Variations in Gut Microbiome are Associated with Prognosis of Hypertriglyceridemia-Associated Acute Pancreatitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Demographic and Clinical Data
2.3. Sample Collection, DNA Extraction, and 16S rRNA Gene Sequencing
2.4. Bioinformatics Analyses
2.5. Statistical Analysis
3. Results
3.1. Clinical Characteristics
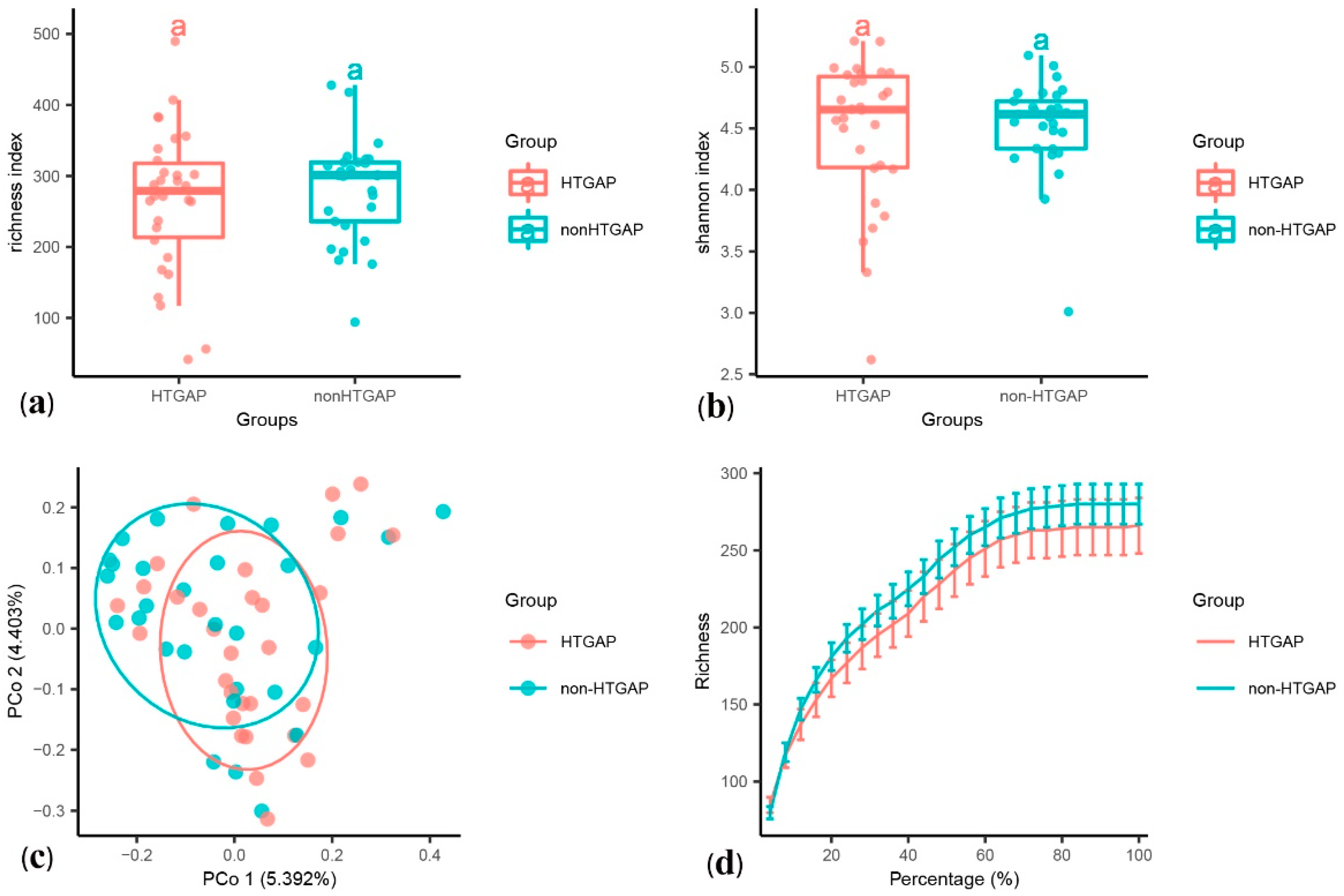
3.2. Complications of HTGAP Patients Affects Gut Microbiome Composition
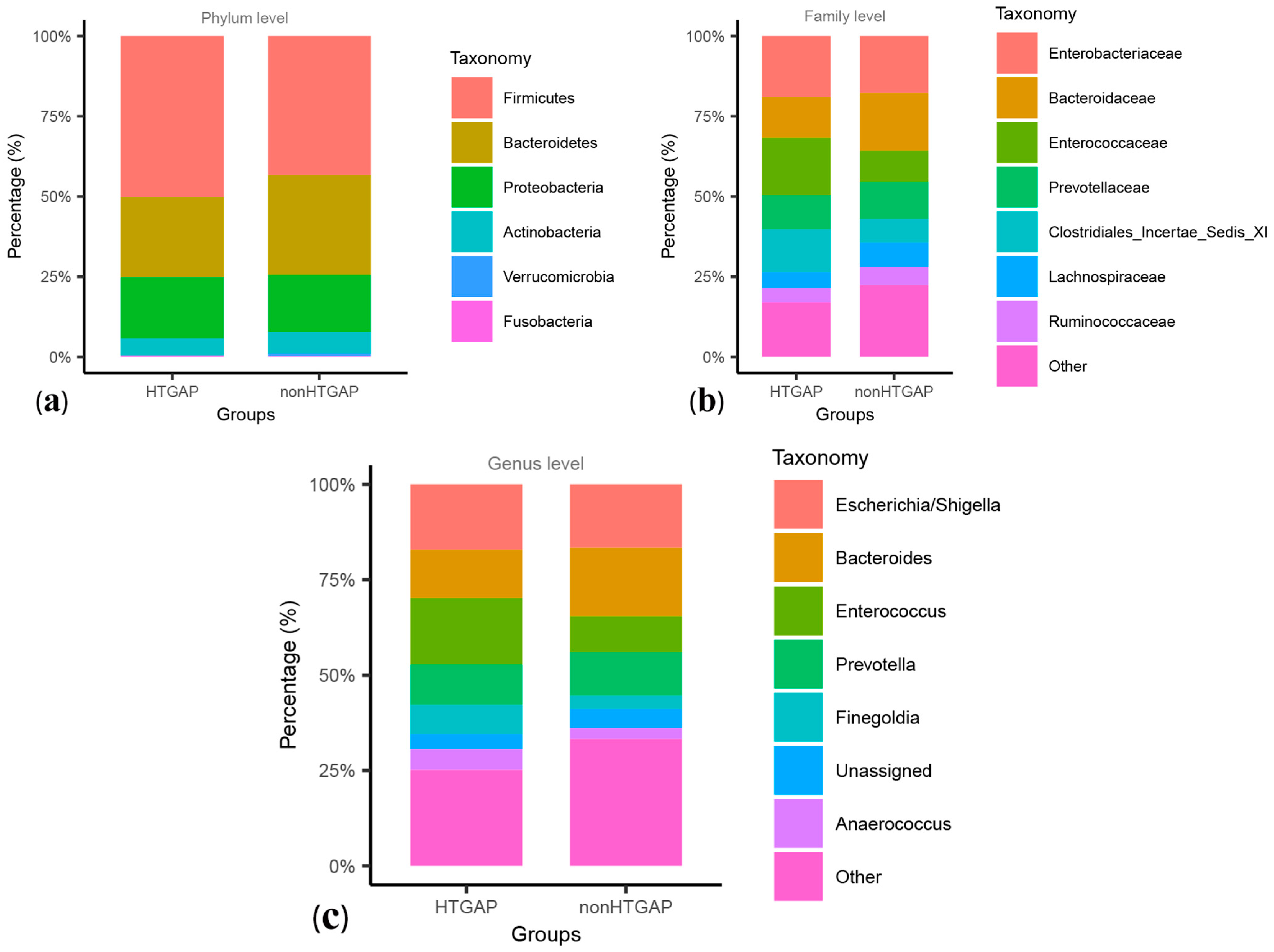
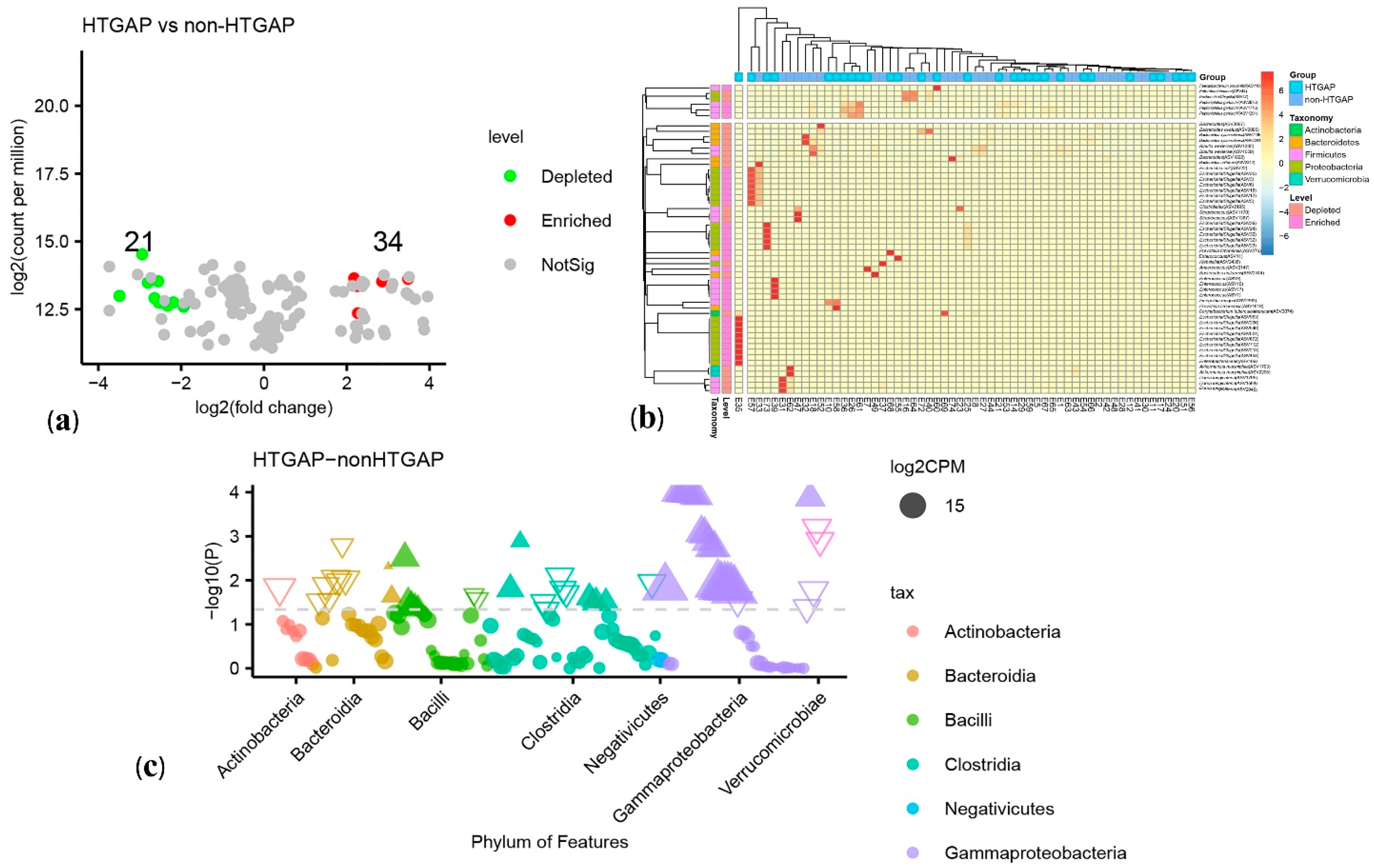
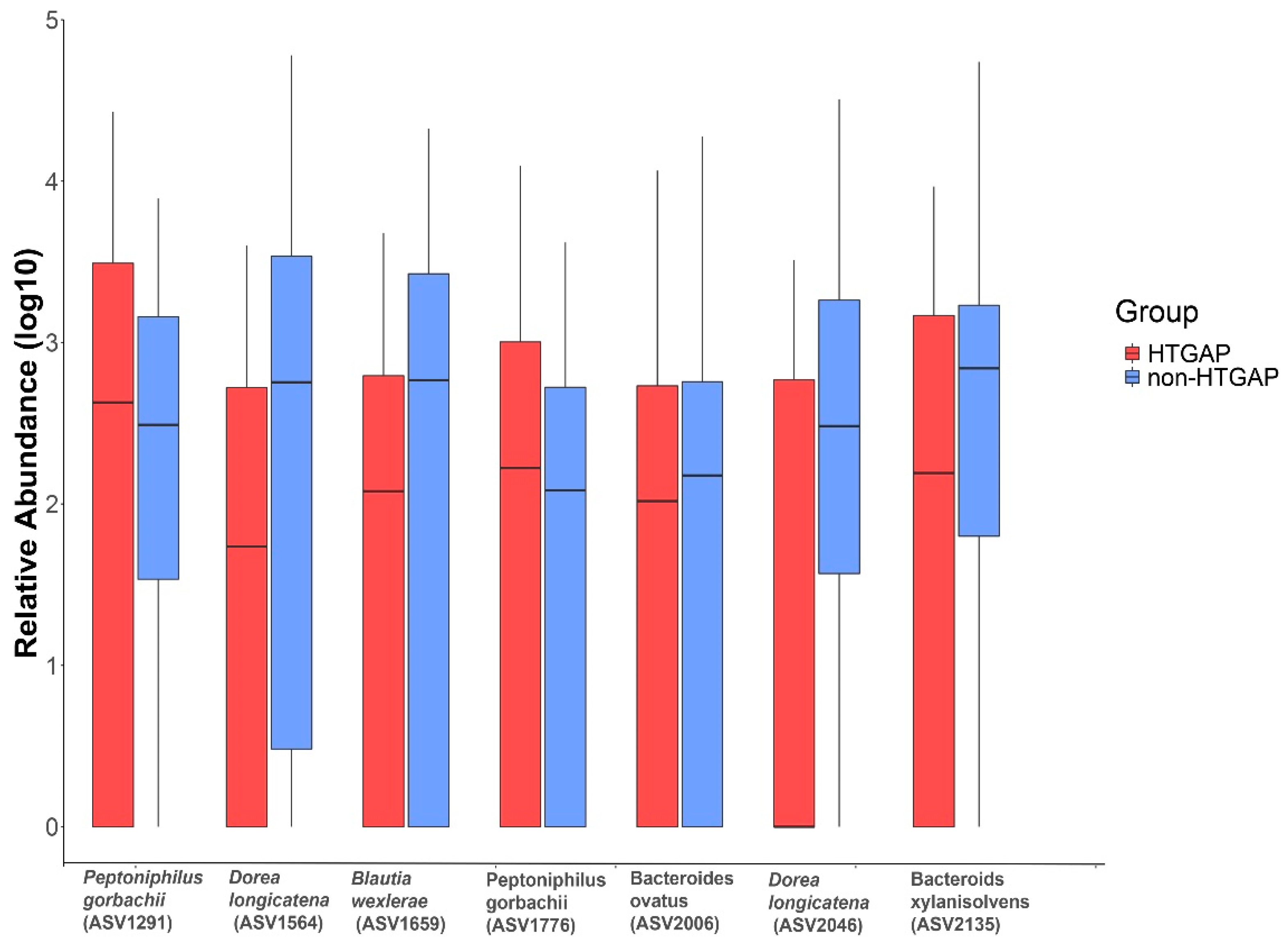
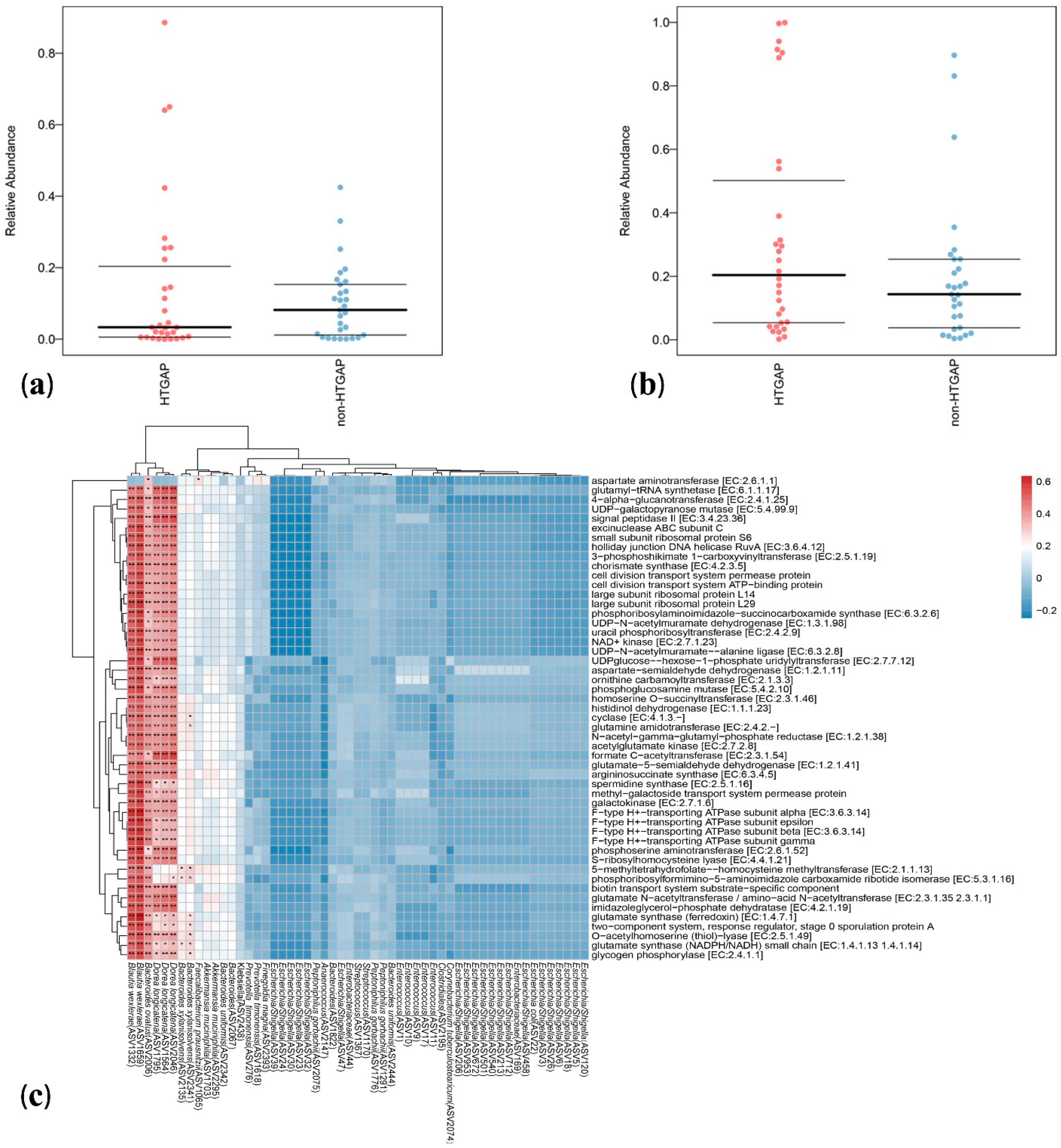
3.3. Taxonomic Features Are Different in Patients with Non-HTGAP and HTGAP
3.4. Microbial Functions Altered in HTGAP Patients
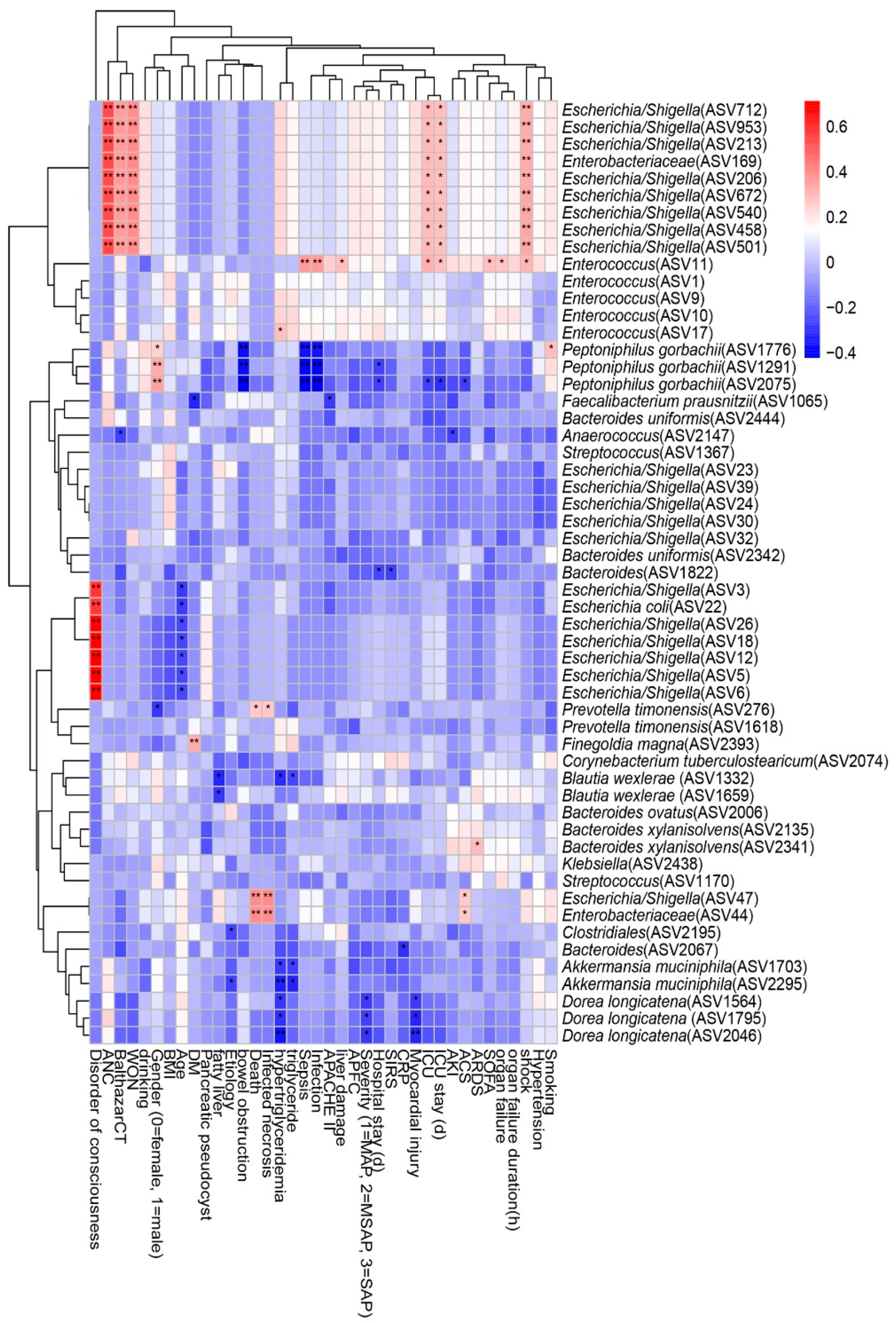
3.5. Associations between Clinical Indicators and Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boxhoorn, L.; Voermans, R.P.; Bouwense, S.A.; Bruno, M.J.; Verdonk, R.C.; Boermeester, M.A.; van Santvoort, H.C.; Besselink, M.G. Acute pancreatitis. Lancet 2020, 396, 726–734. [Google Scholar] [CrossRef]
- Xiao, A.Y.; Tan, M.L.Y.; Wu, L.M.; Asrani, V.M.; Windsor, J.A.; Yadav, D.; Petrov, M.S. Global incidence and mortality of pancreatic diseases: A systematic review, meta-analysis, and meta-regression of population-based cohort studies. Lancet Gastroenterol. Hepatol. 2016, 1, 45–55. [Google Scholar] [CrossRef]
- Banks, P.A.; Bollen, T.L.; Dervenis, C.; Gooszen, H.G.; Johnson, C.D.; Sarr, M.G.; Tsiotos, G.G.; Vege, S.S. Classification of acute pancreatitis—2012: Revision of the Atlanta classification and definitions by international consensus. Gut 2012, 62, 102–111. [Google Scholar] [CrossRef]
- Petrov, M.S.; Shanbhag, S.; Chakraborty, M.; Phillips, A.R.; Windsor, J.A. Organ Failure and Infection of Pancreatic Necrosis as Determinants of Mortality in Patients with Acute Pancreatitis. Gastroenterology 2010, 139, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Speck, L. Fall Von Lipemia. Arch Verin Wissenschaftl Heilkunde 1865; 1: 232. In Quoted in Lipidoses, Diseases of the Intracellular Lipid Metabolism; Thannhauser, S.J., Ed.; Grune & Stratton: New York, NY, USA, 1958. [Google Scholar]
- Jin, M.; Bai, X.; Chen, X.; Zhang, H.; Lu, B.; Li, Y.; Lai, Y.; Qian, J.; Yang, H. A 16-year trend of etiology in acute pancreatitis: The increasing proportion of hypertriglyceridemia-associated acute pancreatitis and its adverse effect on prognosis. J. Clin. Lipidol. 2019, 13, 947–953. [Google Scholar] [CrossRef]
- Olesen, S.S.; Harakow, A.; Krogh, K.; Drewes, A.M.; Handberg, A.; Christensen, P.A. Hypertriglyceridemia is often under recognized as an aetiologic risk factor for acute pancreatitis: A population-based cohort study. Pancreatology 2021, 21, 334–341. [Google Scholar] [CrossRef]
- Yang, A.L.; McNabb-Baltar, J. Hypertriglyceridemia and acute pancreatitis. Pancreatology 2020, 20, 795–800. [Google Scholar] [CrossRef]
- He, W.H.; Zhu, Y.; Liu, P.; Zeng, H.; Xia, L.; Huang, X.; Lei, Y.P.; Lü, N.H. Comparison of severity and clinical outcomes between hypertriglyceridemic pancreatitis and acute pancreatitis due to other causes. Zhonghua yi xue za zhi 2016, 96, 2569–2572. [Google Scholar]
- Nawaz, H.; Koutroumpakis, E.; Easler, J.; Slivka, A.; Whitcomb, D.C.; Singh, V.P.; Yadav, D.; Papachristou, G.I. Elevated serum triglycerides are independently associated with persistent organ failure in acute pancreatitis. Am. J. Gastroenterol. 2015, 110, 1497–1503. [Google Scholar] [CrossRef]
- Pascual, I.; Sanahuja, A.; García, N.; Vázquez, P.; Moreno, O.; Tosca, J.; Peña, A.; Garayoa, A.; Lluch, P.; Mora, F. Association of elevated serum triglyceride levels with a more severe course of acute pancreatitis: Cohort analysis of 1457 patients. Pancreatology 2019, 19, 623–629. [Google Scholar] [CrossRef]
- Tan, C.; Ling, Z.; Huang, Y.; Cao, Y.; Liu, Q.; Cai, T.; Yuan, H.; Liu, C.; Li, Y.; Xu, K. Dysbiosis of intestinal microbiota associated with inflammation involved in the progression of acute pancreatitis. Pancreas 2015, 44, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; He, C.; Zhu, Y.; Lu, N.-H. Role of gut microbiota on intestinal barrier function in acute pancreatitis. World J. Gastroenterol. 2020, 26, 2187–2193. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Xiong, Y.; Xu, J.; Liang, X.; Fu, Y.; Liu, D.; Yu, X.; Wu, D. Identification of Dysfunctional Gut Microbiota Through Rectal Swab in Patients with Different Severity of Acute Pancreatitis. Dig. Dis. Sci. 2020, 65, 3223–3237. [Google Scholar] [CrossRef]
- Yun, K.E.; Kim, J.; Kim, M.-H.; Park, E.; Kim, H.-L.; Chang, Y.; Ryu, S.; Kim, H.-N. Major lipids, apolipoproteins, and alterations of gut microbiota. J. Clin. Med. 2020, 9, 1589. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhang, Y.; Yang, Q.; Lee, P.; Windsor, J.A.; Wu, D. An updated systematic review with meta-analysis. Pancreas 2021, 50, 160–166. [Google Scholar] [CrossRef]
- Carr, R.A.; Rejowski, B.J.; Cote, G.A.; Pitt, H.A.; Zyromski, N.J. Systematic review of hypertriglyceridemia-induced acute pancreatitis: A more virulent etiology? Pancreatology 2016, 16, 469–476. [Google Scholar] [CrossRef]
- Knaus, W.A.; Draper, E.A.; Wagner, D.P.; Zimmerman, J.E. APACHE II: A severity of disease classification system. Crit. Care Med. 1985, 13, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.-L.; De Mendonca, A.; Cantraine, F.; Moreno, R.; Takala, J.; Suter, P.M.; Sprung, C.L.; Colardyn, F.; Blecher, S. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units. Crit. Care Med. 1998, 26, 1793–1800. [Google Scholar] [CrossRef]
- Chatzicostas, C.; Roussomoustakaki, M.; Vardas, E.; Romanos, J.; Kouroumalis, E.A. Balthazar Computed Tomography Severity Index Is Superior to Ranson Criteria and APACHE II and III Scoring Systems in Predicting Acute Pancreatitis Outcome. J. Clin. Gastroenterol. 2003, 36, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Lu, B.; Xue, H.-D.; Yang, H.; Qian, J.-M.; Lee, P.; Windsor, J.A. Validation of Modified Determinant-Based Classification of severity for acute pancreatitis in a tertiary teaching hospital. Pancreatology 2019, 19, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Biehl, L.M.; Garzetti, D.; Farowski, F.; Ring, D.; Koeppel, M.B.; Rohde, H.; Schafhausen, P.; Stecher, B.; Vehreschild, M.J.G.T. Usability of rectal swabs for microbiome sampling in a cohort study of hematological and oncological patients. PLoS ONE 2019, 14, e0215428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debebe, T.; Biagi, E.; Soverini, M.; Holtze, S.; Hildebrandt, T.B.; Birkemeyer, C.; Wyohannis, D.; Lemma, A.; Brigidi, P.; Savkovic, V.; et al. Unraveling the gut microbiome of the long-lived naked mole-rat. Sci. Rep. 2017, 7, 9590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eri, T.; Kawahata, K.; Kanzaki, T.; Imamura, M.; Michishita, K.; Akahira, L.; Bannai, E.; Yoshikawa, N.; Kimura, Y.; Satoh, T.; et al. Intestinal microbiota link lymphopenia to murine autoimmunity via PD-1+CXCR5−/dim B-helper T cell induction. Sci. Rep. 2017, 7, 46037. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Wu, Y.; Wang, J.; Wu, G.; Long, W.; Xue, Z.; Wang, L.; Zhang, X.; Pang, X.; Zhao, Y.; et al. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci. Rep. 2016, 6, 27572. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2020, 1–16. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, T.J.; Ahmed, Y.M.; Zamzami, M.A.; Mohamed, S.A.; Khan, I.; Baothman, O.A.S.; Mehanna, M.G.; Yasir, M. Effect of atorvastatin on the gut microbiota of high fat diet-induced hypercholesterolemic rats. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘vegan’. Community Ecol. Package Version 2013, 2, 1–295. [Google Scholar]
- Pedersen, T.L.; Pedersen, M.T.L.; LazyData, T.; Rcpp, I.; Rcpp, L. Package ‘ggraph’. Retrieved Jan. 2017, 1, 2018. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Ward, T.; Larson, J.; Meulemans, J.; Hillmann, B.; Lynch, J.; Sidiropoulos, D.; Spear, J.R.; Caporaso, G.; Blekhman, R.; Knight, R.; et al. BugBase predicts organism-level microbiome phenotypes. Biorxiv 2017, 133462. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PIC-RUSt2: An improved and customizable approach for metagenome inference. BioRxiv 2020, 672295. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’. R Package 2015, 1, 790. [Google Scholar]
- Deng, L.-H.; Xue, P.; Xia, Q.; Yang, X.-N.; Wan, M.-H. Effect of admission hypertriglyceridemia on the episodes of severe acute pancreatitis. World J. Gastroenterol. 2008, 14, 4558–4561. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ke, L.; Dong, J.; Ye, B.; Meng, L.; Mao, W.; Yang, Q.; Li, W.; Li, J. Significantly different clinical features between hypertriglyceridemia and biliary acute pancreatitis: A retrospective study of 730 patients from a tertiary center. BMC Gastroenterol. 2018, 18, 89. [Google Scholar] [CrossRef]
- Váncsa, S.; Németh, D.; Hegyi, P.; Szakács, Z.; Hegyi, P.J.; Pécsi, D.; Mikó, A.; Erőss, B.; Erős, A.; Pár, G. Fatty liver disease and non-alcoholic fatty liver disease worsen the outcome in acute pancreatitis: A systematic review and meta-analysis. J. Clin. Med. 2020, 9, 2698. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yan, H.; Wang, G.; Qiu, Z.; Bai, B.; Wang, S.; Yu, P.; Feng, Q.; Zhao, Q.; He, X.; et al. RNA sequence analysis of rat acute experimental pancreatitis with and without fatty liver: A gene expression profiling comparative study. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Zhang, X.M.; Zhang, Z.Y.; Zhang, C.H.; Wu, J.; Wang, Y.X.; Zhang, G.X. Intestinal microbial community differs between acute pancreatitis patients and healthy volunteers. Biomed. Environ. Sci. 2018, 31, 81–86. [Google Scholar]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Taras, D.; Simmering, R.; Collins, M.D.; Lawson, P.A.; Blaut, M. Reclassification of eubacterium formicigenerans holdeman and moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2002, 52, 423–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Finegold, S.M.; Song, Y.; Lawson, P.A. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58, 1896–1902. [Google Scholar] [CrossRef]
- Van der Hee, B.; Wells, J.M. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 2021, 1935, 1–13. [Google Scholar] [CrossRef]
- Benítez-Páez, A.; Del Pugar, E.M.G.; López-Almela, I.; Moya-Pérez, Á.; Codoñer-Franch, P.; Sanz, Y.; Jenq, R. Depletion of blautia species in the microbiota of obese children relates to intestinal inflammation and metabolic phenotype worsening. Msystems 2020, 5. [Google Scholar] [CrossRef] [Green Version]
- Mondot, S.; Lepage, P.; Seksik, P.; Allez, M.; Treton, X.; Bouhnik, Y.; Colombel, J.F.; Leclerc, M.; Pochart, P.; Dore, J.; et al. Structural robustness of the gut mucosal microbiota is associated with Crohn’s disease remission after surgery. Gut 2015, 65, 954–962. [Google Scholar] [CrossRef]
- Yang, C.; Mogno, I.; Contijoch, E.J.; Borgerding, J.N.; Aggarwala, V.; Li, Z.; Siu, S.; Grasset, E.K.; Helmus, D.S.; Dubinsky, M.C.; et al. Fecal IgA levels are determined by strain-level differences in bacteroides ovatus and are modifiable by gut microbiota manipulation. Cell Host Microbe 2020, 27, 467–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanagibashi, T.; Hosono, A.; Oyama, A.; Tsuda, M.; Suzuki, A.; Hachimura, S.; Takahashi, Y.; Momose, Y.; Itoh, K.; Hirayama, K.; et al. IgA production in the large intestine is modulated by a different mechanism than in the small intestine: Bacteroides acidifaciens promotes IgA production in the large intestine by inducing germinal center formation and increasing the number of IgA+ B cells. Immunobiology 2013, 218, 645–651. [Google Scholar] [CrossRef] [PubMed]
- López-Almela, I.; Romaní-Pérez, M.; Bullich-Vilarrubias, C.; Benítez-Páez, A.; Del Pulgar, E.M.G.; Francés, R.; Liebisch, G.; Sanz, Y. Bacteroides uniformis combined with fiber amplifies metabolic and immune benefits in obese mice. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Yu, X.; Liu, Y.; Sun, L.; Chen, P.; Ding, Q.; Gao, Y.; Zhang, X.; Yu, M.; Liu, Y.; et al. The baseline gut microbiota directs dieting-induced weight loss trajectories. Gastroenterology 2021. [Google Scholar] [CrossRef] [PubMed]
- Ying, W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: Regulation and biological consequences. Antioxid. Redox Signal. 2008, 10, 179–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Variables | HTGAP (n = 30) | Non-HTGAP (n = 30) | p |
|---|---|---|---|
| Age (years), mean (SD) | 40.8 (11.3) | 51.8 (16.6) | 0.004 |
| Male, n (%) | 17 (56.7) | 14 (46.7) | 0.438 |
| BMI (kg/m2), mean (SD) | 27.6 (3.7) | 24.7 (2.3) | 0.001 |
| Overweight (BMI 25–29.9 kg/m2), n (%) | 19 (63.3) | 13 (43.3) | 0.121 |
| Obesity (BMI ≥ 30 kg/m2), n (%) | 5 (13.3) | 0 (0.0) | 0.020 |
| Smoking, n (%) | 11 (36.7) | 6 (20.0) | 0.152 |
| Drinking, n (%) | 10 (33.3) | 5 (16.7) | 0.136 |
| Comorbid abnormalities, n (%) | |||
| Hypertension | 10 (33.3) | 11 (36.7) | 0.787 |
| Diabetes | 8 (26.7) | 6 (20.0) | 0.542 |
| Fatty liver | 26 (86.7) | 15 (50.0) | 0.002 |
| Laboratory examinations | |||
| Triglyceride (mmol/L), median (IQR) | 18.7 (14.0, 34.9) | 1.2 (0.6, 1.9) | <0.001 |
| CRP (mg/L), median (IQR) | 160.0 (131.8, 232.1) | 106.9 (21.5, 160.0) | 0.002 |
| Etiology, n (%) | |||
| Biliary | 0 (0.0) | 25 (83.3) | <0.001 |
| Hypertriglyceridemia | 30 (100.0) | 0 (0.0) | <0.001 |
| Alcohol consumption | 0 (0.0) | 5 (16.7) | 0.026 |
| APACHE Ⅱ, median (IQR) | 6.0 (3.0, 11.0) | 5.0 (2.0, 7.25) | 0.147 |
| SOFA score, median (IQR) | 2.0 (0.0, 4.3) | 1.0 (0.0, 3.25) | 0.187 |
| Balthazar score E, n (%) | 6 (20.0) | 3 (10.0) | 0.278 |
| Disease severity, n (%) | |||
| MAP | 6 (20.0) | 14 (46.7) | 0.028 |
| MSAP | 10 (33.3) | 10 (33.3) | 1.000 |
| SAP | 14 (46.7) | 6 (20.0) | 0.028 |
| Local complications, n (%) | |||
| APFC | 21 (70.0) | 14 (46.7) | 0.067 |
| Pancreatic pseudocyst | 5 (16.7) | 4 (13.3) | 0.718 |
| ANC | 3 (10.0) | 1 (3.3) | 0.301 |
| WON | 2 (6.7) | 0 (0.0) | 0.150 |
| Infected necrosis | 1 (3.3) | 0 (0.0) | 0.313 |
| Systematic complication, n (%) | |||
| SIRS | 19 (63.3) | 11 (36.7) | 0.039 |
| ARDS | 15 (50.0) | 7 (23.3) | 0.032 |
| AKI | 8 (26.7) | 5 (16.7) | 0.347 |
| Shock | 4 (13.3) | 5 (16.7) | 0.718 |
| ACS | 5 (16.7) | 2 (6.7) | 0.228 |
| Liver damage | 5 (16.7) | 6 (20.0) | 0.739 |
| Myocardial injury | 4 (13.3) | 1 (3.3) | 0.161 |
| Altered mental status | 1 (3.3) | 0 (0.0) | 0.313 |
| Sepsis | 7 (23.3) | 6 (20.0) | 0.754 |
| Bowel obstruction | 8 (26.7) | 3 (10.0) | 0.095 |
| Outcome | |||
| Organ failure, n (%) | 17 (56.7) | 9 (30.0) | 0.037 |
| Organ failure duration (h), median (IQR) | 72.0 (48.0, 276.0) | 50.0 (33.5, 121.0) | 0.465 |
| ICU, n (%) | 16 (53.3) | 6 (20.0) | 0.007 |
| ICU stay (days), median (IQR) | 9.0 (5.3, 14.5) | 6.5 (5.8, 8.8) | 0.395 |
| Hospital stay (days), median (IQR) | 18.0 (9.8, 28.0) | 6.5 (3.0, 14.0) | 0.001 |
| Death, n (%) | 1 (3.3) | 0 (0.0) | 0.313 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Gong, L.; Zhou, R.; Han, Z.; Ji, L.; Zhang, Y.; Zhang, S.; Wu, D. Variations in Gut Microbiome are Associated with Prognosis of Hypertriglyceridemia-Associated Acute Pancreatitis. Biomolecules 2021, 11, 695. https://doi.org/10.3390/biom11050695
Hu X, Gong L, Zhou R, Han Z, Ji L, Zhang Y, Zhang S, Wu D. Variations in Gut Microbiome are Associated with Prognosis of Hypertriglyceridemia-Associated Acute Pancreatitis. Biomolecules. 2021; 11(5):695. https://doi.org/10.3390/biom11050695
Chicago/Turabian StyleHu, Xiaomin, Liang Gong, Ruilin Zhou, Ziying Han, Li Ji, Yan Zhang, Shuyang Zhang, and Dong Wu. 2021. "Variations in Gut Microbiome are Associated with Prognosis of Hypertriglyceridemia-Associated Acute Pancreatitis" Biomolecules 11, no. 5: 695. https://doi.org/10.3390/biom11050695
APA StyleHu, X., Gong, L., Zhou, R., Han, Z., Ji, L., Zhang, Y., Zhang, S., & Wu, D. (2021). Variations in Gut Microbiome are Associated with Prognosis of Hypertriglyceridemia-Associated Acute Pancreatitis. Biomolecules, 11(5), 695. https://doi.org/10.3390/biom11050695








