.; Trovik, J.; Wik, E.; Hapangama, D.; Coenegrachts, L.; Cano, A.; et al. Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Mol. Cancer 2014, 13, 223. [Google Scholar] [CrossRef] [PubMed]
- Cayrefourcq, L.; Mazard, T.; Joosse, S.; Solassol, J.; Ramos, J.; Assenat, E.; Schumacher, U.; Costes, V.; Maudelonde, T.; Pantel, K.; et al. Establishment and Characterization of a Cell Line from Human Circulating Colon Cancer Cells. Cancer Res. 2015, 75, 892–901. [Google Scholar] [CrossRef]
- Hamilton, G.; Rath, B.; Klameth, L.; Hochmair, M.J. Small cell lung cancer: Recruitment of macrophages by circulating tumor cells. OncoImmunology 2016, 5, e1093277. [Google Scholar] [CrossRef]
- Que, Z.; Luo, B.; Zhou, Z.; Dong, C.; Jiang, Y.; Wang, L.; Shi, Q.; Tian, J. Establishment and characterization of a patient-derived circulating lung tumor cell line in vitro and in vivo. Cancer Cell Int. 2019, 19, 21. [Google Scholar] [CrossRef]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Röse, L.; Zollner, T.M.; Krahn, T.; Von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 1–13. [Google Scholar] [CrossRef]
- Hyun, K.-A.; Koo, G.-B.; Han, H.; Sohn, J.; Choi, W.; Kim, S.-I.; Jung, H.-I.; Kim, Y.-S. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 2016, 7, 24677–24687. [Google Scholar] [CrossRef]
- Jojović, M.; Adam, E.; Zangemeister-Wittke, U.; Schumacher, U. Epithelial glycoprotein-2 expression is subject to regulatory processes in epithelial-mesenchymal transitions during metastases: An investigation of human cancers transplanted into severe combined immunodeficient mice. J. Mol. Histol. 1998, 30, 723–729. [Google Scholar] [CrossRef]
- Rao, C.G.; Chianese, D.; Doyle, G.V.; Miller, M.C.; Russell, T.; Sanders, R.A.; Terstappen, L.W. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int. J. Oncol. 2005, 27, 49–57. [Google Scholar] [CrossRef]
- Banyard, J.; Chung, I.; Wilson, A.M.; Vetter, G.; Le Béchec, A.; Bielenberg, D.R.; Zetter, B.R. Regulation of epithelial plasticity by miR-424 and miR-200 in a new prostate cancer metastasis model. Sci. Rep. 2013, 3, 3151. [Google Scholar] [CrossRef]
- Tachtsidis, A.; Le, A.V.-P.; Blick, T.; Gunasinghe, D.; De Sousa, E.; Waltham, M.; Dobrovic, A.; Thompson, E.W. Human-specific RNA analysis shows uncoupled epithelial-mesenchymal plasticity in circulating and disseminated tumour cells from human breast cancer xenografts. Clin. Exp. Metastasis 2019, 36, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Kaigorodova, E.V.; Savelieva, O.E.; Tashireva, L.A.; Tarabanovskaya, N.A.; Simolina, E.I.; Denisov, E.V.; Slonimskaya, E.M.; Choynzonov, E.L.; Perelmuter, V.M. Heterogeneity of Circulating Tumor Cells in Neoadjuvant Chemotherapy of Breast Cancer. Molecules 2018, 23, 727. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-H.; Wang, Z.-R.; Chen, C.-L.; Di, L.; Bi, Z.-F.; Li, Z.-H.; Liu, Y.-M. Molecular detection of epithelial-mesenchymal transition markers in circulating tumor cells from pancreatic cancer patients: Potential role in clinical practice. World J. Gastroenterol. 2019, 25, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Satelli, A.; Xia, X.; Cutrera, J.; Mishra, L.; Li, S. Cell-surface Vimentin: A mislocalized protein for isolating csVimentin+CD133−novel stem-like hepatocellular carcinoma cells expressing EMT markers. Int. J. Cancer 2015, 137, 491–496. [Google Scholar] [CrossRef]
- Zhou, B.-B.S.; Zhang, H.; Damelin, M.; Geles, K.G.; Grindley, J.C.; Dirks, P.B. Tumour-initiating cells: Challenges and opportunities for anticancer drug discovery. Nat. Rev. Drug Discov. 2009, 8, 806–823. [Google Scholar] [CrossRef]
- Ng, V.Y.; Ang, S.N.; Chan, J.X.; Choo, A.B. Characterization of Epithelial Cell Adhesion Molecule as a Surface Marker on Undifferentiated Human Embryonic Stem Cells. Stem Cells 2009, 28, 29–35. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, C.; Becker, S.A.; Hurst, K.; Nogueira, L.M.; Findlay, V.J.; Camp, E.R. miR-145 Antagonizes SNAI1-Mediated Stemness and Radiation Resistance in Colorectal Cancer. Mol. Ther. 2018, 26, 744–754. [Google Scholar] [CrossRef]
- Kim, B.R.; Oh, S.C.; Lee, D.-H.; Kim, J.L.; Lee, S.Y.; Kang, M.H.; Lee, S.I.; Kang, S.; Joung, S.Y.; Min, B.W. BMP-2 induces motility and invasiveness by promoting colon cancer stemness through STAT3 activation. Tumor Biol. 2015, 36, 9475–9486. [Google Scholar] [CrossRef]
- Chen, M.; Baskaran, R.; Lee, N.; Hsu, H.; Ho, T.; Tu, C.; Lin, Y.; Viswanadha, V.P.; Kuo, W.; Huang, C. CXCL2/CXCR2 axis induces cancer stem cell characteristics in CPT-11-resistant LoVo colon cancer cells via Gαi-2 and Gαq/11. J. Cell. Physiol. 2019, 234, 11822–11834. [Google Scholar] [CrossRef]
- Leng, Z.; Xia, Q.; Chen, J.; Li, Y.; Xu, J.; Zhao, E.; Zheng, H.; Ai, W.; Dong, J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell. Physiol. Biochem. 2018, 46, 860–872. [Google Scholar] [CrossRef]
- Münz, M.; Kieu, C.; Mack, B.; Schmitt, B.; Zeidler, R.; Gires, O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene 2004, 23, 5748–5758. [Google Scholar] [CrossRef]
- De Boer, C.J.; van Krieken, J.H.; Janssen-van Rhijn, C.M.; Litvinov, S.V. Expression of Ep-CAM in normal, regenerating, metaplastic, and neoplastic liver. J. Pathol. 1999, 188, 201–206. [Google Scholar] [CrossRef]
- Goldman, O.; Valdes, V.J.; Ezhkova, E.; Gouon-Evans, V. The mesenchymal transcription factor SNAI-1 instructs human liver specification. Stem Cell Res. 2016, 17, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lingala, S.; Khoobyari, S.; Nolta, J.; Zern, M.A.; Wu, J. Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J. Hepatol. 2011, 55, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.-M.; Jing, Y.-Y.; Yu, G.-F.; Kou, X.-R.; Ye, F.; Gao, L.; Li, R.; Zhao, Q.-D.; Yang, Y.; Lu, Z.-H.; et al. Tumor-associated macrophages promote cancer stem cell-like properties via transforming growth factor-beta1-induced epithelial–mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2014, 352, 160–168. [Google Scholar] [CrossRef]
- Nagahara, T.; Shiraha, H.; Sawahara, H.; Uchida, D.; Takeuchi, Y.; Iwamuro, M.; Kataoka, J.; Horiguchi, S.; Kuwaki, T.; Onishi, H.; et al. Hepatic stellate cells promote upregulation of epithelial cell adhesion molecule and epithelial-mesenchymal transition in hepatic cancer cells. Oncol. Rep. 2015, 34, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Choi, G.H.; Na, D.C.; Ahn, E.Y.; Kim, G.I.; Lee, J.E.; Cho, J.Y.; Yoo, J.E.; Choi, J.S.; Park, Y.N. Human hepatocellular carcinomas with “Stemness”-related marker expression: Keratin 19 expression and a poor prognosis. Hepatology 2011, 54, 1707–1717. [Google Scholar] [CrossRef] [PubMed]
- Fernando, J.; Malfettone, A.; Cepeda, E.B.; Vilarrasa-Blasi, R.; Bertran, E.; Raimondi, G.; Fabra, À.; Alvarez-Barrientos, A.; Fernandez-Salguero, P.M.; Rodríguez, E.B.; et al. A mesenchymal-like phenotype and expression of CD44 predict lack of apoptotic response to sorafenib in liver tumor cells. Int. J. Cancer 2014, 136, E161–E172. [Google Scholar] [CrossRef]
- Cirulli, V.; Crisa, L.; Beattie, G.M.; Mally, M.I.; Lopez, A.D.; Fannon, A.; Ptasznik, A.; Inverardi, L.; Ricordi, C.; Deerinck, T.; et al. KSA Antigen Ep-CAM Mediates Cell–Cell Adhesion of Pancreatic Epithelial Cells: Morphoregulatory Roles in Pancreatic Islet Development. J. Cell Biol. 1998, 140, 1519–1534. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of Pancreatic Cancer Stem Cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Palagani, V.; El Khatib, M.; Kossatz, U.; Bozko, P.; Müller, M.R.; Manns, M.P.; Krech, T.; Malek, N.P.; Plentz, R.R. Epithelial Mesenchymal Transition and Pancreatic Tumor Initiating CD44+/EpCAM+ Cells Are Inhibited by γ-Secretase Inhibitor IX. PLoS ONE 2012, 7, e46514. [Google Scholar] [CrossRef]
- Latifi, A.; Abubaker, K.; Castrechini, N.; Ward, A.C.; Liongue, C.; Dobill, F.; Kumar, J.; Thompson, E.W.; Quinn, M.A.; Findlay, J.K.; et al. Cisplatin treatment of primary and metastatic epithelial ovarian carcinomas generates residual cells with mesenchymal stem cell-like profile. J. Cell. Biochem. 2011, 112, 2850–2864. [Google Scholar] [CrossRef] [PubMed]
- Chebouti, I.; Kasimir-Bauer, S.; Buderath, P.; Wimberger, P.; Hauch, S.; Kimmig, R.; Kuhlmann, J.D. EMT-like circulating tumor cells in ovarian cancer patients are enriched by platinum-based chemotherapy. Oncotarget 2017, 8, 48820–48831. [Google Scholar] [CrossRef]
- Bredemeier, M.; Edimiris, P.; Tewes, M.; Mach, P.; Aktas, B.; Schellbach, D.; Wagner, J.; Kimmig, R.; Kasimir-Bauer, S. Establishment of a multimarker qPCR panel for the molecular characterization of circulating tumor cells in blood samples of metastatic breast cancer patients during the course of palliative treatment. Oncotarget 2016, 7, 41677–41690. [Google Scholar] [CrossRef]
- Lenarcic, B.; Bevec, T. Thyropins--new structurally related proteinase inhibitors. Biol. Chem. 1998, 379, 105–111. [Google Scholar]
- Sui, H.; Shi, C.; Yan, Z.; Wu, M. Overexpression of Cathepsin L is associated with chemoresistance and invasion of epithelial ovarian cancer. Oncotarget 2016, 7, 45995–46001. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, S.; Wang, Q.; Yang, Z.; Pan, Z.; Li, L. Overexpression of cysteine cathepsin L is a marker of invasion and metastasis in ovarian cancer. Oncol. Rep. 2014, 31, 1334–1342. [Google Scholar] [CrossRef]
- Tholen, M.; Wolanski, J.; Stolze, B.; Chiabudini, M.; Gajda, M.; Bronsert, P.; Stickeler, E.; Rospert, S.; Reinheckel, T. Stress-resistant Translation of Cathepsin L mRNA in Breast Cancer Progression. J. Biol. Chem. 2015, 290, 15758–15769. [Google Scholar] [CrossRef]
- Sudhan, D.R.; Siemann, D.W. Cathepsin L targeting in cancer treatment. Pharmacol. Ther. 2015, 155, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Seeber, A.; Braicu, I.; Untergasser, G.; Nassir, M.; Fong, D.; Botta, L.; Gastl, G.; Fiegl, H.; Zeimet, A.; Sehouli, J.; et al. Detection of soluble EpCAM (sEpCAM) in malignant ascites predicts poor overall survival in patients treated with catumaxomab. Oncotarget 2015, 6, 25017–25023. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, H.-C.; Wang, Q.; Yang, Z.-J.; Chen, H.; Wang, S.-M.; Pan, Z.-M.; Tang, B.-J.; Li, Q.Q.; Li, L. Clinical value of combined detection of serum matrix metalloproteinase-9, heparanase, and cathepsin for determining ovarian cancer invasion and metastasis. Anticancer Res. 2011, 31, 3423–3428. [Google Scholar]
- Chen, Q.; Fei, J.; Wu, L.; Jiang, Z.; Wu, Y.; Zheng, Y.; Lu, G. Detection of cathepsin B, cathepsin L, cystatin C, urokinase plasminogen activator and urokinase plasminogen activator receptor in the sera of lung cancer patients. Oncol. Lett. 2011, 2, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Sankpal, N.V.; Brown, T.C.; Fleming, T.P.; Herndon, J.M.; Amaravati, A.A.; Loynd, A.N.; Gillanders, W.E. Cancer-associated mutations reveal a novel role for EpCAM as an inhibitor of cathepsin-L and tumor cell invasion. BMC Cancer 2021, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
| Cell Type & Cell Line | Associated Signaling Pathway/Mechanism | Promotes/Inhibits | Reference |
|---|---|---|---|
| Head and Neck Squamous Cell Carcinoma: FaDu and Kyse-30 | EpEX & EGFR | Inhibits | [13] |
| Nasopharyngea S Carcinoma: S18, 6-10B, and HONE1 | PTEN/AKT/mTOR | Promotes | [75] |
| Esophageal Squmous Cell Carinoma: Kyse-30 and Kyse-520 | EMT | Inhibits | [76] |
| Lung Cancer: A549 and NCI-H446 | MTA1 | Promotes | [77] |
| Benign Breast: MCF-10A and Human Mammary Epithelial Cells | ERK & EMT | Inhibits | [37,78] |
| Breast Cancer: MDA-MB-231 | E-Cadherin & α,β-catenin | Promotes | [61] |
| Breast Cancer: MDA-231 and CA1a | AP-1 | Promotes | [79] |
| Breast Cancer: MCF-7 | TGF-β1 | Promotes | [80] |
| Breast Cancer: MDA-MB-231 | NF-κβ | Promotes | [81,82] |
| Breast Cancer: MCF-7, T47D, SkBR3, MDA-MB-231, and Hs578t | EMT | Both | [83] |
| Kideny: MDCK | ERK, Claudin-7, & actomyosin contractility | Inhibits | [84] |
| Colon: HCT116 | EpEX & EpICD | Promotes | [55] |
| Endometrial: Ishikawa and RL95-2 | Adhesion formation & EpICD | Both | [44] |
| Ovarian: SKOV3 & OVCAR4 | ERK | Inhibits | [85] |
| Prostate: KrasG12D & p53L/L mouse knockout prostate cancer model | KRAS & p53 | Promotes | [86] |
| Langerhans cells: Knockout mice model | Decrease adhesiveness | Promotes | [87] |
| Embryonic and Colon: Xenopus laevis and Caco-2 and SW480 | nPKC and myosin regulation | Promotes | [88,89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, T.C.; Sankpal, N.V.; Gillanders, W.E. Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules 2021, 11, 956. https://doi.org/10.3390/biom11070956
Brown TC, Sankpal NV, Gillanders WE. Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules. 2021; 11(7):956. https://doi.org/10.3390/biom11070956
Chicago/Turabian StyleBrown, Taylor C., Narendra V. Sankpal, and William E. Gillanders. 2021. "Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition" Biomolecules 11, no. 7: 956. https://doi.org/10.3390/biom11070956
APA StyleBrown, T. C., Sankpal, N. V., & Gillanders, W. E. (2021). Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules, 11(7), 956. https://doi.org/10.3390/biom11070956






