Identification of Low Molecular Weight Proteins and Peptides from Schistosoma mekongi Worm, Egg and Infected Mouse Sera
Abstract
1. Introduction
2. Methods
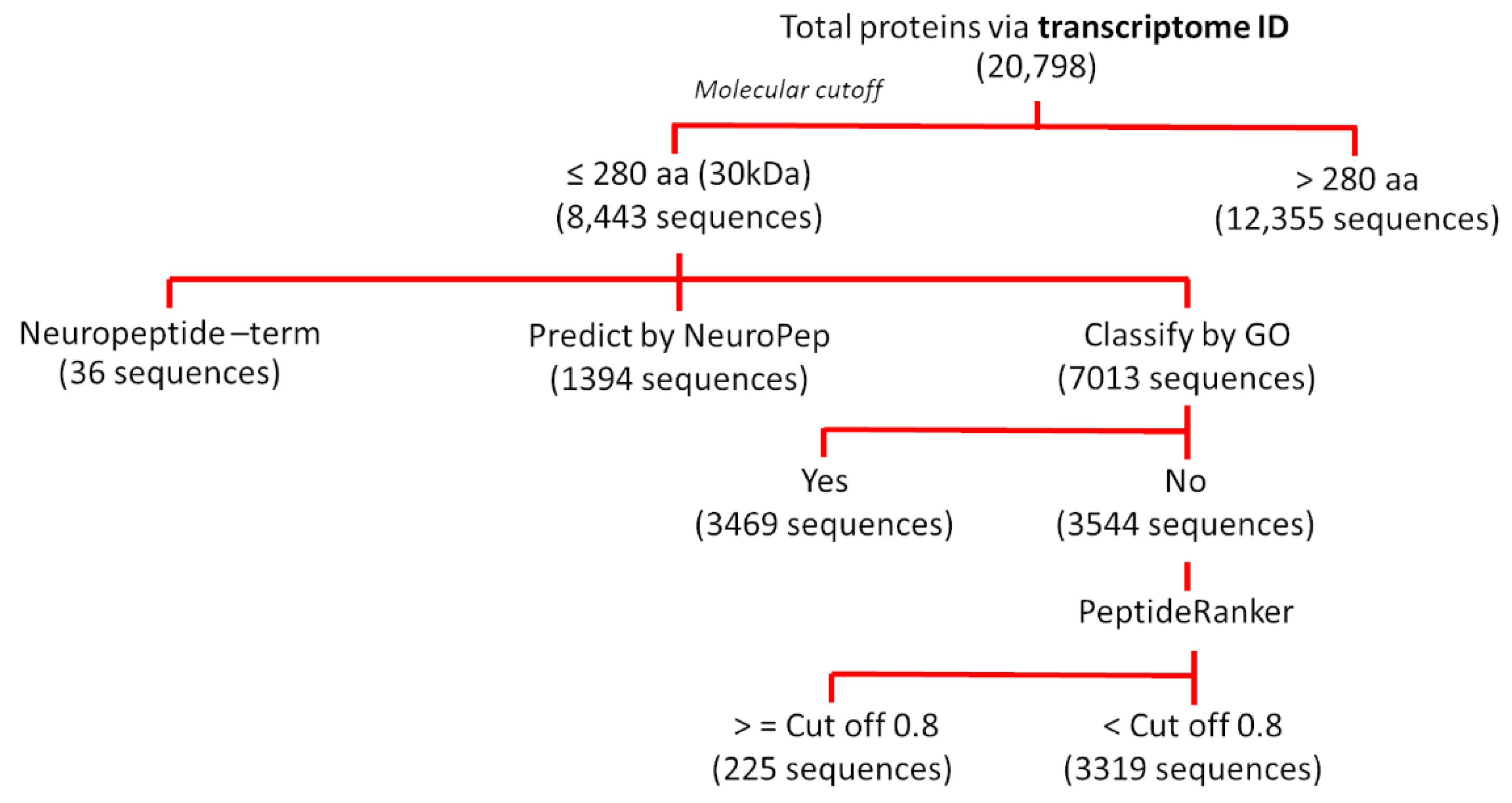
2.1. Identification of Peptidomes from S. mekongi Transcriptome Data
2.2. Preparation of Worms, Eggs, and Infected Mouse Sera
2.3. Peptide Preparation
2.4. Mass spectrometry
2.5. Bioinformatic Analysis
3. Results
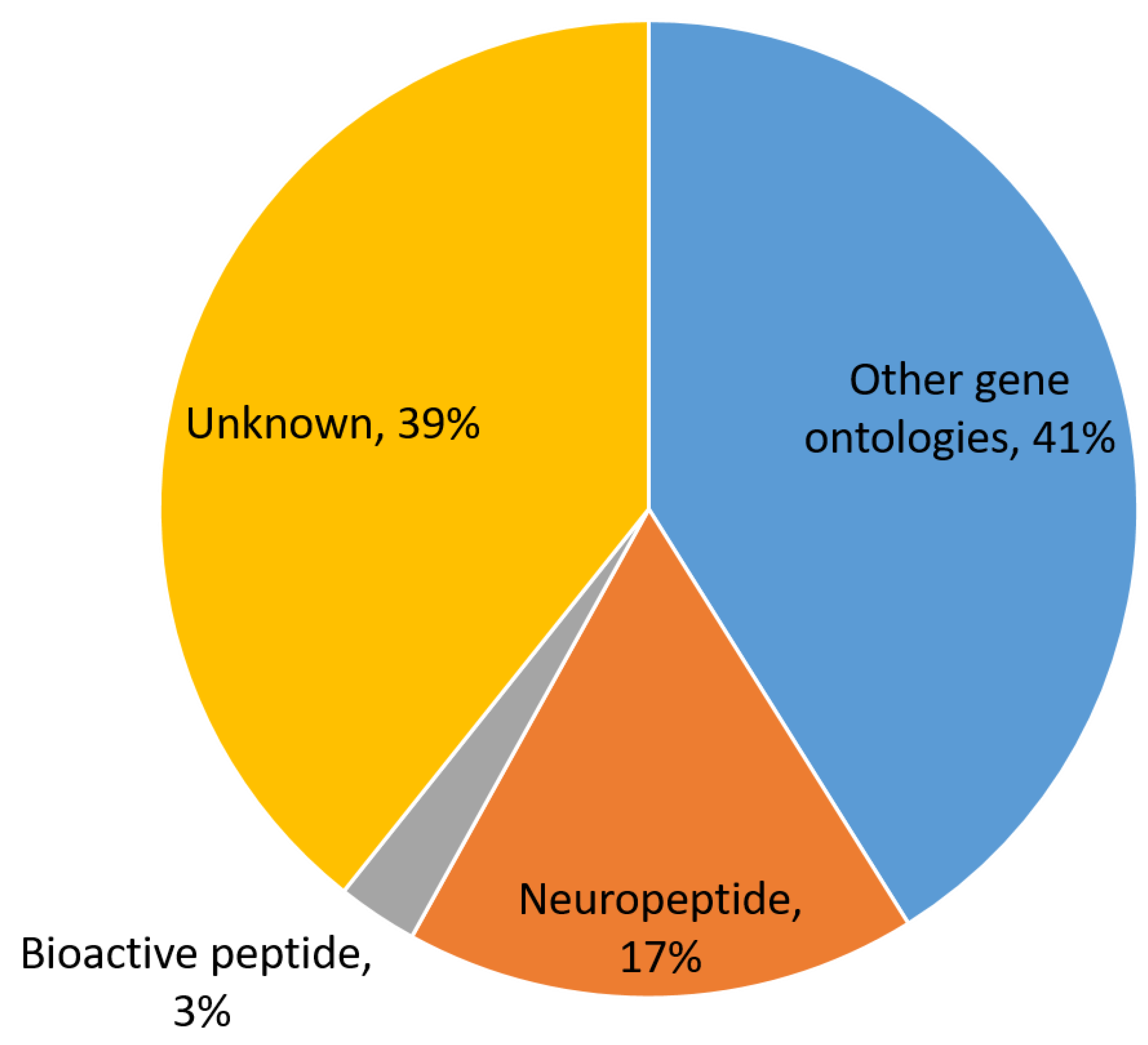
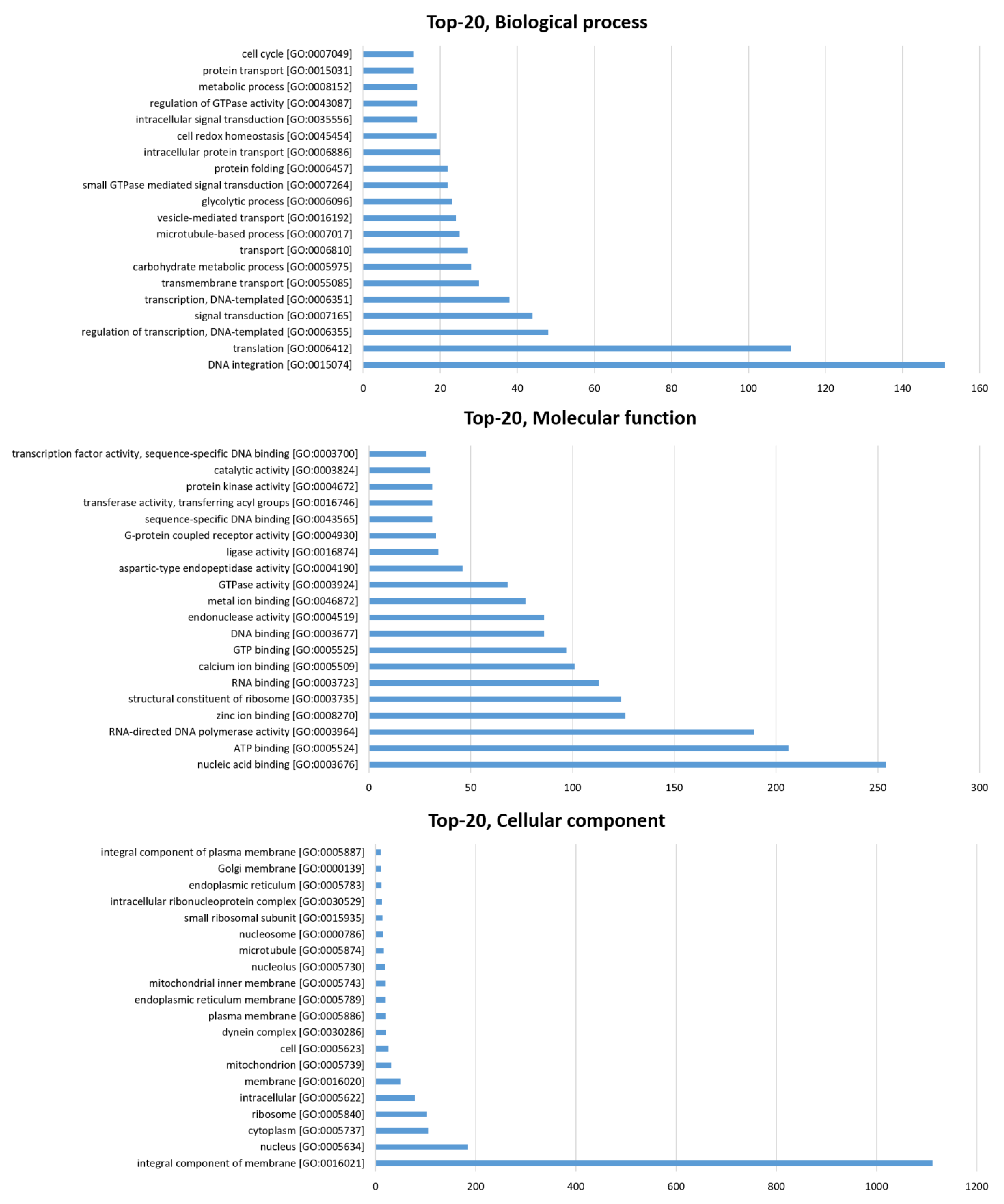
3.1. In Silico Prediction of S. mekongi Peptidomes Using Transcriptome Mining
3.2. Characterization of S. mekongi Adult Worm and Egg Peptidomes by Mass Spectrometry
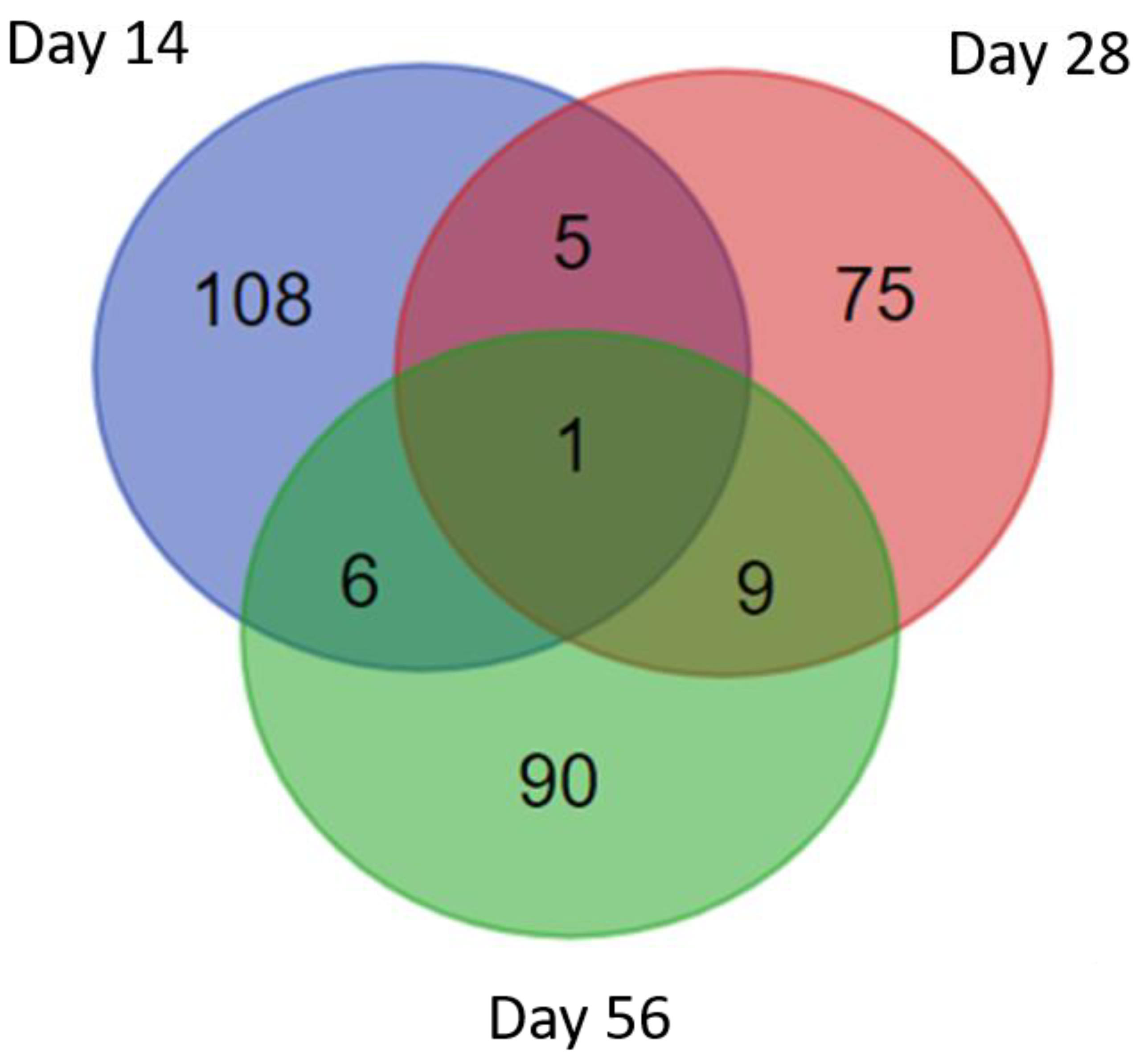
3.3. Identification of S. mekongi Peptides in Infected Mouse Sera
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Muth, S.; Sayasone, S.; Odermatt-Biays, S.; Phompida, S.; Duong, S.; Odermatt, P. Schistosoma mekongi in Cambodia and Lao People’s Democratic Republic. Adv. Parasitol. 2010, 72, 179–203. [Google Scholar] [PubMed]
- World Health Organization. Meeting Report. In: Expert Consultation to Accelerate Elimination of Asian Schistosomiasis. 2017. Available online: https://apps.who.int/iris/handle/10665/259630 (accessed on 17 February 2021).
- Wittes, R.; MacLean, J.D.; Law, C.; Lough, J.O. Three Cases of Schistosomiasis Mekongi from Northern Laos. Am. J. Trop. Med. Hyg. 1984, 33, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Van Bael, S.; Watteyne, J.; Boonen, K.; De Haes, W.; Menschaert, G.; Ringstad, N.; Horvitz, H.R.; Schoofs, L.; Husson, S.J.; Temmerman, L. Mass spectrometric evidence for neuropeptide-amidating enzymes in Caenorhabditis elegans. J. Biol. Chem. 2018, 293, 6052–6063. [Google Scholar] [CrossRef] [PubMed]
- Yew, J.Y.; Kutz, K.K.; Dikler, S.; Messinger, L.; Li, L.; Stretton, A.O. Mass spectrometric map of neuropeptide expression in Ascaris suum. J. Comp. Neurol. 2005, 488, 396–413. [Google Scholar] [CrossRef] [PubMed]
- Quinn, G.A.; Heymans, R.; Rondaj, F.; Shaw, C.; de Jong-Brink, M. Schistosoma mansoni dermaseptin-like peptide: Structural and functional characterization. J. Parasitol. 2005, 91, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- McVeigh, P.; Mair, G.R.; Novozhilova, E.; Day, A.; Zamanian, M.; Marks, N.J.; Kimber, M.J.; Day, T.A.; Maule, A.G. Schistosome I/Lamides—A new family of bioactive helminth neuropeptides. Int. J. Parasitol. 2011, 41, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.J., 3rd; Hou, X.; Romanova, E.V.; Lambrus, B.G.; Miller, C.M.; Saberi, A.; Sweedler, J.V.; Newmark, P.A. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010, 8, e1000509. [Google Scholar] [CrossRef] [PubMed]
- Balog, C.I.; Alexandrov, T.; Derks, R.J.; Hensbergen, P.J.; van Dam, G.J.; Tukahebwa, E.M.; Kabatereine, N.B.; Thiele, H.; Vennervald, B.J.; Mayboroda, O.A.; et al. The feasibility of MS and advanced data processing for monitoring Schistosoma mansoni infection. Proteom. Clin. Appl. 2010, 4, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Balog, C.I.; Hensbergen, P.J.; Derks, R.; Verweij, J.J.; Van Dam, G.J.; Vennervald, B.J.; Deelder, A.M.; Mayboroda, O.A. Novel Automated Biomarker Discovery Work Flow for Urinary Peptidomics. Clin. Chem. 2009, 55, 117–125. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Phuphisut, O.; Ajawatanawong, P.; Limpanont, Y.; Reamtong, O.; Nuamtanong, S.; Ampawong, S.; Chaimon, S.; Dekumyoy, P.; Watthanakulpanich, D.; Swierczewski, B.E.; et al. Transcriptomic analysis of male and female Schistosoma mekongi adult worms. Parasites Vectors 2018, 11, 504. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Yin, S.; Jang, R.; Wang, J.; Xue, Z.; Xu, T. NeuroPep: A comprehensive resource of neuropeptides. Database 2015, 2015, bav038. [Google Scholar] [CrossRef] [PubMed]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed]
- Christie, A.E.; Roncalli, V.; Cieslak, M.C.; Pascual, M.G.; Yu, A.; Lameyer, T.J.; Stanhope, M.E.; Dickinson, P. Prediction of a neuropeptidome for the eyestalk ganglia of the lobster Homarus americanus using a tissue-specific de novo assembled transcriptome. Gen. Comp. Endocrinol. 2017, 243, 96–119. [Google Scholar] [CrossRef] [PubMed]
- Ferro, E.S.; Rioli, V.; Castro, L.M.; Fricker, L.D. Intracellular peptides: From discovery to function. EuPA Open Proteomics 2014, 3, 143–151. [Google Scholar] [CrossRef]
- Gelman, J.S.; Sironi, J.; Castro, L.M.; Ferro, E.S.; Fricker, L.D. Peptidomic Analysis of Human Cell Lines. J. Proteome Res. 2011, 10, 1583–1592. [Google Scholar] [CrossRef] [PubMed]
- Fricker, L.D. Analysis of mouse brain peptides using mass spectrometry-based peptidomics: Implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol. BioSyst. 2010, 6, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Fricker, L.D.; Gelman, J.S.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S. Peptidomic Analysis of HEK293T Cells: Effect of the Proteasome Inhibitor Epoxomicin on Intracellular Peptides. J. Proteome Res. 2012, 11, 1981–1990. [Google Scholar] [CrossRef] [PubMed]
- ChristieElizabeth, A.E.; Stemmler, E.A.; Dickinson, P.S. Crustacean neuropeptides. Cell. Mol. Life Sci. 2010, 67, 4135–4169. [Google Scholar] [CrossRef]
- Chance, M.R.A.; Mansour, T.E. A Contribution to the Pharmacology Of Movement in the Liver Fluke. Br. J. Pharmacol. Chemother. 1953, 8, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, D.; Fairweather, I.; Holden-Dye, L.; Walker, R. Nematode neuropeptides: Localization, isolation and functions. Parasitol. Today 1996, 12, 343–351. [Google Scholar] [CrossRef]
- Bargmann, C.I.; Horvitz, H.R. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 1991, 251, 1243–1246. [Google Scholar] [CrossRef] [PubMed]
- Halton, D.W.; Maule, A.G. Flatworm nerve–muscle: Structural and functional analysis. Can. J. Zool. 2004, 82, 316–333. [Google Scholar] [CrossRef]
- Day, T.A.; Maule, A.G.; Shaw, C.; Pax, R.A. Structure-Activity Relationships of FMRFamide-Related Peptides Contracting Schistosoma mansoni Muscle. Peptides 1997, 18, 917–921. [Google Scholar] [CrossRef]
- Chang, Y.J.; Burton, T.; Ha, L.; Huang, Z.; Olajubelo, A.; Li, C. Modulation of Locomotion and Reproduction by FLP Neuropeptides in the Nematode Caenorhabditis elegans. PLoS ONE 2015, 10, e0135164. [Google Scholar] [CrossRef] [PubMed]
- Novozhilova, E.; Kimber, M.J.; Qian, H.; McVeigh, P.; Robertson, A.P.; Zamanian, M.; Maule, A.G.; Day, T.A. FMRFamide-Like Peptides (FLPs) Enhance Voltage-Gated Calcium Currents to Elicit Muscle Contraction in the Human Parasite Schistosoma mansoni. PLoS Negl. Trop. Dis. 2010, 4, e790. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.; Shaw, C.; Halton, D.; Verhaert, P.; Baguña, J. GYIRFamide: A Novel FMRFAmide-Related Peptide (FaRP) from the Triclad Turbellarian, Dugesia tigrina. Biochem. Biophys. Res. Commun. 1995, 209, 689–697. [Google Scholar] [CrossRef]
- Johnston, R.N.; Shaw, C.; Halton, D.W.; Verhaert, P.; Blair, K.L.; Brennan, G.P.; Price, D.A.; Anderson, P.A.V. Isolation, Localization, and Bioactivity of the FMRFamide-Related Neuropeptides GYIRFamide and YIRFamide from the Marine Turbellarian Bdelloura candida. J. Neurochem. 2002, 67, 814–821. [Google Scholar] [CrossRef]
- Nathoo, A.N.; Moeller, R.A.; Westlund, B.A.; Hart, A.C. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc. Natl. Acad. Sci. USA 2001, 98, 14000–14005. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.E.; Kimber, M.J.; Barton, Y.-W.; Hsu, W.; Marks, N.J.; Greer, B.; Harriott, P.; Maule, A.G.; Day, T.A. Structure and Bioactivity of Neuropeptide F from the Human Parasites Schistosoma mansoni and Schistosoma japonicum. J. Biol. Chem. 2004, 279, 39880–39885. [Google Scholar] [CrossRef]
- McGovern, M.; Yu, L.; Kosinski, M.; Greenstein, D.; Savage-Dunn, C. A role for sperm in regulation of egg-laying in the Nematode C. elegans. BMC Dev. Biol. 2007, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Nuurai, P.; Engsusophon, A.; Poomtong, T.; Sretarugsa, P.; Hanna, P.; Sobhon, P.; Wanichanon, C. Stimulatory Effects of Egg-Laying Hormone and Gonadotropin-Releasing Hormone on Reproduction of the Tropical Abalone, Haliotis asinine Linnaeus. J. Shellfish Res. 2010, 29, 627–635. [Google Scholar] [CrossRef]
- McCormick, S.D.; Bradshaw, D. Hormonal control of salt and water balance in vertebrates. Gen. Comp. Endocrinol. 2006, 147, 3–8. [Google Scholar] [CrossRef]
- Gimpl, G.; Fahrenholz, F. The Oxytocin Receptor System: Structure, Function, and Regulation. Physiol. Rev. 2001, 81, 629–683. [Google Scholar] [CrossRef] [PubMed]
- Beets, I.; Temmerman, L.; Janssen, T.; Schoofs, L. Ancient neuromodulation by vasopressin/oxytocin-related peptides. Worm 2013, 2, e24246. [Google Scholar] [CrossRef]
- Skorupskaite, K.; George, J.T.; Anderson, R.A. The kisspeptin-GnRH pathway in human reproductive health and disease. Hum. Reprod. Update 2014, 20, 485–500. [Google Scholar] [CrossRef]
- Wang, T.; Cao, Z.; Shen, Z.; Yang, J.; Chen, X.; Yang, Z.; Xu, K.; Xiang, X.; Yu, Q.; Song, Y.; et al. Existence and functions of a kisspeptin neuro-peptide signaling system in a non-chordate deuterostome species. Elife 2020, 9, e53370. [Google Scholar] [CrossRef]
- Felip, A.; Espigares, F.; Zanuy, S.; Gomez, A. Differential activation of kiss receptors by Kiss1 and Kiss2 peptides in the sea bass. Reproduction 2015, 150, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Jayasankar, V.; Tomy, S.; Wilder, M.N. Insights on Molecular Mechanisms of Ovarian Development in Decapod Crustacea: Focus on Vitellogenesis-Stimulating Factors and Pathways. Front. Endocrinol. 2020, 11, 577925. [Google Scholar] [CrossRef]
- Zmora, N.; Trant, J.; Zohar, Y.; Chung, J.S. Molt-inhibiting hormone stimulates vitellogenesis at advanced ovarian developmental stages in the female blue crab, Callinectes sapidus 1: An ovarian stage dependent involvement. Saline Syst. 2009, 5, 7. [Google Scholar] [CrossRef]
- Griffiths, E.C. Thyrotrophin releasing hormone: Endocrine and central effects. Psychoneuroendocrinology 1985, 10, 225–235. [Google Scholar] [CrossRef]
- Jékely, G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 8702–8707. [Google Scholar] [CrossRef]
- Van Sinay, E.; Mirabeau, O.; Depuydt, G.; Van Hiel, M.B.; Peymen, K.; Watteyne, J.; Zels, S.; Schoofs, L.; Beets, I. Evolutionarily conserved TRH neuropeptide pathway regulates growth in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2017, 114, E4065–E4074. [Google Scholar] [CrossRef] [PubMed]
- Bhasin, M.; Wu, M.; Tsirka, S.E. Modulation of microglial/macrophage activation by macrophage inhibitory factor (TKP) or tuftsin (TKPR) attenuates the disease course of experimental autoimmune encephalomyelitis. BMC Immunol. 2007, 8, 10. [Google Scholar] [CrossRef]
- Rogove, A.; Tsirka, S. Neurotoxic responses by microglia elicited by excitotoxic injury in the mouse hippocampus. Curr. Biol. 1998, 8, 19–25. [Google Scholar] [CrossRef]
- Tran, H.; Hamada, F.; Schwarz-Romond, T.; Bienz, M. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 2008, 22, 528–542. [Google Scholar] [CrossRef]
- Bienz, M.; Clevers, H. Linking Colorectal Cancer to Wnt Signaling. Cell 2000, 103, 311–320. [Google Scholar] [CrossRef]

| Family | Sequences |
|---|---|
| YGGW-amide related peptide | 474 |
| TRH | 265 |
| Vasopressin/oxytocin | 177 |
| FMRFamide related peptide | 74 |
| KISS1 | 26 |
| Opioid | 18 |
| LWamide neuropeptide | 14 |
| Arthropod CHH/MIH/GIH/VIH hormone | 11 |
| Serpin | 10 |
| Molluscan ELH | 7 |
| Pyrokinin | 3 |
| Neurotensin | 1 |
| NPY | 1 |
| N/A | 491 |
| No. | Precursor Protein | Score | E-Value | Peptide | Modification |
|---|---|---|---|---|---|
| D14 | |||||
| 1 | Putative cation-transporting ATPase worm | 56.76 | 0.00014 | SQFQPHFVVDTMSKGA | Oxidation (M) |
| 2 | Cyclin-dependent kinase 14 | 53.61 | 0.00083 | STPLSLVNM | Oxidation (M) |
| 3 | Uncharacterized protein | 53.46 | 0.00027 | VVYPWTQRYFDSF | |
| 4 | Uncharacterized protein | 53.35 | 0.00011 | VITSKY | |
| 5 | Mitogen-activated protein kinase 15 | 52.79 | 0.00025 | VITSGYA | |
| 6 | Uncharacterized protein | 52.22 | 0.00028 | VITSYQ | |
| 7 | Uncharacterized protein | 52.08 | 0.00032 | VLTSQY | |
| 8 | Uncharacterized protein | 47.46 | 0.001 | HDGTFISSIGD | |
| 9 | Uncharacterized protein | 47.23 | 0.0011 | ETSGTSSRVR | Glu- > pyro-Glu (N-term E) |
| 10 | SJCHGC05429 protein | 44.74 | 0.0019 | VSFITLFM | Oxidation (M) |
| 11 | Dynein heavy chain 1 cytosolic | 44.03 | 0.0022 | IFNIEPIRAKV | |
| 12 | SOSS complex subunit B1 | 43.75 | 0.0022 | AGDSSSTNR | |
| 13 | Vigilin | 43.24 | 0.003 | GRGGSKLTELLEGYKRVQV | |
| 14 | Uncharacterized protein | 42.6 | 0.0027 | LEQENRH | |
| 15 | Uncharacterized protein | 42.6 | 0.0027 | LEQEEYD | |
| 16 | Serine/threonine-protein kinase SIK3 | 42.45 | 0.0028 | PSIPASNNN | |
| 17 | Putative helix-loop-helix zipper protein | 41.88 | 0.0034 | SSNTSSNPT | |
| 18 | Rho GTPase-activating protein 35 | 41.81 | 0.0039 | SAFSAPNHS | |
| 19 | Helicase | 41.29 | 0.0043 | DVGLITGDIKVAPD | |
| 20 | Uncharacterized protein | 41.24 | 0.015 | SPIKKEEVPAGFSPSEYHLIKKMRDILR | Oxidation (M) |
| D28 | |||||
| 1 | Uncharacterized protein | 74.43 | 2.6E-06 | QWANLMEKIQASVATNPIITPVAQENQ | Gln- > pyro-Glu (N-term Q); Oxidation (M) |
| 2 | Putative glycosyltransferase | 60.49 | 0.000051 | IDVMPSIKTPIE | |
| 3 | Putative actin | 56.38 | 0.00013 | VFPSIVGRPR | |
| 4 | Bifunctional protein NCOAT | 52.25 | 0.00034 | NSVAVTLEDL | |
| 5 | Putative DNA polymerase delta small subunit | 51.8 | 0.00039 | FAGSGQVKPGHSM | |
| 6 | Glycosyltransferase 14 family member | 50.1 | 0.00091 | TKRQEFF | |
| 7 | Uncharacterized protein | 45.94 | 0.0013 | THTLTLEN | |
| 8 | Coiled-coil domain-containing protein 170 | 45.9 | 0.0014 | EYVRHNEK | Glu- > pyro-Glu (N-term E) |
| 9 | Coiled-coil domain-containing protein 81 | 45.14 | 0.0018 | EIIFNDIGKLRI | Glu- > pyro-Glu (N-term E) |
| 10 | Uncharacterized protein | 44.23 | 0.002 | THVDIDKT | |
| 11 | Putative multidrug resistance protein 1, 2, 3 | 44.09 | 0.0022 | QSRANLVTGIIALL | |
| 12 | Uncharacterized protein | 44.02 | 0.0044 | CLSVMQII | |
| 13 | Pogo transposable element with ZNF domain | 42.73 | 0.0036 | NIENLDCLECGKCMGD | |
| 14 | Putative organic solute transporter | 42.32 | 0.0031 | KQATLQFCV | |
| 15 | Protein kinase | 42.08 | 0.0037 | TEPTIKRMLAENVS | Oxidation (M) |
| 16 | Uncharacterized protein | 41.47 | 0.0041 | SRQAVQTMGSLFQ | Oxidation (M) |
| 17 | Actin bundling/missing in metastasis-related | 41.18 | 0.0045 | TTVVSNNGI | |
| 18 | Putative Family with sequence similarity 98, member A | 40.69 | 0.0049 | GISDRQWS | |
| 19 | Nuclear factor 1 C-type | 40.67 | 0.008 | LAKENSFF | |
| 20 | Uncharacterized protein | 40.4 | 0.0051 | SRQAVQTMGSLF | Oxidation (M) |
| D56 | |||||
| 1 | Uncharacterized protein | 60.61 | 0.000054 | NEVHTMLGQSTEEIRA | Oxidation (M) |
| 2 | Putative organic solute transporter | 51.52 | 0.00037 | KQATLQFCV | |
| 3 | Uncharacterized protein | 49.52 | 0.00067 | LPFLQELDSDQILR | |
| 4 | Voltage-dependent calcium channel OS = Schistosoma mansoni GN = Smp_197640 PE = 4 SV = 1 | 49.14 | 0.0037 | TTSSPLTLIL | |
| 5 | Putative importin-beta 2 | 46.95 | 0.0015 | MLMPPLFEKWNAL | |
| 6 | Uncharacterized protein | 46.95 | 0.0011 | YDEGKIGIFI | |
| 7 | Zinc finger MIZ domain-containing protein 1 | 46.95 | 0.0015 | PFKCEQPPNGCADAL | |
| 8 | Complement component 1 Q subcomponent-binding protein, mitochondrial | 45.48 | 0.0016 | EAHPDLRI | Glu- > pyro-Glu (N-term E) |
| 9 | 39S ribosomal protein L46, mitochondrial | 45.03 | 0.0018 | RTRSGVNIFPI | |
| 10 | Uncharacterized protein | 44.96 | 0.003 | LQLMVPV | |
| 11 | Proteasome 26S subunit subunit 4 ATPase | 44.88 | 0.0018 | LSFVDKGMLE | Oxidation (M) |
| 12 | Uncharacterized protein | 44.71 | 0.002 | SVATNPIITPVAQENQ | |
| 13 | Teneurin-2 | 44.14 | 0.0024 | ISILILAFLLAL | |
| 14 | Protein kinase | 43.75 | 0.0024 | TQCIAYAAGY | |
| 15 | Tyrosine-protein kinase Abl | 43.57 | 0.0025 | IEAEVALELEKQP | |
| 16 | Uncharacterized protein | 42.4 | 0.0056 | LEEKMLM | 2 Oxidation (M) |
| 17 | Protein kinase | 42.05 | 0.0037 | TEPTIKRMLAENVS | Oxidation (M) |
| 18 | Rhabdoid tumor deletion region protein 1 | 41.97 | 0.0047 | TTNHGRYTTLNAGAI | |
| 19 | Cadherin-related tumor suppressor | 41.64 | 0.0038 | MMLSNDLIDS | Oxidation (M) |
| 20 | Uncharacterized protein | 40.32 | 0.0048 | LTMNTEL | Oxidation (M) |
| Sequence | Score | E-Value | Modification |
|---|---|---|---|
| Day 14 | |||
| SSNESTADINQTTG * | 36.65 | 0.012 | |
| TSYSPYASPRSSSR * | 21.27 | 4.40 × 10−1 | |
| TYTQMPSTNIPLSTPSE * | 28.91 | 0.079 | Oxidation (M) |
| KLSSPLTGNQIHPALQLVFN * | 20.94 | 1.30 | |
| Day 28 | |||
| HSTLPV | 11.45 | 3.10 | |
| DGGAKWPCGV * | 39 | 0.0073 | |
| SSNESTADINQTTG * | 32.69 | 0.031 | |
| Day 56 | |||
| DGGAKWPCGV * | 34.35 | 0.021 | |
| VMCFASSPQPLC * | 24.27 | 2.10 × 10−1 | Oxidation (M) |
| ESPLTSCGGTTLPV * | 26.69 | 1.20 × 10−1 | Glu- > pyro-Glu (N-term E) |
| S. mekongi | S. japonicum | S. mansoni | S. haematobium | |
|---|---|---|---|---|
| S. mekongi | 100 | 93.24 | 81.49 | 81.12 |
| S. japonicum | 93.24 | 100 | 82.02 | 81.66 |
| S. mansoni | 81.49 | 82.02 | 100 | 94.86 |
| S. haematobium | 81.12 | 81.66 | 94.86 | 100 |
| O. felineus | 55.13 | 54.77 | 56.23 | 56.12 |
| C. sinensis | 54.89 | 54.53 | 55.87 | 55.65 |
| O. viverrini | 54.89 | 54.18 | 55.87 | 55.53 |
| M. musculus | 42.70 | 42.70 | 42.25 | 42.51 |
| H. sapiens | 39.52 | 39.84 | 40.22 | 40.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thiangtrongjit, T.; Simanon, N.; Adisakwattana, P.; Limpanont, Y.; Chusongsang, P.; Chusongsang, Y.; Reamtong, O. Identification of Low Molecular Weight Proteins and Peptides from Schistosoma mekongi Worm, Egg and Infected Mouse Sera. Biomolecules 2021, 11, 559. https://doi.org/10.3390/biom11040559
Thiangtrongjit T, Simanon N, Adisakwattana P, Limpanont Y, Chusongsang P, Chusongsang Y, Reamtong O. Identification of Low Molecular Weight Proteins and Peptides from Schistosoma mekongi Worm, Egg and Infected Mouse Sera. Biomolecules. 2021; 11(4):559. https://doi.org/10.3390/biom11040559
Chicago/Turabian StyleThiangtrongjit, Tipparat, Nattapon Simanon, Poom Adisakwattana, Yanin Limpanont, Phiraphol Chusongsang, Yupa Chusongsang, and Onrapak Reamtong. 2021. "Identification of Low Molecular Weight Proteins and Peptides from Schistosoma mekongi Worm, Egg and Infected Mouse Sera" Biomolecules 11, no. 4: 559. https://doi.org/10.3390/biom11040559
APA StyleThiangtrongjit, T., Simanon, N., Adisakwattana, P., Limpanont, Y., Chusongsang, P., Chusongsang, Y., & Reamtong, O. (2021). Identification of Low Molecular Weight Proteins and Peptides from Schistosoma mekongi Worm, Egg and Infected Mouse Sera. Biomolecules, 11(4), 559. https://doi.org/10.3390/biom11040559







