Some CSF Kynurenine Pathway Intermediates Associated with Disease Evolution in Amyotrophic Lateral Sclerosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Controls
2.2. CSF Amino Acid Quantification
2.3. Kynurenine Pathway Exploration
2.4. Statistical Analysis
2.4.1. Univariate Analysis
2.4.2. Multivariate Analysis
3. Results
3.1. Patient Characteristics
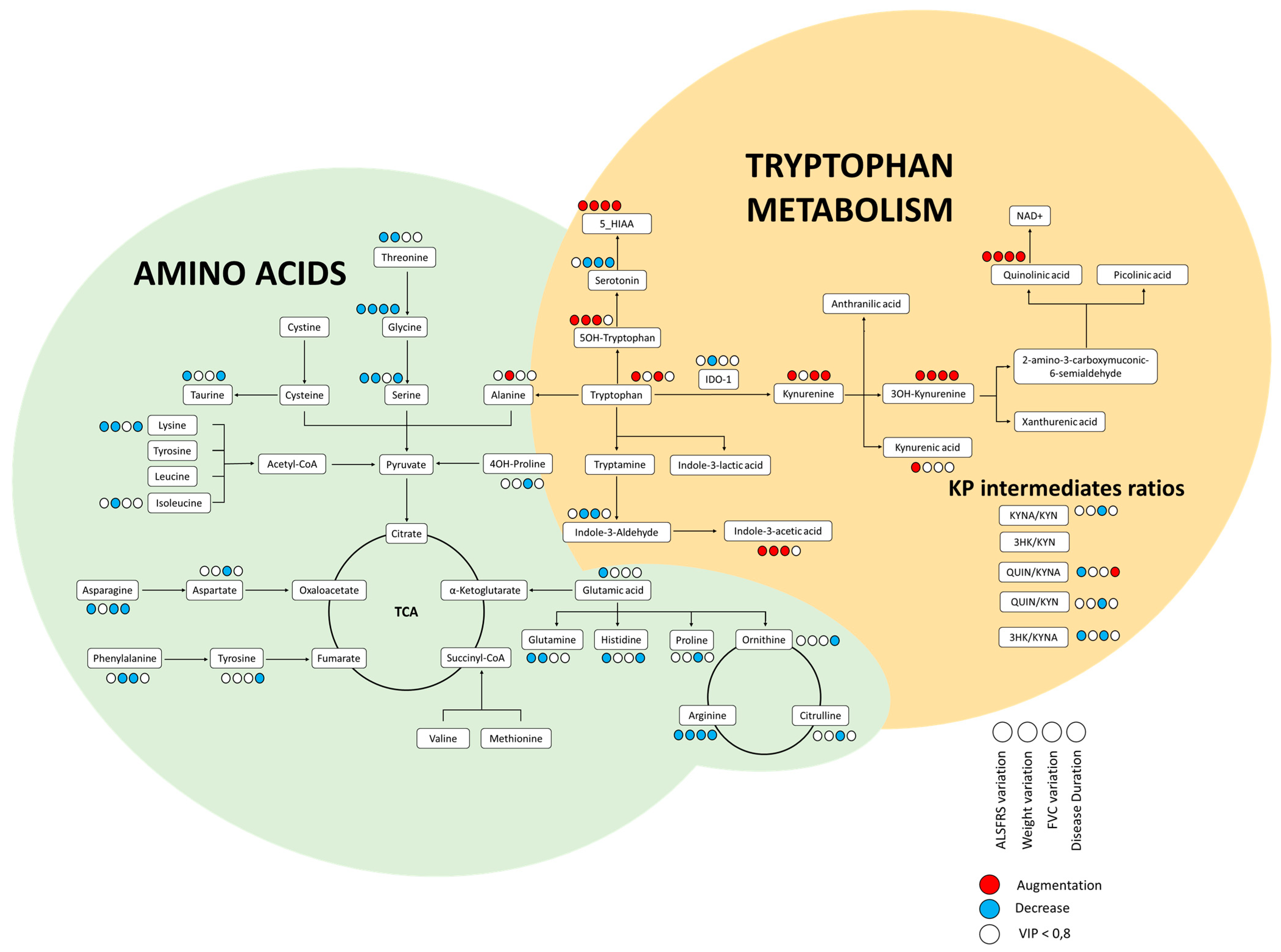
3.2. Amino Acids and Metabolites of the KP
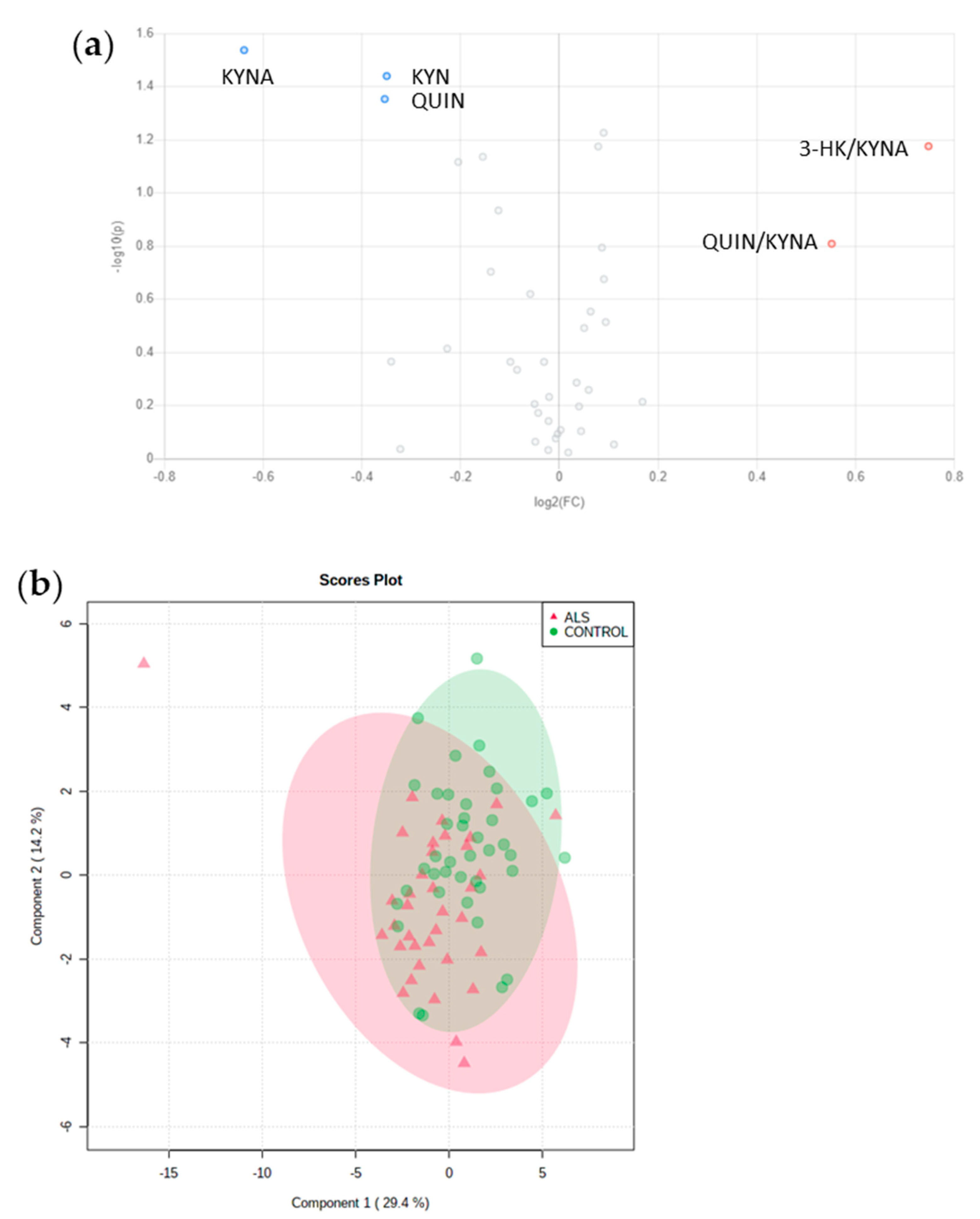
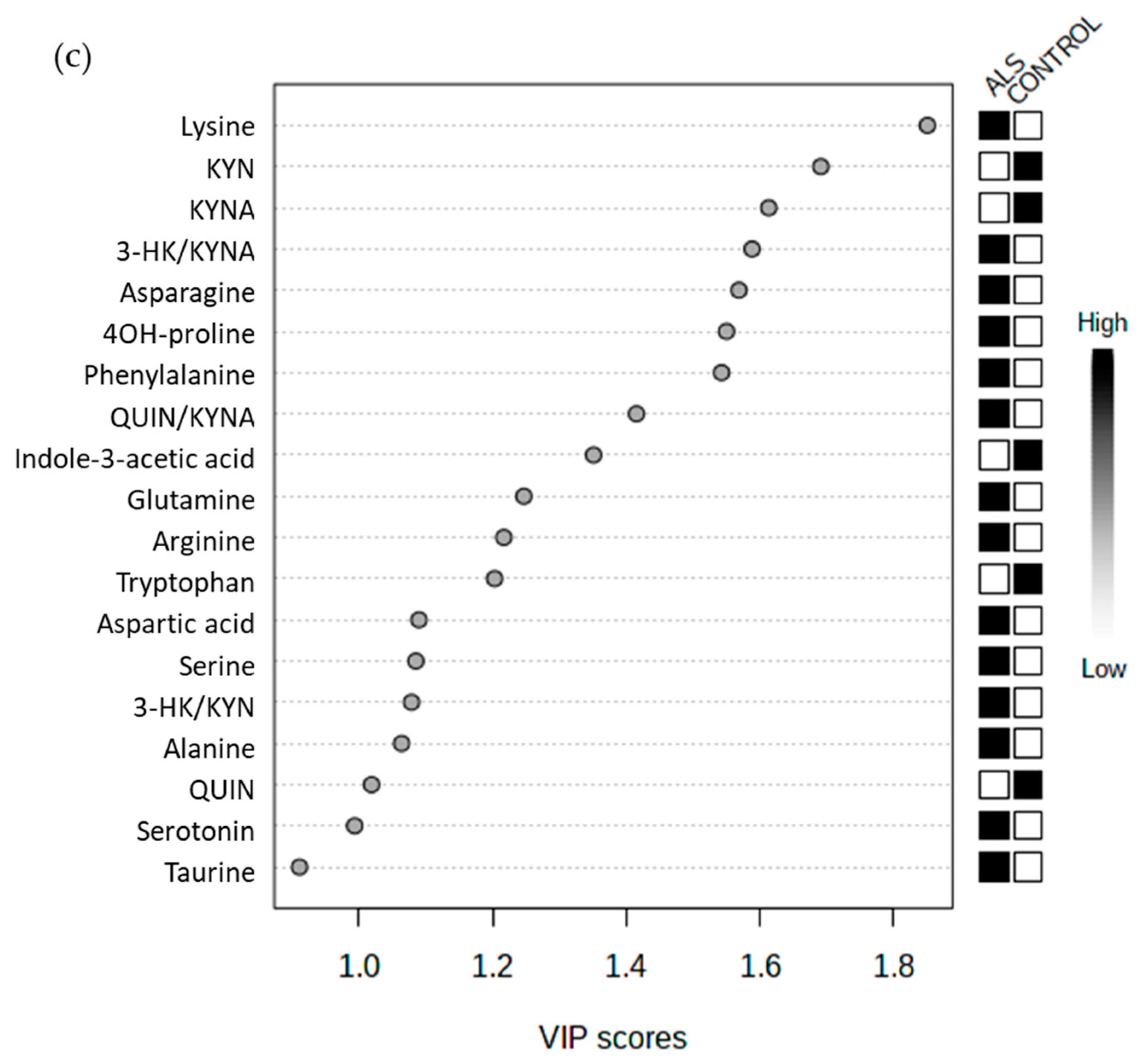
3.3. No relevant Diagnosis Biomarker among Amino Acids or Metabolites from KP
3.4. KP Intermediates Modestly Associated with ALS Characteristics
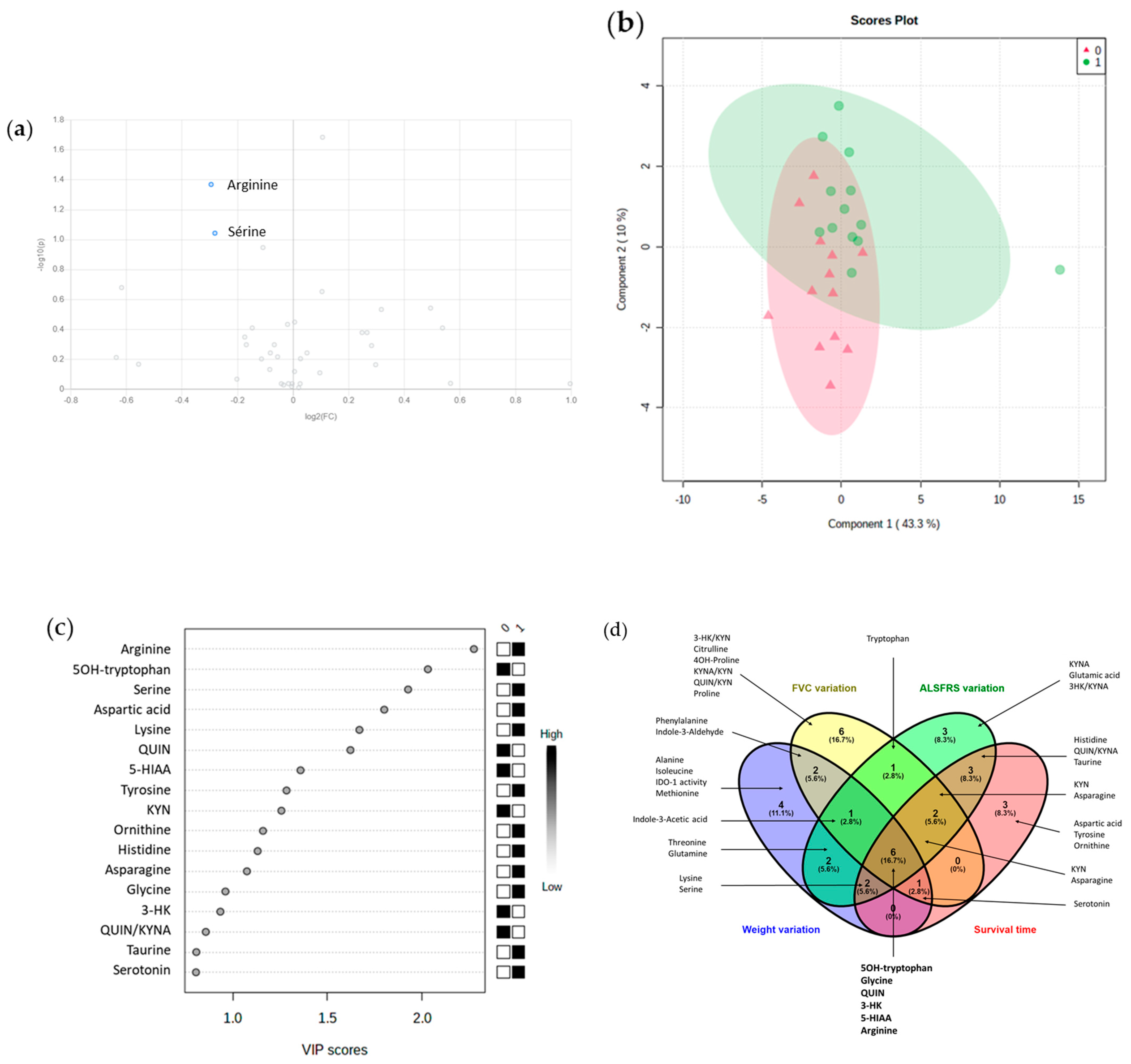
3.5. Some Promising Prognosis Biomarker among Amino Acids or KP Metabolites
3.5.1. Weight Loss at 12 Months (n = 21)
3.5.2. ALSFRS-r at 12 Months (n = 22)
3.5.3. FVC at 12 Months (n = 9)
3.5.4. Duration of Disease (n = 25)
3.5.5. Combined Parameters of Prognosis
4. Discussion
4.1. Validation of Analytical Methods Used in Routine Practice for Research Projects
4.2. No Benefit to Early Exploration of CSF Amino Acids in ALS
4.3. Contradictory Findings on Kynurenine Pathway
4.4. Major Candidates among AA and KP Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, R.H.; Al-Chalabi, A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017, 377, 162–172. [Google Scholar] [CrossRef]
- Hardiman, O.; Al-Chalabi, A.; Chio, A.; Corr, E.M.; Logroscino, G.; Robberecht, W.; Shaw, P.J.; Simmons, Z.; van den Berg, L.H. Amyotrophic Lateral Sclerosis. Nat. Rev. Dis. Primers 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, L.; Pradat, P.-F.; Ludolph, A.C.; Loeffler, J.-P. Energy Metabolism in Amyotrophic Lateral Sclerosis. Lancet Neurol. 2011, 10, 75–82. [Google Scholar] [CrossRef]
- Lee, J.-M.; Tan, V.; Lovejoy, D.; Braidy, N.; Rowe, D.B.; Brew, B.J.; Guillemin, G.J. Involvement of Quinolinic Acid in the Neuropathogenesis of Amyotrophic Lateral Sclerosis. Neuropharmacology 2017, 112, 346–364. [Google Scholar] [CrossRef]
- Tan, V.X.; Guillemin, G.J. Kynurenine Pathway Metabolites as Biomarkers for Amyotrophic Lateral Sclerosis. Front. Neurosci. 2019, 13, 1013. [Google Scholar] [CrossRef] [PubMed]
- Török, N.; Tanaka, M.; Vécsei, L. Searching for Peripheral Biomarkers in Neurodegenerative Diseases: The Tryptophan-Kynurenine Metabolic Pathway. Int. J. Mol. Sci. 2020, 21, 9338. [Google Scholar] [CrossRef] [PubMed]
- Muneer, A. Kynurenine Pathway of Tryptophan Metabolism in Neuropsychiatric Disorders: Pathophysiologic and Therapeutic Considerations. Clin. Psychopharmacol. Neurosci. 2020, 18, 507–526. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Z.-H.; Zabed, H.M.; Yun, J.; Zhang, G.; Qi, X. An Insight into the Roles of Dietary Tryptophan and Its Metabolites in Intestinal Inflammation and Inflammatory Bowel Disease. Mol. Nutr. Food Res. 2020, e2000461. [Google Scholar] [CrossRef]
- Tanaka, M.; Toldi, J.; Vécsei, L. Exploring the Etiological Links behind Neurodegenerative Diseases: Inflammatory Cytokines and Bioactive Kynurenines. Int. J. Mol. Sci. 2020, 21, 2431. [Google Scholar] [CrossRef]
- Chen, Y.; Stankovic, R.; Cullen, K.M.; Meininger, V.; Garner, B.; Coggan, S.; Grant, R.; Brew, B.J.; Guillemin, G.J. The Kynurenine Pathway and Inflammation in Amyotrophic Lateral Sclerosis. Neurotox. Res. 2010, 18, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Iłzecka, J.; Kocki, T.; Stelmasiak, Z.; Turski, W.A. Endogenous Protectant Kynurenic Acid in Amyotrophic Lateral Sclerosis. Acta Neurol. Scand. 2003, 107, 412–418. [Google Scholar] [CrossRef]
- Janssens, J.; Vermeiren, Y.; van Faassen, M.; van der Ley, C.; Kema, I.P.; De Deyn, P.P. Monoaminergic and Kynurenergic Characterization of Frontotemporal Dementia and Amyotrophic Lateral Sclerosis in Cerebrospinal Fluid and Serum. Neurochem. Res. 2020, 45, 1191–1201. [Google Scholar] [CrossRef]
- González-Sánchez, M.; Jiménez, J.; Narváez, A.; Antequera, D.; Llamas-Velasco, S.; Martín, A.H.-S.; Arjona, J.A.M.; de Munain, A.L.; Bisa, A.L.; Marco, M.-P.; et al. Kynurenic Acid Levels Are Increased in the CSF of Alzheimer’s Disease Patients. Biomolecules 2020, 10, 571. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L. World Federation of Neurology Research Group on Motor Neuron Diseases El Escorial Revisited: Revised Criteria for the Diagnosis of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Motor Neuron Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Birner, A.; Platzer, M.; Bengesser, S.A.; Dalkner, N.; Fellendorf, F.T.; Queissner, R.; Pilz, R.; Rauch, P.; Maget, A.; Hamm, C.; et al. Increased Breakdown of Kynurenine towards Its Neurotoxic Branch in Bipolar Disorder. PLoS ONE 2017, 12, e0172699. [Google Scholar] [CrossRef] [PubMed]
- Rothstein, J.D.; Tsai, G.; Kuncl, R.W.; Clawson, L.; Cornblath, D.R.; Drachman, D.B.; Pestronk, A.; Stauch, B.L.; Coyle, J.T. Abnormal Excitatory Amino Acid Metabolism in Amyotrophic Lateral Sclerosis. Ann. Neurol. 1990, 28, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Perry, T.L.; Krieger, C.; Hansen, S.; Eisen, A. Amyotrophic Lateral Sclerosis: Amino Acid Levels in Plasma and Cerebrospinal Fluid. Ann. Neurol. 1990, 28, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Blin, O.; Samuel, D.; Nieoullon, A.; Serratice, G. Changes in CSF Amino Acid Concentrations during the Evolution of Amyotrophic Lateral Sclerosis. J. Neurol. Neurosurg. Psychiatry 1994, 57, 119–120. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meier, D.H.; Schott, K.J. Free Amino Acid Pattern of Cerebrospinal Fluid in Amyotrophic Lateral Sclerosis. Acta Neurol. Scand. 1988, 77, 50–53. [Google Scholar] [CrossRef]
- Valentino, F.; Bivona, G.; Butera, D.; Paladino, P.; Fazzari, M.; Piccoli, T.; Ciaccio, M.; La Bella, V. Elevated Cerebrospinal Fluid and Plasma Homocysteine Levels in ALS. Eur. J. Neurol. 2010, 17, 84–89. [Google Scholar] [CrossRef]
- Blasco, H.; Corcia, P.; Moreau, C.; Veau, S.; Fournier, C.; Vourc’h, P.; Emond, P.; Gordon, P.; Pradat, P.-F.; Praline, J.; et al. 1H-NMR-Based Metabolomic Profiling of CSF in Early Amyotrophic Lateral Sclerosis. PLoS ONE 2010, 5, e13223. [Google Scholar] [CrossRef]
- Wu, J.; Wuolikainen, A.; Trupp, M.; Jonsson, P.; Marklund, S.L.; Andersen, P.M.; Forsgren, L.; Öhman, A. NMR Analysis of the CSF and Plasma Metabolome of Rigorously Matched Amyotrophic Lateral Sclerosis, Parkinson’s Disease and Control Subjects. Metabolomics 2016, 12, 101. [Google Scholar] [CrossRef]
- Wuolikainen, A.; Jonsson, P.; Ahnlund, M.; Antti, H.; Marklund, S.L.; Moritz, T.; Forsgren, L.; Andersen, P.M.; Trupp, M. Multi-Platform Mass Spectrometry Analysis of the CSF and Plasma Metabolomes of Rigorously Matched Amyotrophic Lateral Sclerosis, Parkinson’s Disease and Control Subjects. Mol. Biosyst. 2016, 12, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Bruno, C.; Veyrat-Durebex, C.; Lumbu Lukuntonda, C.H.; Andres, C.R.; Moreau, C.; Bendavid, C.; Homedan, C.; Labarthe, F.; Tardieu, M.; Bigot, A.; et al. Validation of Plasma Amino Acid Profile Using UHPLC-Mass Spectrometer (QDa) as a Screening Method in a Metabolic Disorders Reference Centre: Performance and Accreditation Concerns. Clin. Biochem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ng Kee Kwong, K.C.; Harbham, P.K.; Selvaraj, B.T.; Gregory, J.M.; Pal, S.; Hardingham, G.E.; Chandran, S.; Mehta, A.R. 40 Years of CSF Toxicity Studies in ALS: What Have We Learnt About ALS Pathophysiology? Front. Mol. Neurosci. 2021, 14, 647895. [Google Scholar] [CrossRef]
- Shaw, P.J.; Forrest, V.; Ince, P.G.; Richardson, J.P.; Wastell, H.J. CSF and Plasma Amino Acid Levels in Motor Neuron Disease: Elevation of CSF Glutamate in a Subset of Patients. Neurodegeneration 1995, 4, 209–216. [Google Scholar] [CrossRef]
- Bereman, M.S.; Kirkwood, K.I.; Sabaretnam, T.; Furlong, S.; Rowe, D.B.; Guillemin, G.J.; Mellinger, A.L.; Muddiman, D.C. Metabolite Profiling Reveals Predictive Biomarkers and the Absence of β-Methyl Amino-l-Alanine in Plasma from Individuals Diagnosed with Amyotrophic Lateral Sclerosis. J. Proteome Res. 2020, 19, 3276–3285. [Google Scholar] [CrossRef]
- Thonhoff, J.R.; Simpson, E.P.; Appel, S.H. Neuroinflammatory Mechanisms in Amyotrophic Lateral Sclerosis Pathogenesis. Curr. Opin. Neurol. 2018, 31, 635–639. [Google Scholar] [CrossRef]
- Lyon, M.S.; Wosiski-Kuhn, M.; Gillespie, R.; Caress, J.; Milligan, C. Inflammation, Immunity, and Amyotrophic Lateral Sclerosis: I. Etiology and Pathology. Muscle Nerve 2019, 59, 10–22. [Google Scholar] [CrossRef]
- Béland, L.-C.; Markovinovic, A.; Jakovac, H.; De Marchi, F.; Bilic, E.; Mazzini, L.; Kriz, J.; Munitic, I. Immunity in Amyotrophic Lateral Sclerosis: Blurred Lines between Excessive Inflammation and Inefficient Immune Responses. Brain Commun. 2020, 2, fcaa124. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.M. Microbiota-Brain-Gut Axis and Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef] [PubMed]
- Więdłocha, M.; Marcinowicz, P.; Janoska-Jaździk, M.; Szulc, A. Gut Microbiota, Kynurenine Pathway and Mental Disorders—Review. Prog. Neuropsychopharmacol. Biol. Psychiatry 2020, 110145. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, P.J.; Cryan, J.F.; Dinan, T.G.; Clarke, G. Kynurenine Pathway Metabolism and the Microbiota-Gut-Brain Axis. Neuropharmacology 2017, 112, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Erber, A.C.; Cetin, H.; Berry, D.; Schernhammer, E.S. The Role of Gut Microbiota, Butyrate and Proton Pump Inhibitors in Amyotrophic Lateral Sclerosis: A Systematic Review. Int. J. Neurosci. 2020, 130, 727–735. [Google Scholar] [CrossRef] [PubMed]
| ALS Patients | Control Subjects | p-Value | |
|---|---|---|---|
| N | 40 | 42 | |
| Gender (% female) | 42.5 | 28.6 | 0.28 |
| Age at sample collection (mean ± SD) | 65.3 ± 12.4 | 68.0 ± 12.43 | 0.33 |
| Age at onset (mean ± SD) | 64.4 ± 12.5 | ||
| BMI (kg/m2) (mean ± SD) | |||
| At diagnosis | 23.2 ± 4.2 | ||
| At 12 months | 22.6 ± 5.2 | ||
| Weight loss at diagnosis (%) (mean ± SD) | 4.54 ± 6.5 | ||
| Diagnostic delay (month) (mean ± SD) | 9.0 ± 5.8 | ||
| Site at onset (%) | |||
| bulbar | 27.5 | ||
| spinal | 72.5 | ||
| ALSFRS-r score (mean ± SD) | |||
| At diagnosis | 39.8 ± 5.3 | ||
| At 12 months | 28.2 ± 9.8 | ||
| FVC (%) (mean ± SD) | |||
| At diagnosis | 94.5 ± 26.4 | ||
| At 12 months | 85.2 ± 24.7 | ||
| Disease duration (months) (median, quartiles) | 25.0 (18.1–38.8) |
| ALS Patients | Control Subjects | Raw p-Value | |
| N | 40 | 42 | |
| Amino Acids (Mean ± SD) (µM) | |||
| Aspartic acid | 1.4 ± 1.0 | 1.1 ± 0.2 | 0.88 |
| Asparagine | 7.3 ± 2.1 | 7.0 ± 1.8 | 0.46 |
| 4-OH proline | 3.3 ± 0.6 | 3.0 ± 0.7 | 0.20 |
| Proline | 1.2 ± 0.4 | 1.5 ± 1.3 | 0.62 |
| Ornithine | 3.5 ± 1.8 | 3.4 ± 1.4 | 0.79 |
| Threonine | 29.9 ± 9.2 | 30.0 ± 8.5 | 0.97 |
| Glycine | 5.7 ± 2.9 | 5.4 ± 2.8 | 0.48 |
| Citrulline | 4.4 ± 1.1 | 4.4 ± 1.3 | 0.74 |
| Glutamic acid | 26.8 ± 48.9 | 22.9 ± 45.9 | 0.61 |
| Alanine | 41.3 ± 13.0 | 41.1 ± 12.8 | 0.88 |
| Cystine | <1 | <1 | 1 |
| Methionine | 3.2 ± 1.1 | 3.2 ± 1.0 | 0.75 |
| Tyrosine | 9.4 ± 3.3 | 9.8 ± 3.0 | 0.53 |
| Histidine | 10.8 ± 3.4 | 10.9 ± 3.1 | 0.44 |
| Lysine | 28.3 ± 6.6 | 27.1 ± 7.6 | 0.29 |
| Arginine | 17.2 ± 4.9 | 16.5 ± 5.0 | 0.14 |
| Leucine | 15.8 ± 5.6 | 16.3 ± 4.5 | 0.37 |
| Isoleucine | 5.8 ± 2.5 | 5.9 ± 2.2 | 0.56 |
| Taurine | 8.3 ± 1.9 | 8.2 ± 1.8 | 0.91 |
| Serine | 20.5 ± 5.6 | 19.3 ± 5.0 | 0.40 |
| Homocysteine | <1 | <1 | 1 |
| Phenylalanine | 10.7 ± 2.9 | 10.4 ± 2.4 | 0.52 |
| Glutamine | 523.3 ± 96.3 | 515.7 ± 89.8 | 0.75 |
| Valine | 20.2 ± 7.2 | 21.7 ± 7.1 | 0.34 |
| ALS Patients | Control Subjects | Raw p-Value | |
| N | 36 | 41 | |
| Tryptophan Metabolism Intermediates (Mean ± SD) (nM) | |||
| 3-HK | 35.0 ± 14.0 | 40.9 ± 24.1 | 0.380 |
| QUIN | 27.1 ± 18.9 | 34.6 ± 20.1 | 0.044 |
| Serotonin | 69.9 ± 9.8 | 67.5 ± 7.7 | 0.32 |
| 5-OH tryptophan | 7.4 ± 2.7 | 7.1 ± 2.1 | 0.55 |
| KYN | 44.5 ± 24.0 | 56.7 ± 26.1 | 0.036 |
| Tryptophan | 1671.2 ± 639.2 | 1840.2 ± 586.9 | 0.20 |
| 5-HIAA | 96.9 ± 63.4 | 100.4 ± 52.7 | 0.62 |
| KYNA | 1.1 ± 1.4 | 1.7 ± 1.7 | 0.029 |
| Indole 3 lactic acid | 5.5 ± 0.04 | 6.0 ± 1.8 | 0.11 |
| Indole 3 aldehyde | 2.4 ± 0.3 | 2.4 ± 0.2 | 0.72 |
| Indole 3 acetic acid | 28.6 ± 46.2 | 31.9 ± 22.9 | 0.072 |
| IDO-1 activity | 0.027 ± 0.01 | 0.031 ± 0.01 | 0.076 |
| KYNA/KYN | 0.029 ± 0.038 | 0.030 ± 0.026 | 0.24 |
| 3-HK/KYN | 1.0 ± 0.64 | 0.95 ± 1.17 | 0.059 |
| QUIN/KYNA | 54.5 ± 47.3 | 37.2 ± 32.2 | 0.15 |
| QUIN/KYN | 0.68 ± 0.33 | 0.63 ± 0.28 | 0.88 |
| 3-HK/KYNA | 84.5 ± 79 | 50.3 ± 47.8 | 0.067 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alarcan, H.; Chaumond, R.; Emond, P.; Benz-De Bretagne, I.; Lefèvre, A.; Bakkouche, S.-e.; Veyrat-Durebex, C.; Vourc’h, P.; Andres, C.; Corcia, P.; et al. Some CSF Kynurenine Pathway Intermediates Associated with Disease Evolution in Amyotrophic Lateral Sclerosis. Biomolecules 2021, 11, 691. https://doi.org/10.3390/biom11050691
Alarcan H, Chaumond R, Emond P, Benz-De Bretagne I, Lefèvre A, Bakkouche S-e, Veyrat-Durebex C, Vourc’h P, Andres C, Corcia P, et al. Some CSF Kynurenine Pathway Intermediates Associated with Disease Evolution in Amyotrophic Lateral Sclerosis. Biomolecules. 2021; 11(5):691. https://doi.org/10.3390/biom11050691
Chicago/Turabian StyleAlarcan, Hugo, Romane Chaumond, Patrick Emond, Isabelle Benz-De Bretagne, Antoine Lefèvre, Salah-eddine Bakkouche, Charlotte Veyrat-Durebex, Patrick Vourc’h, Christian Andres, Philippe Corcia, and et al. 2021. "Some CSF Kynurenine Pathway Intermediates Associated with Disease Evolution in Amyotrophic Lateral Sclerosis" Biomolecules 11, no. 5: 691. https://doi.org/10.3390/biom11050691
APA StyleAlarcan, H., Chaumond, R., Emond, P., Benz-De Bretagne, I., Lefèvre, A., Bakkouche, S.-e., Veyrat-Durebex, C., Vourc’h, P., Andres, C., Corcia, P., & Blasco, H. (2021). Some CSF Kynurenine Pathway Intermediates Associated with Disease Evolution in Amyotrophic Lateral Sclerosis. Biomolecules, 11(5), 691. https://doi.org/10.3390/biom11050691








