Molecular Mechanisms behind Inherited Neurodegeneration of the Optic Nerve
Abstract
1. Introduction
2. Complex I Dysfunction: The Paradigm of Leber’s Hereditary Optic Neuropathy (LHON)
3. Mitochondrial Dynamics Failure: From Dominant Optic Atrophy (DOA) to Complex Syndromes
3.1. Mitochondrial Fusion Dysfunction as the Cause of DOA or “Plus” Syndromic Forms
3.2. Mitochondrial Fission Dysfunction as the Cause of DOA and Syndromic Forms
4. Mitochondria-Associated Membranes: The Case of Wolfram Syndrome and More
4.1. MAMs Function in Calcium and Lipids Homeostasis, Autophagy, and Apoptosis
4.2. MAMs Dysfunction as The Cause of Optic Neuropathy and Neurodegenerative Syndromes
4.3. Wolfram Syndrome: The MAMs Perspective
5. Other Emerging Mechanisms: From Oxidative Phosphorylation to Lipid Metabolism
6. Concluding Remarks and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carelli, V.; la Morgia, C.; Valentino, M.L.; Barboni, P.; Ross-Cisneros, F.N.; Sadun, A.A. Retinal Ganglion Cell Neurodegeneration in Mitochondrial Inherited Disorders. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Yu-Wai-Man, P.; Votruba, M.; Burté, F.; la Morgia, C.; Barboni, P.; Carelli, V. A Neurodegenerative Perspective on Mitochondrial Optic Neuropathies. Acta Neuropathol. 2016, 132, 789–806. [Google Scholar] [CrossRef]
- Carelli, V.; Ross-Cisneros, F.N.; Sadun, A.A. Mitochondrial Dysfunction as a Cause of Optic Neuropathies. Prog. Retin. Eye Res. 2004, 23, 53–89. [Google Scholar] [CrossRef]
- Yu-Wai-Man, P.; Griffiths, P.G.; Chinnery, P.F. Mitochondrial Optic Neuropathies—Disease Mechanisms and Therapeutic Strategies. Prog. Retin. Eye Res. 2011, 30, 81–114. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, S.; Schon, E.A.; Carelli, V.; Hirano, M. The Clinical Maze of Mitochondrial Neurology. Nat. Rev. Neurol. 2013, 9, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Gorman, G.S.; Chinnery, P.F.; DiMauro, S.; Hirano, M.; Koga, Y.; McFarland, R.; Suomalainen, A.; Thorburn, D.R.; Zeviani, M.; Turnbull, D.M. Mitochondrial Diseases. Nat. Rev. Dis. Primers 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. Mitochondrial Genetic Medicine. Nat. Genet. 2018, 50, 1642–1649. [Google Scholar] [CrossRef]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An Updated Mitochondrial Proteome Now with Sub-Organelle Localization and Pathway Annotations. Nucleic Acids Res. 2021, 49, D1541–D1547. [Google Scholar] [CrossRef]
- Lenaers, G.; Neutzner, A.; le Dantec, Y.; Jüschke, C.; Xiao, T.; Decembrini, S.; Swirski, S.; Kieninger, S.; Agca, C.; Kim, U.S.; et al. Dominant Optic Atrophy: Culprit Mitochondria in the Optic Nerve. Prog. Retin. Eye Res. 2020, 100935. [Google Scholar] [CrossRef]
- Carelli, V.; Chan, D.C. Mitochondrial DNA: Impacting Central and Peripheral Nervous Systems. Neuron 2014, 84, 1126–1142. [Google Scholar] [CrossRef]
- Amati-Bonneau, P.; Valentino, M.L.; Reynier, P.; Gallardo, M.E.; Bornstein, B.; Boissiere, A.; Campos, Y.; Rivera, H.; de la Aleja, J.G.; Carroccia, R.; et al. OPA1 Mutations Induce Mitochondrial DNA Instability and Optic Atrophy “plus” Phenotypes. Brain 2008, 131, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Jurkute, N.; Leu, C.; Pogoda, H.; Arno, G.; Robson, A.G.; Nürnberg, G.; Altmüller, J.; Thiele, H.; Motameny, S.; Toliat, M.R.; et al. SSBP1 Mutations in Dominant Optic Atrophy with Variable Retinal Degeneration. Ann. Neurol. 2019, 86, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Del Dotto, V.; Ullah, F.; di Meo, I.; Magini, P.; Gusic, M.; Maresca, A.; Caporali, L.; Palombo, F.; Tagliavini, F.; Baugh, E.H.; et al. SSBP1 Mutations Cause MtDNA Depletion Underlying a Complex Optic Atrophy Disorder. J. Clin. Investig. 2019, 130, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Piro-Mégy, C.; Sarzi, E.; Tarrés-Solé, A.; Péquignot, M.; Hensen, F.; Quilès, M.; Manes, G.; Chakraborty, A.; Sénéchal, A.; Bocquet, B.; et al. Dominant Mutations in MtDNA Maintenance Gene SSBP1 Cause Optic Atrophy and Foveopathy. J. Clin. Investig. 2019, 130, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Burté, F.; Carelli, V.; Chinnery, P.F.; Yu-Wai-Man, P. Disturbed Mitochondrial Dynamics and Neurodegenerative Disorders. Nat. Rev. Neurol. 2015, 11, 11–24. [Google Scholar] [CrossRef]
- Maresca, A.; la Morgia, C.; Caporali, L.; Valentino, M.L.; Carelli, V. The Optic Nerve: A “Mito-Window” on Mitochondrial Neurodegeneration. Mol. Cell. Neurosci. 2013, 55, 62–76. [Google Scholar] [CrossRef]
- Li, L.; Venkataraman, L.; Chen, S.; Fu, H. Function of WFS1 and WFS2 in the Central Nervous System: Implications for Wolfram Syndrome and Alzheimer’s Disease. Neurosci. Biobehav. Rev. 2020, 118, 775–783. [Google Scholar] [CrossRef]
- Angebault, C.; Fauconnier, J.; Patergnani, S.; Rieusset, J.; Danese, A.; Affortit, C.A.; Jagodzinska, J.; Mégy, C.; Quiles, M.; Cazevieille, C.; et al. ER-Mitochondria Cross-Talk Is Regulated by the Ca2+ Sensor NCS1 and Is Impaired in Wolfram Syndrome. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef]
- La Morgia, C.; Maresca, A.; Amore, G.; Gramegna, L.L.; Carbonelli, M.; Scimonelli, E.; Danese, A.; Patergnani, S.; Caporali, L.; Tagliavini, F.; et al. Calcium Mishandling in Absence of Primary Mitochondrial Dysfunction Drives Cellular Pathology in Wolfram Syndrome. Sci Rep 2020, 10, 4785. [Google Scholar] [CrossRef]
- Krols, M.; van Isterdael, G.; Asselbergh, B.; Kremer, A.; Lippens, S.; Timmerman, V.; Janssens, S. Mitochondria-Associated Membranes as Hubs for Neurodegeneration. Acta Neuropathol. 2016, 131, 505–523. [Google Scholar] [CrossRef]
- Lewis, S.C.; Uchiyama, L.F.; Nunnari, J. ER-Mitochondria Contacts Couple MtDNA Synthesis with Mitochondrial Division in Human Cells. Science 2016, 353. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Tamura, N.; Kono, N.; Shimanaka, Y.; Arai, H.; Yamamoto, H.; Mizushima, N. Autophagosome Formation Is Initiated at Phosphatidylinositol Synthase-enriched ER Subdomains. EMBO J. 2017, 36, 1719–1735. [Google Scholar] [CrossRef] [PubMed]
- Carelli, V.; la Morgia, C.; Ross-Cisneros, F.N.; Sadun, A.A. Optic Neuropathies: The Tip of the Neurodegeneration Iceberg|Human Molecular Genetics|Oxford Academic. Available online: https://academic.oup.com/hmg/article/26/R2/R139/4036433 (accessed on 6 February 2021).
- Gerber, S.; Ding, M.G.; Gérard, X.; Zwicker, K.; Zanlonghi, X.; Rio, M.; Serre, V.; Hanein, S.; Munnich, A.; Rotig, A.; et al. Compound Heterozygosity for Severe and Hypomorphic NDUFS2 Mutations Cause Non-Syndromic LHON-like Optic Neuropathy. J. Med. Genet. 2017, 54, 346–356. [Google Scholar] [CrossRef]
- Stenton, S.L.; Sheremet, N.L.; Catarino, C.B.; Andreeva, N.; Assouline, Z.; Barboni, P.; Barel, O.; Berutti, R.; Bychkov, I.O.; Caporali, L.; et al. Impaired Complex I Repair Causes Recessive Leber’s Hereditary Optic Neuropathy. J. Clin. Invest. 2021. [Google Scholar] [CrossRef] [PubMed]
- Agip, A.-N.A.; Blaza, J.N.; Fedor, J.G.; Hirst, J. Mammalian Respiratory Complex I Through the Lens of Cryo-EM. Annu. Rev. Biophys. 2019, 48, 165–184. [Google Scholar] [CrossRef]
- Fiedorczuk, K.; Sazanov, L.A. Mammalian Mitochondrial Complex I Structure and Disease-Causing Mutations. Trends Cell Biol. 2018, 28, 835–867. [Google Scholar] [CrossRef]
- Majander, A. Electron Transfer Properties of NADH: Ubiquinone Reductase in the ND1/3460 and the ND4/11778 Mutations of the Leber Hereditary Optic Neuroretinopathy (LHON). FEBS Lett. 1991, 292, 289–292. [Google Scholar] [CrossRef]
- Carelli, V.; Ghelli, A.; Ratta, M.; Bacchilega, E.; Sangiorgi, S.; Mancini, R.; Leuzzi, V.; Cortelli, P.; Montagna, P.; Lugaresi, E.; et al. Leber’s Hereditary Optic Neuropathy: Biochemical Effect of 11778/ND4 and 3460/ND1 Mutations and Correlation with the Mitochondrial Genotype. Neurology 1997, 48, 1623–1632. [Google Scholar] [CrossRef]
- Grba, D.N.; Hirst, J. Mitochondrial Complex I Structure Reveals Ordered Water Molecules for Catalysis and Proton Translocation. Nat. Struct. Mol. Biol. 2020, 27, 892–900. [Google Scholar] [CrossRef]
- Larsson, N.-G.; Andersen, O.; Holme, E.; Oldfors, A.; Wahlström, J. Leber’s Hereditary Optic Neuropathy and Complex I Deficiency in Muscle. Ann. Neurol. 1991, 30, 701–708. [Google Scholar] [CrossRef]
- Degli-Esposti, M.; Carelli, V.; Ghelli, A.; Ratta, M.; Crimi, M.; Sangiorgi, S.; Montagna, P.; Lenaz, G.; Lugaresi, E.; Cortelli, P. Functional Alterations of the Mitochondrially Encoded ND4 Subunit Associated with Leber’s Hereditary Optic Neuropathy. FEBS Lett. 1994, 352, 375–379. [Google Scholar] [CrossRef]
- Carelli, V.; Ghelli, A.; Bucchi, L.; Montagna, P.; De Negri, A.; Leuzzi, V.; Carducci, C.; Lenaz, G.; Lugaresi, E.; Degli Esposti, M. Biochemical Features of MtDNA 14484 (ND6/M64V) Point Mutation Associated with Leber’s Hereditary Optic Neuropathy. Ann. Neurol. 1999, 45, 320–328. [Google Scholar] [CrossRef]
- Brown, M.D.; Trounce, I.A.; Jun, A.S.; Allen, J.C.; Wallace, D.C. Functional Analysis of Lymphoblast and Cybrid Mitochondria Containing the 3460, 11778, or 14484 Leber’s Hereditary Optic Neuropathy Mitochondrial DNA Mutation. J. Biol. Chem. 2000, 275, 39831–39836. [Google Scholar] [CrossRef]
- Brown, M.D. The Enigmatic Relationship between Mitochondrial Dysfunction and Leber’s Hereditary Optic Neuropathy. J. Neurol. Sci. 1999, 165, 1–5. [Google Scholar] [CrossRef]
- Baracca, A.; Solaini, G.; Sgarbi, G.; Lenaz, G.; Baruzzi, A.; Schapira, A.H.V.; Martinuzzi, A.; Carelli, V. Severe Impairment of Complex I–Driven Adenosine Triphosphate Synthesis in Leber Hereditary Optic Neuropathy Cybrids. Arch. Neurol. 2005, 62, 730. [Google Scholar] [CrossRef] [PubMed]
- Kampjut, D.; Sazanov, L.A. The Coupling Mechanism of Mammalian Respiratory Complex I. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Danielson, S.R. Cells Bearing Mutations Causing Leber’s Hereditary Optic Neuropathy Are Sensitized to Fas-Induced Apoptosis. J. Biol. Chem. 2002, 277, 5810–5815. [Google Scholar] [CrossRef]
- Ghelli, A.; Zanna, C.; Porcelli, A.M.; Schapira, A.H.V.; Martinuzzi, A.; Carelli, V.; Rugolo, M. Leber’s Hereditary Optic Neuropathy (LHON) Pathogenic Mutations Induce Mitochondrial-Dependent Apoptotic Death in Transmitochondrial Cells Incubated with Galactose Medium. J. Biol. Chem. 2003, 278, 4145–4150. [Google Scholar] [CrossRef] [PubMed]
- Zanna, C.; Ghelli, A.; Porcelli, A.M.; Martinuzzi, A.; Carelli, V.; Rugolo, M. Caspase-Independent Death of Leber’s Hereditary Optic Neuropathy Cybrids Is Driven by Energetic Failure and Mediated by AIF and Endonuclease G. Apoptosis 2005, 10, 997–1007. [Google Scholar] [CrossRef]
- Caporali, L.; Maresca, A.; Capristo, M.; del Dotto, V.; Tagliavini, F.; Valentino, M.L.; la Morgia, C.; Carelli, V. Incomplete Penetrance in Mitochondrial Optic Neuropathies. Mitochondrion 2017, 36, 130–137. [Google Scholar] [CrossRef]
- Torroni, A.; Petrozzi, M.; D’Urbano, L.; Sellitto, D.; Zeviani, M.; Carrara, F.; Carducci, C.; Leuzzi, V.; Carelli, V.; Barboni, P.; et al. Haplotype and Phylogenetic Analyses Suggest That One European-Specific MtDNA Background Plays a Role in the Expression of Leber Hereditary Optic Neuropathy by Increasing the Penetrance of the Primary Mutations 11778 and 14484. Am. J. Hum. Genet. 1997, 60, 1107–1121. [Google Scholar] [PubMed]
- Carelli, V.; Achilli, A.; Valentino, M.L.; Rengo, C.; Semino, O.; Pala, M.; Olivieri, A.; Mattiazzi, M.; Pallotti, F.; Carrara, F.; et al. Haplogroup Effects and Recombination of Mitochondrial DNA: Novel Clues from the Analysis of Leber Hereditary Optic Neuropathy Pedigrees. Am. J. Hum. Genet. 2006, 78, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Hudson, G.; Amati-Bonneau, P.; Blakely, E.L.; Stewart, J.D.; He, L.; Schaefer, A.M.; Griffiths, P.G.; Ahlqvist, K.; Suomalainen, A.; Reynier, P.; et al. Mutation of OPA1 Causes Dominant Optic Atrophy with External Ophthalmoplegia, Ataxia, Deafness and Multiple Mitochondrial DNA Deletions: A Novel Disorder of MtDNA Maintenance. Brain 2008, 131, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Pello, R.; Martín, M.A.; Carelli, V.; Nijtmans, L.G.; Achilli, A.; Pala, M.; Torroni, A.; Gómez-Durán, A.; Ruiz-Pesini, E.; Martinuzzi, A.; et al. Mitochondrial DNA Background Modulates the Assembly Kinetics of OXPHOS Complexes in a Cellular Model of Mitochondrial Disease. Hum. Mol. Genet. 2008, 17, 4001–4011. [Google Scholar] [CrossRef]
- Ghelli, A.; Porcelli, A.M.; Zanna, C.; Vidoni, S.; Mattioli, S.; Barbieri, A.; Iommarini, L.; Pala, M.; Achilli, A.; Torroni, A.; et al. The Background of Mitochondrial DNA Haplogroup J Increases the Sensitivity of Leber’s Hereditary Optic Neuropathy Cells to 2,5-Hexanedione Toxicity. PLoS ONE 2009, 4, e7922. [Google Scholar] [CrossRef]
- Carelli, V.; Franceschini, F.; Venturi, S.; Barboni, P.; Savini, G.; Barbieri, G.; Pirro, E.; la Morgia, C.; Valentino, M.L.; Zanardi, F.; et al. Grand Rounds: Could Occupational Exposure to n-Hexane and Other Solvents Precipitate Visual Failure in Leber Hereditary Optic Neuropathy? Environ. Health Perspect. 2007, 115, 113–115. [Google Scholar] [CrossRef]
- Strobbe, D.; Caporali, L.; Iommarini, L.; Maresca, A.; Montopoli, M.; Martinuzzi, A.; Achilli, A.; Olivieri, A.; Torroni, A.; Carelli, V.; et al. Haplogroup J Mitogenomes Are the Most Sensitive to the Pesticide Rotenone: Relevance for Human Diseases. Neurobiol. Dis. 2018, 114, 129–139. [Google Scholar] [CrossRef]
- Giordano, C.; Montopoli, M.; Perli, E.; Orlandi, M.; Fantin, M.; Ross-Cisneros, F.N.; Caparrotta, L.; Martinuzzi, A.; Ragazzi, E.; Ghelli, A.; et al. Oestrogens Ameliorate Mitochondrial Dysfunction in Leber’s Hereditary Optic Neuropathy. Brain 2011, 134, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Iommarini, L.; Giordano, L.; Maresca, A.; Pisano, A.; Valentino, M.L.; Caporali, L.; Liguori, R.; Deceglie, S.; Roberti, M.; et al. Efficient Mitochondrial Biogenesis Drives Incomplete Penetrance in Leber’s Hereditary Optic Neuropathy. Brain 2014, 137, 335–353. [Google Scholar] [CrossRef]
- Kirkman, M.A.; Yu-Wai-Man, P.; Korsten, A.; Leonhardt, M.; Dimitriadis, K.; de Coo, I.F.; Klopstock, T.; Chinnery, P.F. Gene-Environment Interactions in Leber Hereditary Optic Neuropathy. Brain 2009, 132, 2317–2326. [Google Scholar] [CrossRef]
- Carelli, V.; d’Adamo, P.; Valentino, M.L.; la Morgia, C.; Ross-Cisneros, F.N.; Caporali, L.; Maresca, A.; Loguercio-Polosa, P.; Barboni, P.; de Negri, A.; et al. Parsing the Differences in Affected with LHON: Genetic versus Environmental Triggers of Disease Conversion. Brain 2016, 139, e17. [Google Scholar] [CrossRef] [PubMed]
- Giordano, L.; Deceglie, S.; d’Adamo, P.; Valentino, M.L.; la Morgia, C.; Fracasso, F.; Roberti, M.; Cappellari, M.; Petrosillo, G.; Ciaravolo, S.; et al. Cigarette Toxicity Triggers Leber’s Hereditary Optic Neuropathy by Affecting MtDNA Copy Number, Oxidative Phosphorylation and ROS Detoxification Pathways. Cell Death Dis. 2015, 6, e2021. [Google Scholar] [CrossRef] [PubMed]
- Hanein, S.; Perrault, I.; Roche, O.; Gerber, S.; Khadom, N.; Rio, M.; Boddaert, N.; Jean-Pierre, M.; Brahimi, N.; Serre, V.; et al. TMEM126A, Encoding a Mitochondrial Protein, Is Mutated in Autosomal-Recessive Nonsyndromic Optic Atrophy. Am. J. Hum. Genet. 2009, 84, 493–498. [Google Scholar] [CrossRef]
- D’Angelo, L.; Astro, E.; Luise, M.D.; Kurelac, I.; Umesh-Ganesh, N.; Ding, S.; Fearnley, I.M.; Zeviani, M.; Gasparre, G.; Porcelli, A.M.; et al. Biogenesis of NDUFS3-Less Complex I Indicates TMEM126A/OPA7 as an Assembly Factor of the ND4-Module. bioRxiv 2020. [Google Scholar] [CrossRef]
- Formosa, L.E.; Reljic, B.; Sharpe, A.J.; Muellner-Wong, L.; Stroud, D.A.; Ryan, M.T. Optic Atrophy-Associated TMEM126A Is an Assembly Factor for the ND4-Module of Mitochondrial Complex I. bioRxiv 2020. [Google Scholar] [CrossRef]
- Angebault, C.; Guichet, P.-O.; Talmat-Amar, Y.; Charif, M.; Gerber, S.; Fares-Taie, L.; Gueguen, N.; Halloy, F.; Moore, D.; Amati-Bonneau, P.; et al. Recessive Mutations in RTN4IP1 Cause Isolated and Syndromic Optic Neuropathies. Am. J. Hum. Genet. 2015, 97, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Charif, M.; Nasca, A.; Thompson, K.; Gerber, S.; Makowski, C.; Mazaheri, N.; Bris, C.; Goudenège, D.; Legati, A.; Maroofian, R.; et al. Neurologic Phenotypes Associated With Mutations in RTN4IP1 (OPA10) in Children and Young Adults. JAMA Neurol. 2018, 75, 105–113. [Google Scholar] [CrossRef]
- Tilokani, L.; Nagashima, S.; Paupe, V.; Prudent, J. Mitochondrial Dynamics: Overview of Molecular Mechanisms. Essays Biochem. 2018, 62, 341–360. [Google Scholar] [CrossRef]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The Cell Biology of Mitochondrial Membrane Dynamics. Nat. Rev. Mol. Cell Biol. 2020, 21, 204–224. [Google Scholar] [CrossRef]
- Del Dotto, V.; Fogazza, M.; Carelli, V.; Rugolo, M.; Zanna, C. Eight Human OPA1 Isoforms, Long and Short: What Are They For? Biochim. Biophys. Acta Bioenerg. 2018, 1859, 263–269. [Google Scholar] [CrossRef]
- Griparic, L.; Kanazawa, T.; van der Bliek, A.M. Regulation of the Mitochondrial Dynamin-like Protein Opa1 by Proteolytic Cleavage. J. Cell Biol. 2007, 178, 757–764. [Google Scholar] [CrossRef]
- Ehses, S.; Raschke, I.; Mancuso, G.; Bernacchia, A.; Geimer, S.; Tondera, D.; Martinou, J.-C.; Westermann, B.; Rugarli, E.I.; Langer, T. Regulation of OPA1 Processing and Mitochondrial Fusion by M-AAA Protease Isoenzymes and OMA1. J. Cell Biol. 2009, 187, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.; Wai, T.; Baker, M.J.; Kladt, N.; Schauss, A.C.; Rugarli, E.; Langer, T. The I-AAA Protease YME1L and OMA1 Cleave OPA1 to Balance Mitochondrial Fusion and Fission. J. Cell Biol. 2014, 204, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Chen, H.; Fiket, M.; Alexander, C.; Chan, D.C. OPA1 Processing Controls Mitochondrial Fusion and Is Regulated by MRNA Splicing, Membrane Potential, and Yme1L. J. Cell Biol. 2007, 178, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Dotto, V.D.; Mishra, P.; Vidoni, S.; Fogazza, M.; Maresca, A.; Caporali, L.; McCaffery, J.M.; Cappelletti, M.; Baruffini, E.; Lenaers, G.; et al. OPA1 Isoforms in the Hierarchical Organization of Mitochondrial Functions. Cell Rep. 2017, 19, 2557–2571. [Google Scholar] [CrossRef]
- Ban, T.; Ishihara, T.; Kohno, H.; Saita, S.; Ichimura, A.; Maenaka, K.; Oka, T.; Mihara, K.; Ishihara, N. Molecular Basis of Selective Mitochondrial Fusion by Heterotypic Action between OPA1 and Cardiolipin. Nat. Cell Biol. 2017, 19, 856–863. [Google Scholar] [CrossRef]
- Wang, R.; Mishra, P.; Garbis, S.D.; Moradian, A.; Sweredoski, M.J.; Chan, D.C. Identification of New OPA1 Cleavage Site Reveals That Short Isoforms Regulate Mitochondrial Fusion. Mol. Biol. Cell 2021, 32, 157–168. [Google Scholar] [CrossRef]
- MacVicar, T.; Langer, T. OPA1 Processing in Cell Death and Disease—The Long and Short of It. J Cell Sci 2016, 129, 2297–2306. [Google Scholar] [CrossRef]
- Baker, M.J.; Lampe, P.A.; Stojanovski, D.; Korwitz, A.; Anand, R.; Tatsuta, T.; Langer, T. Stress-Induced OMA1 Activation and Autocatalytic Turnover Regulate OPA1-Dependent Mitochondrial Dynamics. EMBO J. 2014, 33, 578–593. [Google Scholar] [CrossRef]
- Zhang, K.; Li, H.; Song, Z. Membrane Depolarization Activates the Mitochondrial Protease OMA1 by Stimulating Self-Cleavage. EMBO Rep. 2014, 15, 576–585. [Google Scholar] [CrossRef]
- Rainbolt, T.K.; Saunders, J.M.; Wiseman, R.L. YME1L Degradation Reduces Mitochondrial Proteolytic Capacity during Oxidative Stress. EMBO Rep. 2015, 16, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Consolato, F.; Maltecca, F.; Tulli, S.; Sambri, I.; Casari, G. M-AAA and i-AAA Complexes Coordinate to Regulate OMA1, the Stress-Activated Supervisor of Mitochondrial Dynamics. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [PubMed]
- Richter, U.; Ng, K.Y.; Suomi, F.; Marttinen, P.; Turunen, T.; Jackson, C.; Suomalainen, A.; Vihinen, H.; Jokitalo, E.; Nyman, T.A.; et al. Mitochondrial Stress Response Triggered by Defects in Protein Synthesis Quality Control. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef] [PubMed]
- Tulli, S.; Bondio, A.D.; Baderna, V.; Mazza, D.; Codazzi, F.; Pierson, T.M.; Ambrosi, A.; Nolte, D.; Goizet, C.; Toro, C.; et al. Pathogenic Variants in the AFG3L2 Proteolytic Domain Cause SCA28 through Haploinsufficiency and Proteostatic Stress-Driven OMA1 Activation. J. Med. Genet. 2019, 56, 499–511. [Google Scholar] [CrossRef]
- Tondera, D.; Grandemange, S.; Jourdain, A.; Karbowski, M.; Mattenberger, Y.; Herzig, S.; Da Cruz, S.; Clerc, P.; Raschke, I.; Merkwirth, C.; et al. SLP-2 Is Required for Stress-Induced Mitochondrial Hyperfusion. EMBO J. 2009, 28, 1589–1600. [Google Scholar] [CrossRef]
- Wai, T.; Saita, S.; Nolte, H.; Müller, S.; König, T.; Richter-Dennerlein, R.; Sprenger, H.-G.; Madrenas, J.; Mühlmeister, M.; Brandt, U.; et al. The Membrane Scaffold SLP2 Anchors a Proteolytic Hub in Mitochondria Containing PARL and the I-AAA Protease YME1L. EMBO Rep. 2016, 17, 1844–1856. [Google Scholar] [CrossRef]
- Olichon, A.; ElAchouri, G.; Baricault, L.; Delettre, C.; Belenguer, P.; Lenaers, G. OPA1 Alternate Splicing Uncouples an Evolutionary Conserved Function in Mitochondrial Fusion from a Vertebrate Restricted Function in Apoptosis. Cell Death Differ. 2007, 14, 682–692. [Google Scholar] [CrossRef]
- Lee, H.; Smith, S.B.; Yoon, Y. The Short Variant of the Mitochondrial Dynamin OPA1 Maintains Mitochondrial Energetics and Cristae Structure. J. Biol. Chem. 2017, 292, 7115–7130. [Google Scholar] [CrossRef]
- Del Dotto, V.; Fogazza, M.; Musiani, F.; Maresca, A.; Aleo, S.J.; Caporali, L.; la Morgia, C.; Nolli, C.; Lodi, T.; Goffrini, P.; et al. Deciphering OPA1 Mutations Pathogenicity by Combined Analysis of Human, Mouse and Yeast Cell Models. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3496–3514. [Google Scholar] [CrossRef] [PubMed]
- Zanna, C.; Ghelli, A.; Porcelli, A.M.; Karbowski, M.; Youle, R.J.; Schimpf, S.; Wissinger, B.; Pinti, M.; Cossarizza, A.; Vidoni, S.; et al. OPA1 Mutations Associated with Dominant Optic Atrophy Impair Oxidative Phosphorylation and Mitochondrial Fusion. Brain 2008, 131, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Chevrollier, A.; Guillet, V.; Loiseau, D.; Gueguen, N.; de Crescenzo, M.-A.P.; Verny, C.; Ferre, M.; Dollfus, H.; Odent, S.; Milea, D.; et al. Hereditary Optic Neuropathies Share a Common Mitochondrial Coupling Defect. Ann. Neurol. 2008, 63, 794–798. [Google Scholar] [CrossRef]
- Agier, V.; Oliviero, P.; Lainé, J.; L’Hermitte-Stead, C.; Girard, S.; Fillaut, S.; Jardel, C.; Bouillaud, F.; Bulteau, A.L.; Lombès, A. Defective Mitochondrial Fusion, Altered Respiratory Function, and Distorted Cristae Structure in Skin Fibroblasts with Heterozygous OPA1 Mutations. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 1570–1580. [Google Scholar] [CrossRef]
- Alavi, M.V.; Bette, S.; Schimpf, S.; Schuettauf, F.; Schraermeyer, U.; Wehrl, H.F.; Ruttiger, L.; Beck, S.C.; Tonagel, F.; Pichler, B.J.; et al. A Splice Site Mutation in the Murine Opa1 Gene Features Pathology of Autosomal Dominant Optic Atrophy. Brain 2007, 130, 1029–1042. [Google Scholar] [CrossRef]
- Davies, V.J.; Hollins, A.J.; Piechota, M.J.; Yip, W.; Davies, J.R.; White, K.E.; Nicols, P.P.; Boulton, M.E.; Votruba, M. Opa1 Deficiency in a Mouse Model of Autosomal Dominant Optic Atrophy Impairs Mitochondrial Morphology, Optic Nerve Structure and Visual Function. Hum. Mol. Genet. 2007, 16, 1307–1318. [Google Scholar] [CrossRef]
- Sarzi, E.; Angebault, C.; Seveno, M.; Gueguen, N.; Chaix, B.; Bielicki, G.; Boddaert, N.; Mausset-Bonnefont, A.-L.; Cazevieille, C.; Rigau, V.; et al. The Human OPA1delTTAG Mutation Induces Premature Age-Related Systemic Neurodegeneration in Mouse. Brain 2012, 135, 3599–3613. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.A.; Morgan, J.E.; Votruba, M. Mouse Models of Dominant Optic Atrophy: What Do They Tell Us about the Pathophysiology of Visual Loss? Vis. Res. 2011, 51, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Carelli, V.; Musumeci, O.; Caporali, L.; Zanna, C.; Morgia, C.L.; Dotto, V.D.; Porcelli, A.M.; Rugolo, M.; Valentino, M.L.; Iommarini, L.; et al. Syndromic Parkinsonism and Dementia Associated with OPA1 Missense Mutations. Ann. Neurol. 2015, 78, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Yu-Wai-Man, P.; Sitarz, K.S.; Samuels, D.C.; Griffiths, P.G.; Reeve, A.K.; Bindoff, L.A.; Horvath, R.; Chinnery, P.F. OPA1 Mutations Cause Cytochrome c Oxidase Deficiency Due to Loss of Wild-Type MtDNA Molecules. Hum Mol. Genet. 2010, 19, 3043–3052. [Google Scholar] [CrossRef] [PubMed]
- Iannielli, A.; Bido, S.; Folladori, L.; Segnali, A.; Cancellieri, C.; Maresca, A.; Massimino, L.; Rubio, A.; Morabito, G.; Caporali, L.; et al. Pharmacological Inhibition of Necroptosis Protects from Dopaminergic Neuronal Cell Death in Parkinson’s Disease Models. Cell Rep. 2018, 22, 2066–2079. [Google Scholar] [CrossRef]
- Jonikas, M.; Madill, M.; Mathy, A.; Zekoll, T.; Zois, C.E.; Wigfield, S.; Kurzawa-Akanbi, M.; Browne, C.; Sims, D.; Chinnery, P.F.; et al. Stem Cell Modeling of Mitochondrial Parkinsonism Reveals Key Functions of OPA1. Ann. Neurol. 2018, 83, 915–925. [Google Scholar] [CrossRef]
- Iannielli, A.; Ugolini, G.S.; Cordiglieri, C.; Bido, S.; Rubio, A.; Colasante, G.; Valtorta, M.; Cabassi, T.; Rasponi, M.; Broccoli, V. Reconstitution of the Human Nigro-Striatal Pathway on-a-Chip Reveals OPA1-Dependent Mitochondrial Defects and Loss of Dopaminergic Synapses. Cell Rep. 2019, 29, 4646–4656.e4. [Google Scholar] [CrossRef]
- Kane, M.S.; Alban, J.; Desquiret-Dumas, V.; Gueguen, N.; Ishak, L.; Ferre, M.; Amati-Bonneau, P.; Procaccio, V.; Bonneau, D.; Lenaers, G.; et al. Autophagy Controls the Pathogenicity of OPA1 Mutations in Dominant Optic Atrophy. J. Cell. Mol. Med. 2017, 21, 2284–2297. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Ashley, N.; Diot, A.; Morten, K.; Phadwal, K.; Williams, A.; Fearnley, I.; Rosser, L.; Lowndes, J.; Fratter, C.; et al. Dysregulated Mitophagy and Mitochondrial Organization in Optic Atrophy Due to OPA1 Mutations. Neurology 2017, 88, 131–142. [Google Scholar] [CrossRef] [PubMed]
- White, K.E.; Davies, V.J.; Hogan, V.E.; Piechota, M.J.; Nichols, P.P.; Turnbull, D.M.; Votruba, M. OPA1 Deficiency Associated with Increased Autophagy in Retinal Ganglion Cells in a Murine Model of Dominant Optic Atrophy. Invest. Ophthalmol. Vis. Sci. 2009, 50, 2567–2571. [Google Scholar] [CrossRef]
- Diot, A.; Agnew, T.; Sanderson, J.; Liao, C.; Carver, J.; das Neves, R.P.; Gupta, R.; Guo, Y.; Waters, C.; Seto, S.; et al. Validating the RedMIT/GFP-LC3 Mouse Model by Studying Mitophagy in Autosomal Dominant Optic Atrophy Due to the OPA1Q285STOP Mutation. Front. Cell Dev. Biol. 2018, 6, 103. [Google Scholar] [CrossRef]
- Zaninello, M.; Palikaras, K.; Naon, D.; Iwata, K.; Herkenne, S.; Quintana-Cabrera, R.; Semenzato, M.; Grespi, F.; Ross-Cisneros, F.N.; Carelli, V.; et al. Inhibition of Autophagy Curtails Visual Loss in a Model of Autosomal Dominant Optic Atrophy. Nat. Commun. 2020, 11, 4029. [Google Scholar] [CrossRef]
- Bocca, C.; Nzoughet, J.K.; Leruez, S.; Amati-Bonneau, P.; Ferré, M.; Kane, M.-S.; Veyrat-Durebex, C.; de la Barca, J.M.C.; Chevrollier, A.; Homedan, C.; et al. A Plasma Metabolomic Signature Involving Purine Metabolism in Human Optic Atrophy 1 (OPA1)-Related Disorders. Invest. Ophthalmol. Vis. Sci. 2018, 59, 185–195. [Google Scholar] [CrossRef]
- Bocca, C.; Kane, M.S.; Veyrat-Durebex, C.; Nzoughet, J.K.; Chao de la Barca, J.M.; Chupin, S.; Alban, J.; Procaccio, V.; Bonneau, D.; Simard, G.; et al. Lipidomics Reveals Triacylglycerol Accumulation Due to Impaired Fatty Acid Flux in Opa1-Disrupted Fibroblasts. J. Proteome Res. 2019, 18, 2779–2790. [Google Scholar] [CrossRef] [PubMed]
- Chao de la Barca, J.M.; Fogazza, M.; Rugolo, M.; Chupin, S.; del Dotto, V.; Ghelli, A.M.; Carelli, V.; Simard, G.; Procaccio, V.; Bonneau, D.; et al. Metabolomics Hallmarks OPA1 Variants Correlating with Their in Vitro Phenotype and Predicting Clinical Severity. Hum. Mol. Genet. 2020, 29, 1319–1329. [Google Scholar] [CrossRef]
- Chao de la Barca, J.M.; Simard, G.; Sarzi, E.; Chaumette, T.; Rousseau, G.; Chupin, S.; Gadras, C.; Tessier, L.; Ferré, M.; Chevrollier, A.; et al. Targeted Metabolomics Reveals Early Dominant Optic Atrophy Signature in Optic Nerves of Opa1delTTAG/+ Mice. Invest. Ophthalmol. Vis. Sci. 2017, 58, 812–820. [Google Scholar] [CrossRef]
- Buzkova, J.; Nikkanen, J.; Ahola, S.; Hakonen, A.H.; Sevastianova, K.; Hovinen, T.; Yki-Järvinen, H.; Pietiläinen, K.H.; Lönnqvist, T.; Velagapudi, V.; et al. Metabolomes of Mitochondrial Diseases and Inclusion Body Myositis Patients: Treatment Targets and Biomarkers. EMBO Mol. Med. 2018, 10, e9091. [Google Scholar] [CrossRef]
- Chen, Q.; Kirk, K.; Shurubor, Y.I.; Zhao, D.; Arreguin, A.J.; Shahi, I.; Valsecchi, F.; Primiano, G.; Calder, E.L.; Carelli, V.; et al. Rewiring of Glutamine Metabolism Is a Bioenergetic Adaptation of Human Cells with Mitochondrial DNA Mutations. Cell Metab. 2018, 27, 1007–1025.e5. [Google Scholar] [CrossRef]
- Nikkanen, J.; Forsström, S.; Euro, L.; Paetau, I.; Kohnz, R.A.; Wang, L.; Chilov, D.; Viinamäki, J.; Roivainen, A.; Marjamäki, P.; et al. Mitochondrial DNA Replication Defects Disturb Cellular DNTP Pools and Remodel One-Carbon Metabolism. Cell Metab. 2016, 23, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Maresca, A.; del Dotto, V.; Capristo, M.; Scimonelli, E.; Tagliavini, F.; Morandi, L.; Tropeano, C.V.; Caporali, L.; Mohamed, S.; Roberti, M.; et al. DNMT1 Mutations Leading to Neurodegeneration Paradoxically Reflect on Mitochondrial Metabolism. Hum. Mol. Genet. 2020, 29, 1864–1881. [Google Scholar] [CrossRef] [PubMed]
- Di Bella, D.; Lazzaro, F.; Brusco, A.; Plumari, M.; Battaglia, G.; Pastore, A.; Finardi, A.; Cagnoli, C.; Tempia, F.; Frontali, M.; et al. Mutations in the Mitochondrial Protease Gene AFG3L2 Cause Dominant Hereditary Ataxia SCA28. Nat. Genet. 2010, 42, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Pierson, T.M.; Adams, D.; Bonn, F.; Martinelli, P.; Cherukuri, P.F.; Teer, J.K.; Hansen, N.F.; Cruz, P.; Program, J.C.M. for the N.C.S.; Blakesley, R.W.; et al. Whole-Exome Sequencing Identifies Homozygous AFG3L2 Mutations in a Spastic Ataxia-Neuropathy Syndrome Linked to Mitochondrial m-AAA Proteases. PLoS Genet. 2011, 7, e1002325. [Google Scholar] [CrossRef]
- Casari, G.; de Fusco, M.; Ciarmatori, S.; Zeviani, M.; Mora, M.; Fernandez, P.; de Michele, G.; Filla, A.; Cocozza, S.; Marconi, R.; et al. Spastic Paraplegia and OXPHOS Impairment Caused by Mutations in Paraplegin, a Nuclear-Encoded Mitochondrial Metalloprotease. Cell 1998, 93, 973–983. [Google Scholar] [CrossRef]
- Caporali, L.; Magri, S.; Legati, A.; Dotto, V.D.; Tagliavini, F.; Balistreri, F.; Nasca, A.; Morgia, C.L.; Carbonelli, M.; Valentino, M.L.; et al. ATPase Domain AFG3L2 Mutations Alter OPA1 Processing and Cause Optic Neuropathy. Ann. Neurol. 2020, 88, 18–32. [Google Scholar] [CrossRef]
- Charif, M.; Chevrollier, A.; Gueguen, N.; Bris, C.; Goudenège, D.; Desquiret-Dumas, V.; Leruez, S.; Colin, E.; Meunier, A.; Vignal, C.; et al. Mutations in the M-AAA Proteases AFG3L2 and SPG7 Are Causing Isolated Dominant Optic Atrophy. Neurol. Genet. 2020, 6. [Google Scholar] [CrossRef]
- Baderna, V.; Schultz, J.; Kearns, L.S.; Fahey, M.; Thompson, B.A.; Ruddle, J.B.; Huq, A.; Maltecca, F. A Novel AFG3L2 Mutation Close to AAA Domain Leads to Aberrant OMA1 and OPA1 Processing in a Family with Optic Atrophy | Acta Neuropathologica Communications | Full Text. Available online: https://actaneurocomms.biomedcentral.com/articles/10.1186/s40478-020-00975-w (accessed on 16 February 2021).
- Magri, S.; Fracasso, V.; Plumari, M.; Alfei, E.; Ghezzi, D.; Gellera, C.; Rusmini, P.; Poletti, A.; Bella, D.D.; Elia, A.E.; et al. Concurrent AFG3L2 and SPG7 Mutations Associated with Syndromic Parkinsonism and Optic Atrophy with Aberrant OPA1 Processing and Mitochondrial Network Fragmentation. Hum. Mutat. 2018, 39, 2060–2071. [Google Scholar] [CrossRef]
- Klebe, S.; Depienne, C.; Gerber, S.; Challe, G.; Anheim, M.; Charles, P.; Fedirko, E.; Lejeune, E.; Cottineau, J.; Brusco, A.; et al. Spastic Paraplegia Gene 7 in Patients with Spasticity and/or Optic Neuropathy. Brain 2012, 135, 2980–2993. [Google Scholar] [CrossRef]
- Pfeffer, G.; Gorman, G.S.; Griffin, H.; Kurzawa-Akanbi, M.; Blakely, E.L.; Wilson, I.; Sitarz, K.; Moore, D.; Murphy, J.L.; Alston, C.L.; et al. Mutations in the SPG7 Gene Cause Chronic Progressive External Ophthalmoplegia through Disordered Mitochondrial DNA Maintenance. Brain 2014, 137, 1323–1336. [Google Scholar] [CrossRef]
- Hartmann, B.; Wai, T.; Hu, H.; MacVicar, T.; Musante, L.; Fischer-Zirnsak, B.; Stenzel, W.; Gräf, R.; van den Heuvel, L.; Ropers, H.-H.; et al. Homozygous YME1L1 Mutation Causes Mitochondriopathy with Optic Atrophy and Mitochondrial Network Fragmentation. eLife 2016, 5, e16078. [Google Scholar] [CrossRef]
- Sprenger, H.-G.; Wani, G.; Hesseling, A.; König, T.; Patron, M.; MacVicar, T.; Ahola, S.; Wai, T.; Barth, E.; Rugarli, E.I.; et al. Loss of the Mitochondrial I-AAA Protease YME1L Leads to Ocular Dysfunction and Spinal Axonopathy. EMBO Mol. Med. 2019, 11, e9288. [Google Scholar] [CrossRef]
- Wai, T.; García-Prieto, J.; Baker, M.J.; Merkwirth, C.; Benit, P.; Rustin, P.; Rupérez, F.J.; Barbas, C.; Ibañez, B.; Langer, T. Imbalanced OPA1 Processing and Mitochondrial Fragmentation Cause Heart Failure in Mice. Science 2015, 350. [Google Scholar] [CrossRef] [PubMed]
- MacVicar, T.; Ohba, Y.; Nolte, H.; Mayer, F.C.; Tatsuta, T.; Sprenger, H.-G.; Lindner, B.; Zhao, Y.; Li, J.; Bruns, C.; et al. Lipid Signalling Drives Proteolytic Rewiring of Mitochondria by YME1L. Nature 2019, 575, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 Coordinately Regulate Mitochondrial Fusion and Are Essential for Embryonic Development. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Eura, Y.; Ishihara, N.; Yokota, S.; Mihara, K. Two Mitofusin Proteins, Mammalian Homologues of FZO, with Distinct Functions Are Both Required for Mitochondrial Fusion. J. Biochem. 2003, 134, 333–344. [Google Scholar] [CrossRef]
- Ishihara, N.; Eura, Y.; Mihara, K. Mitofusin 1 and 2 Play Distinct Roles in Mitochondrial Fusion Reactions via GTPase Activity. J. Cell Sci. 2004, 117, 6535–6546. [Google Scholar] [CrossRef] [PubMed]
- de Brito, O.M.; Scorrano, L. Mitofusin 2 Tethers Endoplasmic Reticulum to Mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Naon, D.; Zaninello, M.; Giacomello, M.; Varanita, T.; Grespi, F.; Lakshminaranayan, S.; Serafini, A.; Semenzato, M.; Herkenne, S.; Hernández-Alvarez, M.I.; et al. Critical Reappraisal Confirms That Mitofusin 2 Is an Endoplasmic Reticulum–Mitochondria Tether. PNAS 2016, 113, 11249–11254. [Google Scholar] [CrossRef]
- Filadi, R.; Greotti, E.; Turacchio, G.; Luini, A.; Pozzan, T.; Pizzo, P. Mitofusin 2 Ablation Increases Endoplasmic Reticulum–Mitochondria Coupling. PNAS 2015, 112, E2174–E2181. [Google Scholar] [CrossRef]
- Giorgi, C.; Missiroli, S.; Patergnani, S.; Duszynski, J.; Wieckowski, M.R.; Pinton, P. Mitochondria-Associated Membranes: Composition, Molecular Mechanisms, and Physiopathological Implications. Antioxid. Redox Signal. 2015, 22, 995–1019. [Google Scholar] [CrossRef]
- Züchner, S.; Mersiyanova, I.V.; Muglia, M.; Bissar-Tadmouri, N.; Rochelle, J.; Dadali, E.L.; Zappia, M.; Nelis, E.; Patitucci, A.; Senderek, J.; et al. Mutations in the Mitochondrial GTPase Mitofusin 2 Cause Charcot-Marie-Tooth Neuropathy Type 2A. Nat. Genet. 2004, 36, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Züchner, S.; de Jonghe, P.; Jordanova, A.; Claeys, K.G.; Guergueltcheva, V.; Cherninkova, S.; Hamilton, S.R.; van Stavern, G.; Krajewski, K.M.; Stajich, J.; et al. Axonal Neuropathy with Optic Atrophy Is Caused by Mutations in Mitofusin 2. Ann. Neurol. 2006, 59, 276–281. [Google Scholar] [CrossRef]
- Rouzier, C.; Bannwarth, S.; Chaussenot, A.; Chevrollier, A.; Verschueren, A.; Bonello-Palot, N.; Fragaki, K.; Cano, A.; Pouget, J.; Pellissier, J.-F.; et al. The MFN2 Gene Is Responsible for Mitochondrial DNA Instability and Optic Atrophy “plus” Phenotype. Brain 2012, 135, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Guillet, V.; Gueguen, N.; Cartoni, R.; Chevrollier, A.; Desquiret, V.; Angebault, C.; Amati-Bonneau, P.; Procaccio, V.; Bonneau, D.; Martinou, J.-C.; et al. Bioenergetic Defect Associated with MKATP Channel Opening in a Mouse Model Carrying a Mitofusin 2 Mutation. FASEB J. 2011, 25, 1618–1627. [Google Scholar] [CrossRef]
- Misko, A.L.; Sasaki, Y.; Tuck, E.; Milbrandt, J.; Baloh, R.H. Mitofusin2 Mutations Disrupt Axonal Mitochondrial Positioning and Promote Axon Degeneration. J. Neurosci. 2012, 32, 4145–4155. [Google Scholar] [CrossRef] [PubMed]
- Baloh, R.H.; Schmidt, R.E.; Pestronk, A.; Milbrandt, J. Altered Axonal Mitochondrial Transport in the Pathogenesis of Charcot-Marie-Tooth Disease from Mitofusin 2 Mutations. J. Neurosci. 2007, 27, 422–430. [Google Scholar] [CrossRef]
- Wolf, C.; Zimmermann, R.; Thaher, O.; Bueno, D.; Wüllner, V.; Schäfer, M.K.E.; Albrecht, P.; Methner, A. The Charcot–Marie Tooth Disease Mutation R94Q in MFN2 Decreases ATP Production but Increases Mitochondrial Respiration under Conditions of Mild Oxidative Stress. Cells 2019, 8, 1289. [Google Scholar] [CrossRef]
- Amiott, E.A.; Lott, P.; Soto, J.; Kang, P.B.; McCaffery, J.M.; DiMauro, S.; Abel, E.D.; Flanigan, K.M.; Lawson, V.H.; Shaw, J.M. Mitochondrial Fusion and Function in Charcot–Marie–Tooth Type 2A Patient Fibroblasts with Mitofusin 2 Mutations. Exp. Neurol. 2008, 211, 115–127. [Google Scholar] [CrossRef]
- Codron, P.; Chevrollier, A.; Kane, M.S.; Echaniz-Laguna, A.; Latour, P.; Reynier, P.; Bonneau, D.; Verny, C.; Procaccio, V.; Lenaers, G.; et al. Increased Mitochondrial Fusion in a Autosomal Recessive CMT2A Family with Mitochondrial GTPase Mitofusin 2 Mutations. J. Peripher. Nerv. Syst. 2016, 21, 365–369. [Google Scholar] [CrossRef]
- Vielhaber, S.; Debska-Vielhaber, G.; Peeva, V.; Schoeler, S.; Kudin, A.P.; Minin, I.; Schreiber, S.; Dengler, R.; Kollewe, K.; Zuschratter, W.; et al. Mitofusin 2 Mutations Affect Mitochondrial Function by Mitochondrial DNA Depletion. Acta Neuropathol. 2013, 125, 245–256. [Google Scholar] [CrossRef]
- Rizzo, F.; Ronchi, D.; Salani, S.; Nizzardo, M.; Fortunato, F.; Bordoni, A.; Stuppia, G.; Del Bo, R.; Piga, D.; Fato, R.; et al. Selective Mitochondrial Depletion, Apoptosis Resistance, and Increased Mitophagy in Human Charcot-Marie-Tooth 2A Motor Neurons. Hum. Mol. Genet. 2016, 25, 4266–4281. [Google Scholar] [CrossRef]
- Bernard-Marissal, N.; van Hameren, G.; Juneja, M.; Pellegrino, C.; Louhivuori, L.; Bartesaghi, L.; Rochat, C.; Mansour, O.E.; Médard, J.-J.; Croisier, M.; et al. Altered Interplay between Endoplasmic Reticulum and Mitochondria in Charcot–Marie–Tooth Type 2A Neuropathy. PNAS 2019, 116, 2328–2337. [Google Scholar] [CrossRef] [PubMed]
- Larrea, D.; Pera, M.; Gonnelli, A.; Quintana–Cabrera, R.; Akman, H.O.; Guardia-Laguarta, C.; Velasco, K.R.; Area-Gomez, E.; dal Bello, F.; de Stefani, D.; et al. MFN2 Mutations in Charcot–Marie–Tooth Disease Alter Mitochondria-Associated ER Membrane Function but Do Not Impair Bioenergetics. Hum. Mol. Genet. 2019, 28, 1782–1800. [Google Scholar] [CrossRef]
- Nasca, A.; Scotton, C.; Zaharieva, I.; Neri, M.; Selvatici, R.; Magnusson, O.T.; Gal, A.; Weaver, D.; Rossi, R.; Armaroli, A.; et al. Recessive Mutations in MSTO1 Cause Mitochondrial Dynamics Impairment, Leading to Myopathy and Ataxia. Hum. Mutat. 2017, 38, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Gal, A.; Balicza, P.; Weaver, D.; Naghdi, S.; Joseph, S.K.; Várnai, P.; Gyuris, T.; Horváth, A.; Nagy, L.; Seifert, E.L.; et al. MSTO1 Is a Cytoplasmic Pro-Mitochondrial Fusion Protein, Whose Mutation Induces Myopathy and Ataxia in Humans. EMBO Mol. Med. 2017, 9, 967–984. [Google Scholar] [CrossRef] [PubMed]
- Donkervoort, S.; Sabouny, R.; Yun, P.; Gauquelin, L.; Chao, K.R.; Hu, Y.; Al-Khatib, I.; Töpf, A.; Mohassel, P.; Cummings, B.B.; et al. MSTO1 Mutations Cause MtDNA Depletion, Manifesting as Muscular Dystrophy with Cerebellar Involvement. Acta Neuropathol. 2019, 138, 1013–1031. [Google Scholar] [CrossRef]
- Li, K.; Jin, R.; Wu, X. Whole-Exome Sequencing Identifies Rare Compound Heterozygous Mutations in the MSTO1 Gene Associated with Cerebellar Ataxia and Myopathy. Eur. J. Med. Genet. 2020, 63, 103623. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, C.; Grabiger, S.; Schwefel, D.; Faelber, K.; Rosenbaum, E.; Mears, J.; Rocks, O.; Daumke, O. Structural Insights into Oligomerization and Mitochondrial Remodelling of Dynamin 1-like Protein. EMBO J. 2013, 32, 1280–1292. [Google Scholar] [CrossRef]
- Losón, O.C.; Song, Z.; Chen, H.; Chan, D.C. Fis1, Mff, MiD49, and MiD51 Mediate Drp1 Recruitment in Mitochondrial Fission. Mol. Biol. Cell 2013, 24, 659–667. [Google Scholar] [CrossRef]
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER Tubules Mark Sites of Mitochondrial Division. Science 2011, 334, 358–362. [Google Scholar] [CrossRef]
- Manor, U.; Bartholomew, S.; Golani, G.; Christenson, E.; Kozlov, M.; Higgs, H.; Spudich, J.; Lippincott-Schwartz, J. A Mitochondria-Anchored Isoform of the Actin-Nucleating Spire Protein Regulates Mitochondrial Division. eLife 2015, 4, e08828. [Google Scholar] [CrossRef]
- Mears, J.A.; Lackner, L.L.; Fang, S.; Ingerman, E.; Nunnari, J.; Hinshaw, J.E. Conformational Changes in Dnm1 Support a Contractile Mechanism for Mitochondrial Fission. Nat. Struct. Mol. Biol. 2011, 18, 20–26. [Google Scholar] [CrossRef]
- Ban-Ishihara, R.; Ishihara, T.; Sasaki, N.; Mihara, K.; Ishihara, N. Dynamics of Nucleoid Structure Regulated by Mitochondrial Fission Contributes to Cristae Reformation and Release of Cytochrome c. PNAS 2013, 110, 11863–11868. [Google Scholar] [CrossRef] [PubMed]
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An Actin-Dependent Step in Mitochondrial Fission Mediated by the ER-Associated Formin INF2. Science 2013, 339, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, S.; Roelofs, B.A.; Boyman, L.; Lederer, W.J.; Sesaki, H.; Karbowski, M. Transient Assembly of F-Actin on the Outer Mitochondrial Membrane Contributes to Mitochondrial Fission. J. Cell Biol. 2014, 208, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Korobova, F.; Gauvin, T.J.; Higgs, H.N. A Role for Myosin II in Mammalian Mitochondrial Fission. Curr. Biol. 2014, 24, 409–414. [Google Scholar] [CrossRef]
- Li, G.; Zhou, J.; Budhraja, A.; Hu, X.; Chen, Y.; Cheng, Q.; Liu, L.; Zhou, T.; Li, P.; Liu, E.; et al. Mitochondrial Translocation and Interaction of Cofilin and Drp1 Are Required for Erucin-Induced Mitochondrial Fission and Apoptosis. Oncotarget 2014, 6, 1834–1849. [Google Scholar] [CrossRef] [PubMed]
- Rehklau, K.; Hoffmann, L.; Gurniak, C.B.; Ott, M.; Witke, W.; Scorrano, L.; Culmsee, C.; Rust, M.B. Cofilin1-Dependent Actin Dynamics Control DRP1-Mediated Mitochondrial Fission. Cell Death Dis. 2017, 8, e3063. [Google Scholar] [CrossRef]
- Pagliuso, A.; Tham, T.N.; Stevens, J.K.; Lagache, T.; Persson, R.; Salles, A.; Olivo-Marin, J.-C.; Oddos, S.; Spang, A.; Cossart, P.; et al. A Role for Septin 2 in Drp1-Mediated Mitochondrial Fission. EMBO Rep. 2016, 17, 858–873. [Google Scholar] [CrossRef]
- Yoon, Y.; Pitts, K.R.; McNiven, M.A. Mammalian Dynamin-like Protein DLP1 Tubulates Membranes. Mol. Biol. Cell 2001, 12, 2894–2905. [Google Scholar] [CrossRef]
- Lee, J.E.; Westrate, L.M.; Wu, H.; Page, C.; Voeltz, G.K. Multiple Dynamin Family Members Collaborate to Drive Mitochondrial Division. Nature 2016, 540, 139–143. [Google Scholar] [CrossRef]
- Cho, B.; Cho, H.M.; Jo, Y.; Kim, H.D.; Song, M.; Moon, C.; Kim, H.; Kim, K.; Sesaki, H.; Rhyu, I.J.; et al. Constriction of the Mitochondrial Inner Compartment Is a Priming Event for Mitochondrial Division. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, R.; Ji, W.-K.; Stan, R.V.; de Juan-Sanz, J.; Ryan, T.A.; Higgs, H.N. INF2-Mediated Actin Polymerization at the ER Stimulates Mitochondrial Calcium Uptake, Inner Membrane Constriction, and Division. J. Cell Biol. 2017, 217, 251–268. [Google Scholar] [CrossRef] [PubMed]
- Waterham, H.R.; Koster, J.; van Roermund, C.W.T.; Mooyer, P.A.W.; Wanders, R.J.A.; Leonard, J.V. A Lethal Defect of Mitochondrial and Peroxisomal Fission N. Engl. J. Med. 2007, 356:1736-1741, doi/10.1056/NEJMoa064436. 2007, 356, 1736–1741. [Google Scholar] [PubMed]
- Koch, A.; Thiemann, M.; Grabenbauer, M.; Yoon, Y.; McNiven, M.A.; Schrader, M. Dynamin-like Protein 1 Is Involved in Peroxisomal Fission. J. Biol. Chem. 2003, 278, 8597–8605. [Google Scholar] [CrossRef]
- Li, X.; Gould, S.J. The Dynamin-like GTPase DLP1 Is Essential for Peroxisome Division and Is Recruited to Peroxisomes in Part by PEX11. J. Biol. Chem. 2003, 278, 17012–17020. [Google Scholar] [CrossRef] [PubMed]
- Pitts, K.R.; Yoon, Y.; Krueger, E.W.; McNiven, M.A. The Dynamin-like Protein DLP1 Is Essential for Normal Distribution and Morphology of the Endoplasmic Reticulum and Mitochondria in Mammalian Cells. MBoC 1999, 10, 4403–4417. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.; Griparic, L.; Shurland, D.L.; van der Bliek, A.M. Dynamin-Related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells. Mol. Biol. Cell 2001, 12, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Vanstone, J.R.; Smith, A.M.; McBride, S.; Naas, T.; Holcik, M.; Antoun, G.; Harper, M.-E.; Michaud, J.; Sell, E.; Chakraborty, P.; et al. DNM1L- Related Mitochondrial Fission Defect Presenting as Refractory Epilepsy. Eur. J. Hum. Genet. 2016, 24, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Sheffer, R.; Douiev, L.; Edvardson, S.; Shaag, A.; Tamimi, K.; Soiferman, D.; Meiner, V.; Saada, A. Postnatal Microcephaly and Pain Insensitivity Due to a de Novo Heterozygous DNM1L Mutation Causing Impaired Mitochondrial Fission and Function. Am. J. Med. Genet. Part A 2016, 170, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, J.A.; Liu, R.; Perry, M.S.; Klein, J.; Chan, D.C. A Novel de Novo Dominant Negative Mutation in DNM1L Impairs Mitochondrial Fission and Presents as Childhood Epileptic Encephalopathy. Am. J. Med. Genet. A 2016, 170, 2002–2011. [Google Scholar] [CrossRef]
- Vandeleur, D.; Chen, C.V.; Huang, E.J.; Connolly, A.J.; Sanchez, H.; Moon-Grady, A.J. Novel and Lethal Case of Cardiac Involvement in DNM1L Mitochondrial Encephalopathy. Am. J. Med. Genet. Part A 2019, 179, 2486–2489. [Google Scholar] [CrossRef] [PubMed]
- Verrigni, D.; di Nottia, M.; Ardissone, A.; Baruffini, E.; Nasca, A.; Legati, A.; Bellacchio, E.; Fagiolari, G.; Martinelli, D.; Fusco, L.; et al. Clinical-Genetic Features and Peculiar Muscle Histopathology in Infantile DNM1L-Related Mitochondrial Epileptic Encephalopathy. Hum. Mutat. 2019, 40, 601–618. [Google Scholar] [CrossRef]
- Longo, F.; Benedetti, S.; Zambon, A.A.; Sora, M.G.N.; di Resta, C.; de Ritis, D.; Quattrini, A.; Maltecca, F.; Ferrari, M.; Previtali, S.C. Impaired Turnover of Hyperfused Mitochondria in Severe Axonal Neuropathy Due to a Novel DRP1 Mutation. Hum. Mo.l Genet. 2020, 29, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Yoon, G.; Malam, Z.; Paton, T.; Marshall, C.R.; Hyatt, E.; Ivakine, Z.; Scherer, S.W.; Lee, K.-S.; Hawkins, C.; Cohn, R.D.; et al. Lethal Disorder of Mitochondrial Fission Caused by Mutations in DNM1L. J. Pediatrics 2016, 171, 313–316.e2. [Google Scholar] [CrossRef]
- Nasca, A.; Legati, A.; Baruffini, E.; Nolli, C.; Moroni, I.; Ardissone, A.; Goffrini, P.; Ghezzi, D. Biallelic Mutations in DNM1L Are Associated with a Slowly Progressive Infantile Encephalopathy. Hum. Mutat. 2016, 37, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.; Charif, M.; Chevrollier, A.; Chaumette, T.; Angebault, C.; Kane, M.S.; Paris, A.; Alban, J.; Quiles, M.; Delettre, C.; et al. Mutations in DNM1L, as in OPA1, Result in Dominant Optic Atrophy despite Opposite Effects on Mitochondrial Fusion and Fission. Brain 2017, 140, 2586–2596. [Google Scholar] [CrossRef] [PubMed]
- Shamseldin, H.E.; Alshammari, M.; Al-Sheddi, T.; Salih, M.A.; Alkhalidi, H.; Kentab, A.; Repetto, G.M.; Hashem, M.; Alkuraya, F.S. Genomic Analysis of Mitochondrial Diseases in a Consanguineous Population Reveals Novel Candidate Disease Genes. J. Med. Genet. 2012, 49, 234–241. [Google Scholar] [CrossRef]
- Koch, J.; Feichtinger, R.G.; Freisinger, P.; Pies, M.; Schrödl, F.; Iuso, A.; Sperl, W.; Mayr, J.A.; Prokisch, H.; Haack, T.B. Disturbed Mitochondrial and Peroxisomal Dynamics Due to Loss of MFF Causes Leigh-like Encephalopathy, Optic Atrophy and Peripheral Neuropathy. J. Med. Genet. 2016, 53, 270–278. [Google Scholar] [CrossRef]
- Charif, M.; Wong, Y.C.; Kim, S.; Guichet, A.; Vignal, C.; Zanlonghi, X.; Bensaid, P.; Procaccio, V.; Bonneau, D.; Amati-Bonneau, P.; et al. Dominant Mutations in MIEF1 Affect Mitochondrial Dynamics and Cause a Singular Late Onset Optic Neuropathy. Mol. Neurodegener. 2021, 16, 12. [Google Scholar] [CrossRef]
- Marco, A.; Cuesta, A.; Pedrola, L.; Palau, F.; Marín, I. Evolutionary and Structural Analyses of GDAP1, Involved in Charcot-Marie-Tooth Disease, Characterize a Novel Class of Glutathione Transferase-Related Genes. Mol. Biol. Evol. 2004, 21, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Niemann, A.; Ruegg, M.; la Padula, V.; Schenone, A.; Suter, U. Ganglioside-Induced Differentiation Associated Protein 1 Is a Regulator of the Mitochondrial Network: New Implications for Charcot-Marie-Tooth Disease. J. Cell Biol. 2005, 170, 1067–1078. [Google Scholar] [CrossRef]
- Baxter, R.V.; Ben-Othmane, K.; Rochelle, J.M.; Stajich, J.E.; Hulette, C.; Dew-Knight, S.; Hentati, F.; Ben Hamida, M.; Bel, S.; Stenger, J.E.; et al. Ganglioside-Induced Differentiation-Associated Protein-1 Is Mutant in Charcot-Marie-Tooth Disease Type 4A/8q21. Nat. Genet. 2002, 30, 21–22. [Google Scholar] [CrossRef]
- Claramunt, R.; Pedrola, L.; Sevilla, T.; de Munain, A.L.; Berciano, J.; Cuesta, A.; Sánchez-Navarro, B.; Millán, J.M.; Saifi, G.M.; Lupski, J.R.; et al. Genetics of Charcot-Marie-Tooth Disease Type 4A: Mutations, Inheritance, Phenotypic Variability, and Founder Effect. J. Med. Genet. 2005, 42, 358–365. [Google Scholar] [CrossRef]
- Niemann, A.; Wagner, K.M.; Ruegg, M.; Suter, U. GDAP1 Mutations Differ in Their Effects on Mitochondrial Dynamics and Apoptosis Depending on the Mode of Inheritance. Neurobiol. Dis. 2009, 36, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Pla-Martín, D.; Rueda, C.B.; Estela, A.; Sánchez-Piris, M.; González-Sánchez, P.; Traba, J.; de la Fuente, S.; Scorrano, L.; Renau-Piqueras, J.; Alvarez, J.; et al. Silencing of the Charcot–Marie–Tooth Disease-Associated Gene GDAP1 Induces Abnormal Mitochondrial Distribution and Affects Ca2+ Homeostasis by Reducing Store-Operated Ca2+ Entry. Neurobiol. Dis. 2013, 55, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Civera-Tregón, A.; Domínguez, L.; Martínez-Valero, P.; Serrano, C.; Vallmitjana, A.; Benítez, R.; Hoenicka, J.; Satrústegui, J.; Palau, F. Mitochondria and Calcium Defects Correlate with Axonal Dysfunction in GDAP1-Related Charcot-Marie-Tooth Mouse Model. Neurobiol. Dis. 2021, 105300. [Google Scholar] [CrossRef] [PubMed]
- Barneo-Muñoz, M.; Juárez, P.; Civera-Tregón, A.; Yndriago, L.; Pla-Martin, D.; Zenker, J.; Cuevas-Martín, C.; Estela, A.; Sánchez-Aragó, M.; Forteza-Vila, J.; et al. Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy. PLoS Genet. 2015, 11, e1005115. [Google Scholar] [CrossRef] [PubMed]
- Bartsakoulia, M.; Pyle, A.; Troncoso-Chandía, D.; Vial-Brizzi, J.; Paz-Fiblas, M.V.; Duff, J.; Griffin, H.; Boczonadi, V.; Lochmüller, H.; Kleinle, S.; et al. A Novel Mechanism Causing Imbalance of Mitochondrial Fusion and Fission in Human Myopathies. Hum. Mol. Genet. 2018, 27, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Bitoun, M.; Maugenre, S.; Jeannet, P.-Y.; Lacène, E.; Ferrer, X.; Laforêt, P.; Martin, J.-J.; Laporte, J.; Lochmüller, H.; Beggs, A.H.; et al. Mutations in Dynamin 2 Cause Dominant Centronuclear Myopathy. Nat. Genet. 2005, 37, 1207–1209. [Google Scholar] [CrossRef]
- Züchner, S.; Noureddine, M.; Kennerson, M.; Verhoeven, K.; Claeys, K.; Jonghe, P.D.; Merory, J.; Oliveira, S.A.; Speer, M.C.; Stenger, J.E.; et al. Mutations in the Pleckstrin Homology Domain of Dynamin 2 Cause Dominant Intermediate Charcot-Marie-Tooth Disease. Nat. Genet. 2005, 37, 289–294. [Google Scholar] [CrossRef]
- Claeys, K.G.; Züchner, S.; Kennerson, M.; Berciano, J.; Garcia, A.; Verhoeven, K.; Storey, E.; Merory, J.R.; Bienfait, H.M.E.; Lammens, M.; et al. Phenotypic Spectrum of Dynamin 2 Mutations in Charcot-Marie-Tooth Neuropathy. Brain 2009, 132, 1741–1752. [Google Scholar] [CrossRef]
- Koutsopoulos, O.S.; Kretz, C.; Weller, C.M.; Roux, A.; Mojzisova, H.; Böhm, J.; Koch, C.; Toussaint, A.; Heckel, E.; Stemkens, D.; et al. Dynamin 2 Homozygous Mutation in Humans with a Lethal Congenital Syndrome. Eur. J. Hum. Genet. 2013, 21, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.J.; Schlöndorff, J.S.; Becker, D.J.; Tsukaguchi, H.; Tonna, S.J.; Uscinski, A.L.; Higgs, H.N.; Henderson, J.M.; Pollak, M.R. Mutations in the Formin Gene INF2 Cause Focal Segmental Glomerulosclerosis. Nat. Genet. 2009, 42, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Boyer, O.; Nevo, F.; Plaisier, E.; Funalot, B.; Gribouval, O.; Benoit, G.; Cong, E.H.; Arrondel, C.; Tête, M.-J.; Montjean, R.; et al. INF2 Mutations in Charcot–Marie–Tooth Disease with Glomerulopathy. N. Engl. J. Med. 2011, 365, 2377–2388. [Google Scholar] [CrossRef]
- Csordás, G.; Renken, C.; Várnai, P.; Walter, L.; Weaver, D.; Buttle, K.F.; Balla, T.; Mannella, C.A.; Hajnóczky, G. Structural and Functional Features and Significance of the Physical Linkage between ER and Mitochondria. J. Cell Biol. 2006, 174, 915–921. [Google Scholar] [CrossRef]
- Vance, J.E. MAM (Mitochondria-Associated Membranes) in Mammalian Cells: Lipids and Beyond. Biochim. Biophys. Acta 2014, 1841, 595–609. [Google Scholar] [CrossRef]
- Kornmann, B.; Currie, E.; Collins, S.R.; Schuldiner, M.; Nunnari, J.; Weissman, J.S.; Walter, P. An ER-Mitochondria Tethering Complex Revealed by a Synthetic Biology Screen. Science 2009, 325, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, A.; Nagashima, S.; Tokuyama, T.; Amo, T.; Matsuki, Y.; Ishido, S.; Kudo, Y.; McBride, H.M.; Fukuda, T.; Matsushita, N.; et al. MITOL Regulates Endoplasmic Reticulum-Mitochondria Contacts via Mitofusin2. Mol. Cell 2013, 51, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Szabadkai, G.; Bianchi, K.; Várnai, P.; de Stefani, D.; Wieckowski, M.R.; Cavagna, D.; Nagy, A.I.; Balla, T.; Rizzuto, R. Chaperone-Mediated Coupling of Endoplasmic Reticulum and Mitochondrial Ca2+ Channels. J. Cell Biol. 2006, 175, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Simmen, T.; Aslan, J.E.; Blagoveshchenskaya, A.D.; Thomas, L.; Wan, L.; Xiang, Y.; Feliciangeli, S.F.; Hung, C.-H.; Crump, C.M.; Thomas, G. PACS-2 Controls Endoplasmic Reticulum–Mitochondria Communication and Bid-Mediated Apoptosis. EMBO J. 2005, 24, 717–729. [Google Scholar] [CrossRef]
- Rizzuto, R.; Pinton, P.; Carrington, W.; Fay, F.S.; Fogarty, K.E.; Lifshitz, L.M.; Tuft, R.A.; Pozzan, T. Close Contacts with the Endoplasmic Reticulum as Determinants of Mitochondrial Ca2+ Responses. Science 1998, 280, 1763–1766. [Google Scholar] [CrossRef] [PubMed]
- Patergnani, S.; Suski, J.M.; Agnoletto, C.; Bononi, A.; Bonora, M.; de Marchi, E.; Giorgi, C.; Marchi, S.; Missiroli, S.; Poletti, F.; et al. Calcium Signaling around Mitochondria Associated Membranes (MAMs). Cell Commun. Signal. 2011, 9, 19. [Google Scholar] [CrossRef]
- Honrath, B.; Metz, I.; Bendridi, N.; Rieusset, J.; Culmsee, C.; Dolga, A.M. Glucose-Regulated Protein 75 Determines ER–Mitochondrial Coupling and Sensitivity to Oxidative Stress in Neuronal Cells. Cell Death Discov. 2017, 3, 1–13. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.-P. Sigma-1 Receptor Chaperones at the ER- Mitochondrion Interface Regulate Ca2+ Signaling and Cell Survival. Cell 2007, 131, 596–610. [Google Scholar] [CrossRef]
- Kumar, V.; Maity, S. ER Stress-Sensor Proteins and ER-Mitochondrial Crosstalk—Signaling Beyond (ER) Stress Response. Biomolecules 2021, 11, 173. [Google Scholar] [CrossRef] [PubMed]
- De Stefani, D.; Raffaello, A.; Teardo, E.; Szabò, I.; Rizzuto, R. A Forty-Kilodalton Protein of the Inner Membrane Is the Mitochondrial Calcium Uniporter. Nature 2011, 476, 336–340. [Google Scholar] [CrossRef]
- Baughman, J.M.; Perocchi, F.; Girgis, H.S.; Plovanich, M.; Belcher-Timme, C.A.; Sancak, Y.; Bao, X.R.; Strittmatter, L.; Goldberger, O.; Bogorad, R.L.; et al. Integrative Genomics Identifies MCU as an Essential Component of the Mitochondrial Calcium Uniporter. Nature 2011, 476, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Mabalirajan, U. Mitochondrial Calcium in Command of Juggling Myriads of Cellular Functions. Mitochondrion 2021, 57, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Denton, R.M.; Randle, P.J.; Martin, B.R. Stimulation by Calcium Ions of Pyruvate Dehydrogenase Phosphate Phosphatase. Biochem J. 1972, 128, 161–163. [Google Scholar] [CrossRef] [PubMed]
- McCormack, J.G.; Halestrap, A.P.; Denton, R.M. Role of Calcium Ions in Regulation of Mammalian Intramitochondrial Metabolism. Physiol. Rev. 1990, 70, 391–425. [Google Scholar] [CrossRef]
- Giorgi, C.; Marchi, S.; Pinton, P. The Machineries, Regulation and Cellular Functions of Mitochondrial Calcium. Nat. Rev. Mol. Cell Biol. 2018, 19, 713–730. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Hernandez, A.; Verkhratsky, A. Calcium Signalling in Diabetes. Cell Calcium 2014, 56, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Gilon, P.; Chae, H.-Y.; Rutter, G.A.; Ravier, M.A. Calcium Signaling in Pancreatic β-Cells in Health and in Type 2 Diabetes. Cell Calcium 2014, 56, 340–361. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, T.; Scharwey, M.; Langer, T. Mitochondrial Lipid Trafficking. Trends Cell Biol. 2014, 24, 44–52. [Google Scholar] [CrossRef]
- Zhang, Q.; Tamura, Y.; Roy, M.; Adachi, Y.; Iijima, M.; Sesaki, H. Biosynthesis and Roles of Phospholipids in Mitochondrial Fusion, Division and Mitophagy. Cell Mol. Life Sci 2014, 71, 3767–3778. [Google Scholar] [CrossRef]
- Potting, C.; Tatsuta, T.; König, T.; Haag, M.; Wai, T.; Aaltonen, M.J.; Langer, T. TRIAP1/PRELI Complexes Prevent Apoptosis by Mediating Intramitochondrial Transport of Phosphatidic Acid. Cell Metab. 2013, 18, 287–295. [Google Scholar] [CrossRef]
- Miliara, X.; Tatsuta, T.; Berry, J.-L.; Rouse, S.L.; Solak, K.; Chorev, D.S.; Wu, D.; Robinson, C.V.; Matthews, S.; Langer, T. Structural Determinants of Lipid Specificity within Ups/PRELI Lipid Transfer Proteins. Nat. Commun. 2019, 10, 1130. [Google Scholar] [CrossRef]
- Hernández-Alvarez, M.I.; Sebastián, D.; Vives, S.; Ivanova, S.; Bartoccioni, P.; Kakimoto, P.; Plana, N.; Veiga, S.R.; Hernández, V.; Vasconcelos, N.; et al. Deficient Endoplasmic Reticulum-Mitochondrial Phosphatidylserine Transfer Causes Liver Disease. Cell 2019, 177, 881–895.e17. [Google Scholar] [CrossRef]
- Horibata, Y.; Sugimoto, H. StarD7 Mediates the Intracellular Trafficking of Phosphatidylcholine to Mitochondria*. J. Biol. Chem. 2010, 285, 7358–7365. [Google Scholar] [CrossRef] [PubMed]
- Saita, S.; Tatsuta, T.; Lampe, P.A.; König, T.; Ohba, Y.; Langer, T. PARL Partitions the Lipid Transfer Protein STARD7 between the Cytosol and Mitochondria. EMBO J. 2018, 37, e97909. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Tamura, Y. Shuttle Mission in the Mitochondrial Intermembrane Space. EMBO J. 2018, 37, e98993. [Google Scholar] [CrossRef] [PubMed]
- Schug, Z.T.; Gottlieb, E. Cardiolipin Acts as a Mitochondrial Signalling Platform to Launch Apoptosis. Biochim. Et Biophys. Acta Biomembr. 2009, 1788, 2022–2031. [Google Scholar] [CrossRef] [PubMed]
- Janikiewicz, J.; Szymański, J.; Malinska, D.; Patalas-Krawczyk, P.; Michalska, B.; Duszyński, J.; Giorgi, C.; Bonora, M.; Dobrzyn, A.; Wieckowski, M.R. Mitochondria-Associated Membranes in Aging and Senescence: Structure, Function, and Dynamics. Cell Death Dis. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes Form at ER–Mitochondria Contact Sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef]
- Hsu, P.; Shi, Y. Regulation of Autophagy by Mitochondrial Phospholipids in Health and Diseases. Biochim Biophys Acta Mol. Cell Biol. Lipids 2017, 1862, 114–129. [Google Scholar] [CrossRef]
- Sawyer, S.L.; Cheuk-Him Ng, A.; Innes, A.M.; Wagner, J.D.; Dyment, D.A.; Tetreault, M.; Care4Rare Canada Consortium; Majewski, J.; Boycott, K.M.; Screaton, R.A.; et al. Homozygous Mutations in MFN2 Cause Multiple Symmetric Lipomatosis Associated with Neuropathy. Hum. Mol. Genet. 2015, 24, 5109–5114. [Google Scholar] [CrossRef]
- Rocha, N.; Bulger, D.A.; Frontini, A.; Titheradge, H.; Gribsholt, S.B.; Knox, R.; Page, M.; Harris, J.; Payne, F.; Adams, C.; et al. Human Biallelic MFN2 Mutations Induce Mitochondrial Dysfunction, Upper Body Adipose Hyperplasia, and Suppression of Leptin Expression. eLife 2017, 6, e23813. [Google Scholar] [CrossRef]
- Capel, E.; Vatier, C.; Cervera, P.; Stojkovic, T.; Disse, E.; Cottereau, A.-S.; Auclair, M.; Verpont, M.-C.; Mosbah, H.; Gourdy, P.; et al. MFN2-Associated Lipomatosis: Clinical Spectrum and Impact on Adipose Tissue. J. Clin. Lipidol. 2018, 12, 1420–1435. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.-P.; Hsu, C.-L.; Fan, L.-C.; Huang, Z.; Bhatia, D.; Chen, Y.-J.; Hisata, S.; Cho, S.J.; Nakahira, K.; Imamura, M.; et al. Mitofusins Regulate Lipid Metabolism to Mediate the Development of Lung Fibrosis. Nat. Commun. 2019, 10, 3390. [Google Scholar] [CrossRef]
- Boutant, M.; Kulkarni, S.S.; Joffraud, M.; Ratajczak, J.; Valera-Alberni, M.; Combe, R.; Zorzano, A.; Cantó, C. Mfn2 Is Critical for Brown Adipose Tissue Thermogenic Function. EMBO J. 2017, 36, 1543–1558. [Google Scholar] [CrossRef]
- Ding, Y.; Gao, H.; Zhao, L.; Wang, X.; Zheng, M. Mitofusin 2-Deficiency Suppresses Cell Proliferation through Disturbance of Autophagy. PLoS ONE 2015, 10, e0121328. [Google Scholar] [CrossRef]
- Zhao, T.; Huang, X.; Han, L.; Wang, X.; Cheng, H.; Zhao, Y.; Chen, Q.; Chen, J.; Cheng, H.; Xiao, R.; et al. Central Role of Mitofusin 2 in Autophagosome-Lysosome Fusion in Cardiomyocytes. J. Biol. Chem 2012, 287, 23615–23625. [Google Scholar] [CrossRef]
- Li, J.; Liu, X.; Wang, H.; Zhang, W.; Chan, D.C.; Shi, Y. Lysocardiolipin Acyltransferase 1 (ALCAT1) Controls Mitochondrial DNA Fidelity and Biogenesis through Modulation of MFN2 Expression. Proc. Natl. Acad. Sci. USA 2012, 109, 6975–6980. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Romestaing, C.; Han, X.; Li, Y.; Hao, X.; Wu, Y.; Sun, C.; Liu, X.; Jefferson, L.S.; Xiong, J.; et al. Cardiolipin Remodeling by ALCAT1 Links Oxidative Stress and Mitochondrial Dysfunction to Obesity. Cell Metab. 2010, 12, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Nie, J.; Zhang, J.; Kimball, S.R.; Zhang, H.; Zhang, W.J.; Jefferson, L.S.; Cheng, Z.; Ji, Q.; et al. ALCAT1 Controls Mitochondrial Etiology of Fatty Liver Diseases, Linking Defective Mitophagy to Steatosis. Hepatology 2015, 61, 486–496. [Google Scholar] [CrossRef]
- Bilguvar, K.; Tyagi, N.K.; Ozkara, C.; Tuysuz, B.; Bakircioglu, M.; Choi, M.; Delil, S.; Caglayan, A.O.; Baranoski, J.F.; Erturk, O.; et al. Recessive Loss of Function of the Neuronal Ubiquitin Hydrolase UCHL1 Leads to Early-Onset Progressive Neurodegeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 3489–3494. [Google Scholar] [CrossRef]
- Rydning, S.L.; Backe, P.H.; Sousa, M.M.L.; Iqbal, Z.; Øye, A.-M.; Sheng, Y.; Yang, M.; Lin, X.; Slupphaug, G.; Nordenmark, T.H.; et al. Novel UCHL1 Mutations Reveal New Insights into Ubiquitin Processing. Hum. Mol. Genet. 2017, 26, 1031–1040. [Google Scholar] [CrossRef]
- Yamazaki, K.; Wakasugi, N.; Tomita, T.; Kikuchi, T.; Mukoyama, M.; Ando, K. Gracile Axonal Dystrophy (GAD), a New Neurological Mutant in the Mouse. Proc. Soc. Exp. Biol. Med. 1988, 187, 209–215. [Google Scholar] [CrossRef]
- Chen, F.; Sugiura, Y.; Myers, K.G.; Liu, Y.; Lin, W. Ubiquitin Carboxyl-Terminal Hydrolase L1 Is Required for Maintaining the Structure and Function of the Neuromuscular Junction. PNAS 2010, 107, 1636–1641. [Google Scholar] [CrossRef]
- Cerqueira, F.M.; von Stockum, S.; Giacomello, M.; Goliand, I.; Kakimoto, P.; Marchesan, E.; de Stefani, D.; Kowaltowski, A.J.; Ziviani, E.; Shirihai, O.S. A New Target for an Old DUB: UCH-L1 Regulates Mitofusin-2 Levels, Altering Mitochondrial Morphology, Function and Calcium Uptake. Redox Biol. 2020, 37, 101676. [Google Scholar] [CrossRef] [PubMed]
- Poston, C.N.; Krishnan, S.C.; Bazemore-Walker, C.R. In-Depth Proteomic Analysis of Mammalian Mitochondria-Associated Membranes (MAM). J. Proteom. 2013, 79, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Meray, R.K.; Grammatopoulos, T.N.; Fredenburg, R.A.; Cookson, M.R.; Liu, Y.; Logan, T.; Lansbury, P.T. Membrane-Associated Farnesylated UCH-L1 Promotes α-Synuclein Neurotoxicity and Is a Therapeutic Target for Parkinson’s Disease. PNAS 2009, 106, 4635–4640. [Google Scholar] [CrossRef]
- Costes, S.; Huang, C.; Gurlo, T.; Daval, M.; Matveyenko, A.V.; Rizza, R.A.; Butler, A.E.; Butler, P.C. β-Cell Dysfunctional ERAD/Ubiquitin/Proteasome System in Type 2 Diabetes Mediated by Islet Amyloid Polypeptide-Induced UCH-L1 Deficiency. Diabetes 2011, 60, 227–238. [Google Scholar] [CrossRef]
- Petrinovic, M.M.; Duncan, C.S.; Bourikas, D.; Weinman, O.; Montani, L.; Schroeter, A.; Maerki, D.; Sommer, L.; Stoeckli, E.T.; Schwab, M.E. Neuronal Nogo-A Regulates Neurite Fasciculation, Branching and Extension in the Developing Nervous System. Development 2010, 137, 2539–2550. [Google Scholar] [CrossRef]
- Rigoli, L.; Bramanti, P.; di Bella, C.; de Luca, F. Genetic and Clinical Aspects of Wolfram Syndrome 1, a Severe Neurodegenerative Disease. Pediatric Res. 2018, 83, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Tanizawa, Y.; Wasson, J.; Behn, P.; Kalidas, K.; Bernal-Mizrachi, E.; Mueckler, M.; Marshall, H.; Donis-Keller, H.; Crock, P.; et al. A Gene Encoding a Transmembrane Protein Is Mutated in Patients with Diabetes Mellitus and Optic Atrophy (Wolfram Syndrome). Nat. Genet. 1998, 20, 143–148. [Google Scholar] [CrossRef]
- Strom, T.M.; Hörtnagel, K.; Hofmann, S.; Gekeler, F.; Scharfe, C.; Rabl, W.; Gerbitz, K.D.; Meitinger, T. Diabetes Insipidus, Diabetes Mellitus, Optic Atrophy and Deafness (DIDMOAD) Caused by Mutations in a Novel Gene (Wolframin) Coding for a Predicted Transmembrane Protein. Hum. Mol. Genet. 1998, 7, 2021–2028. [Google Scholar] [CrossRef]
- Osman, A.A.; Saito, M.; Makepeace, C.; Permutt, M.A.; Schlesinger, P.; Mueckler, M. Wolframin Expression Induces Novel Ion Channel Activity in Endoplasmic Reticulum Membranes and Increases Intracellular Calcium. J. Biol. Chem. 2003, 278, 52755–52762. [Google Scholar] [CrossRef]
- Fonseca, S.G.; Fukuma, M.; Lipson, K.L.; Nguyen, L.X.; Allen, J.R.; Oka, Y.; Urano, F. WFS1 Is a Novel Component of the Unfolded Protein Response and Maintains Homeostasis of the Endoplasmic Reticulum in Pancreatic β-Cells. J. Biol. Chem. 2005, 280, 39609–39615. [Google Scholar] [CrossRef]
- Fonseca, S.G.; Ishigaki, S.; Oslowski, C.M.; Lu, S.; Lipson, K.L.; Ghosh, R.; Hayashi, E.; Ishihara, H.; Oka, Y.; Permutt, M.A.; et al. Wolfram Syndrome 1 Gene Negatively Regulates ER Stress Signaling in Rodent and Human Cells. J. Clin. Invest. 2010, 120, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Delprat, B.; Maurice, T.; Delettre, C. Wolfram Syndrome: MAMs’ Connection? Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Grenier, J.; Meunier, I.; Daien, V.; Baudoin, C.; Halloy, F.; Bocquet, B.; Blanchet, C.; Delettre, C.; Esmenjaud, E.; Roubertie, A.; et al. WFS1 in Optic Neuropathies: Mutation Findings in Nonsyndromic Optic Atrophy and Assessment of Clinical Severity. Ophthalmology 2016, 123, 1989–1998. [Google Scholar] [CrossRef]
- Amr, S.; Heisey, C.; Zhang, M.; Xia, X.-J.; Shows, K.H.; Ajlouni, K.; Pandya, A.; Satin, L.S.; El-Shanti, H.; Shiang, R. A Homozygous Mutation in a Novel Zinc-Finger Protein, ERIS, Is Responsible for Wolfram Syndrome 2. Am. J. Hum. Genet. 2007, 81, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Mozzillo, E.; Delvecchio, M.; Carella, M.; Grandone, E.; Palumbo, P.; Salina, A.; Aloi, C.; Buono, P.; Izzo, A.; D’Annunzio, G.; et al. A Novel CISD2 Intragenic Deletion, Optic Neuropathy and Platelet Aggregation Defect in Wolfram Syndrome Type 2. BMC Med. Genet. 2014, 15, 88. [Google Scholar] [CrossRef]
- Wiley, S.E.; Andreyev, A.Y.; Divakaruni, A.S.; Karisch, R.; Perkins, G.; Wall, E.A.; van der Geer, P.; Chen, Y.-F.; Tsai, T.-F.; Simon, M.I.; et al. Wolfram Syndrome Protein, Miner1, Regulates Sulphydryl Redox Status, the Unfolded Protein Response, and Ca2+ Homeostasis. EMBO Mol. Med. 2013, 5, 904–918. [Google Scholar] [CrossRef] [PubMed]
- El-Shanti, H.; Lidral, A.C.; Jarrah, N.; Druhan, L.; Ajlouni, K. Homozygosity Mapping Identifies an Additional Locus for Wolfram Syndrome on Chromosome 4q. Am. J. Hum. Genet. 2000, 66, 1229–1236. [Google Scholar] [CrossRef]
- Rouzier, C.; Moore, D.; Delorme, C.; Lacas-Gervais, S.; Ait-El-Mkadem, S.; Fragaki, K.; Burté, F.; Serre, V.; Bannwarth, S.; Chaussenot, A.; et al. A Novel CISD2 Mutation Associated with a Classical Wolfram Syndrome Phenotype Alters Ca2+ Homeostasis and ER-Mitochondria Interactions. Hum. Mol. Genet. 2017, 26, 1599–1611. [Google Scholar] [CrossRef]
- Cagalinec, M.; Liiv, M.; Hodurova, Z.; Hickey, M.A.; Vaarmann, A.; Mandel, M.; Zeb, A.; Choubey, V.; Kuum, M.; Safiulina, D.; et al. Role of Mitochondrial Dynamics in Neuronal Development: Mechanism for Wolfram Syndrome. PLoS Biol. 2016, 14, e1002511. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Mahadevan, J.; Kanekura, K.; Hara, M.; Lu, S.; Urano, F. Calcium Efflux from the Endoplasmic Reticulum Leads to β-Cell Death. Endocrinology 2014, 155, 758–768. [Google Scholar] [CrossRef]
- Lu, S.; Kanekura, K.; Hara, T.; Mahadevan, J.; Spears, L.D.; Oslowski, C.M.; Martinez, R.; Yamazaki-Inoue, M.; Toyoda, M.; Neilson, A.; et al. A Calcium-Dependent Protease as a Potential Therapeutic Target for Wolfram Syndrome. PNAS 2014, 111, E5292–E5301. [Google Scholar] [CrossRef] [PubMed]
- Abreu, D.; Asada, R.; Revilla, J.M.P.; Lavagnino, Z.; Kries, K.; Piston, D.W.; Urano, F. Wolfram Syndrome 1 Gene Regulates Pathways Maintaining Beta-Cell Health and Survival. Lab. Invest. 2020, 100, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Wersinger, D.B.; Benkafadar, N.; Jagodzinska, J.; Hamel, C.; Tanizawa, Y.; Lenaers, G.; Delettre, C. Impairment of Visual Function and Retinal ER Stress Activation in Wfs1-Deficient Mice. PLoS ONE 2014, 9, e97222. [Google Scholar] [CrossRef]
- Kanekura, K.; Ma, X.; Murphy, J.T.; Zhu, L.J.; Diwan, A.; Urano, F. IRE1 Prevents Endoplasmic Reticulum Membrane Permeabilization and Cell Death under Pathological Conditions. Sci. Signal. 2015, 8, ra62. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Kao, C.-H.; Chen, Y.-T.; Wang, C.-H.; Wu, C.-Y.; Tsai, C.-Y.; Liu, F.-C.; Yang, C.-W.; Wei, Y.-H.; Hsu, M.-T.; et al. Cisd2 Deficiency Drives Premature Aging and Causes Mitochondria-Mediated Defects in Mice. Genes Dev. 2009, 23, 1183–1194. [Google Scholar] [CrossRef]
- Lugar, H.M.; Koller, J.M.; Rutlin, J.; Marshall, B.A.; Kanekura, K.; Urano, F.; Bischoff, A.N.; Shimony, J.S.; Hershey, T.; Washington University Wolfram Syndrome Research Study Group. Neuroimaging Evidence of Deficient Axon Myelination in Wolfram Syndrome. Sci Rep. 2016, 6, 21167. [Google Scholar] [CrossRef]
- Samara, A.; Rahn, R.; Neyman, O.; Park, K.Y.; Samara, A.; Marshall, B.; Dougherty, J.; Hershey, T. Developmental Hypomyelination in Wolfram Syndrome: New Insights from Neuroimaging and Gene Expression Analyses. Orphanet J. Rare Dis 2019, 14, 279. [Google Scholar] [CrossRef]
- Anikster, Y.; Kleta, R.; Shaag, A.; Gahl, W.A.; Elpeleg, O. Type III 3-Methylglutaconic Aciduria (Optic Atrophy plus Syndrome, or Costeff Optic Atrophy Syndrome): Identification of the OPA3 Gene and Its Founder Mutation in Iraqi Jews. Am. J. Hum. Genet. 2001, 69, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Abrams, A.J.; Hufnagel, R.B.; Rebelo, A.; Zanna, C.; Patel, N.; Gonzalez, M.A.; Campeanu, I.J.; Griffin, L.B.; Groenewald, S.; Strickland, A.V.; et al. Mutations in SLC25A46, Encoding a UGO1-like Protein, Cause an Optic Atrophy Spectrum Disorder. Nat. Genet. 2015, 47, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Janer, A.; Prudent, J.; Paupe, V.; Fahiminiya, S.; Majewski, J.; Sgarioto, N.; des Rosiers, C.; Forest, A.; Lin, Z.; Gingras, A.; et al. SLC25A46 Is Required for Mitochondrial Lipid Homeostasis and Cristae Maintenance and Is Responsible for Leigh Syndrome. EMBO Mol. Med. 2016, 8, 1019–1038. [Google Scholar] [CrossRef] [PubMed]
- Reynier, P.; Amati-Bonneau, P.; Verny, C.; Olichon, A.; Simard, G.; Guichet, A.; Bonnemains, C.; Malecaze, F.; Malinge, M.C.; Pelletier, J.B.; et al. OPA3 Gene Mutations Responsible for Autosomal Dominant Optic Atrophy and Cataract. J. Med. Genet. 2004, 41, e110. [Google Scholar] [CrossRef]
- Sergouniotis, P.I.; Perveen, R.; Thiselton, D.L.; Giannopoulos, K.; Sarros, M.; Davies, J.R.; Biswas, S.; Ansons, A.M.; Ashworth, J.L.; Lloyd, I.C.; et al. Clinical and Molecular Genetic Findings in Autosomal Dominant OPA3-Related Optic Neuropathy. Neurogenetics 2015, 16, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Horga, A.; Bugiardini, E.; Manole, A.; Bremner, F.; Jaunmuktane, Z.; Dankwa, L.; Rebelo, A.P.; Woodward, C.E.; Hargreaves, I.P.; Cortese, A.; et al. Autosomal Dominant Optic Atrophy and Cataract “plus” Phenotype Including Axonal Neuropathy. Neurol Genet. 2019, 5, e322. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Steffen, J.; Yourshaw, M.; Mamsa, H.; Andersen, E.; Rudnik-Schöneborn, S.; Pope, K.; Howell, K.B.; McLean, C.A.; Kornberg, A.J.; et al. Loss of Function of SLC25A46 Causes Lethal Congenital Pontocerebellar Hypoplasia. Brain 2016, 139, 2877–2890. [Google Scholar] [CrossRef]
- van Dijk, T.; Rudnik-Schöneborn, S.; Senderek, J.; Hajmousa, G.; Mei, H.; Dusl, M.; Aronica, E.; Barth, P.; Baas, F. Pontocerebellar Hypoplasia with Spinal Muscular Atrophy (PCH1): Identification of SLC25A46 Mutations in the Original Dutch PCH1 Family. Brain 2017, 140, e46. [Google Scholar] [CrossRef][Green Version]
- Braunisch, M.C.; Gallwitz, H.; Abicht, A.; Diebold, I.; Holinski-Feder, E.; Maldergem, L.V.; Lammens, M.; Kovács-Nagy, R.; Alhaddad, B.; Strom, T.M.; et al. Extension of the Phenotype of Biallelic Loss-of-Function Mutations in SLC25A46 to the Severe Form of Pontocerebellar Hypoplasia Type I. Clin. Genet. 2018, 93, 255–265. [Google Scholar] [CrossRef]
- Bitetto, G.; Malaguti, M.C.; Ceravolo, R.; Monfrini, E.; Straniero, L.; Morini, A.; di Giacopo, R.; Frosini, D.; Palermo, G.; Biella, F.; et al. SLC25A46 Mutations in Patients with Parkinson’s Disease and Optic Atrophy. Parkinsonism. Relat. Disord. 2020, 74, 1–5. [Google Scholar] [CrossRef]
- Huizing, M.; Dorward, H.; Ly, L.; Klootwijk, E.; Kleta, R.; Skovby, F.; Pei, W.; Feldman, B.; Gahl, W.A.; Anikster, Y. OPA3, Mutated in 3-Methylglutaconic Aciduria Type III, Encodes Two Transcripts Targeted Primarily to Mitochondria. Mol. Genet. Metab. 2010, 100, 149–154. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ryu, S.-W.; Jeong, H.J.; Choi, M.; Karbowski, M.; Choi, C. Optic Atrophy 3 as a Protein of the Mitochondrial Outer Membrane Induces Mitochondrial Fragmentation. Cell Mol. Life Sci 2010, 67, 2839–2850. [Google Scholar] [CrossRef]
- Ryu, S.-W.; Yoon, J.; Yim, N.; Choi, K.; Choi, C. Downregulation of OPA3 Is Responsible for Transforming Growth Factor-β-Induced Mitochondrial Elongation and F-Actin Rearrangement in Retinal Pigment Epithelial ARPE-19 Cells. PLoS ONE 2013, 8, e63495. [Google Scholar] [CrossRef]
- Wells, T.; Davies, J.R.; Guschina, I.A.; Ball, D.J.; Davies, J.S.; Davies, V.J.; Evans, B.A.J.; Votruba, M. Opa3, a Novel Regulator of Mitochondrial Function, Controls Thermogenesis and Abdominal Fat Mass in a Mouse Model for Costeff Syndrome. Hum. Mol. Genet. 2012, 21, 4836–4844. [Google Scholar] [CrossRef]
- Bourne, S.C.; Townsend, K.N.; Shyr, C.; Matthews, A.; Lear, S.A.; Attariwala, R.; Lehman, A.; Wasserman, W.W.; van Karnebeek, C.; Sinclair, G.; et al. Optic Atrophy, Cataracts, Lipodystrophy/Lipoatrophy, and Peripheral Neuropathy Caused by a de Novo OPA3 Mutation. Cold Spring Harb Mol. Case Stud. 2017, 3, a001156. [Google Scholar] [CrossRef]
- Steffen, J.; Vashisht, A.A.; Wan, J.; Jen, J.C.; Claypool, S.M.; Wohlschlegel, J.A.; Koehler, C.M. Rapid Degradation of Mutant SLC25A46 by the Ubiquitin-Proteasome System Results in MFN1/2-Mediated Hyperfusion of Mitochondria. Mol. Biol. Cell 2017, 28, 600–612. [Google Scholar] [CrossRef]
- Mueller, N.; Sassa, T.; Morales-Gonzalez, S.; Schneider, J.; Salchow, D.J.; Seelow, D.; Knierim, E.; Stenzel, W.; Kihara, A.; Schuelke, M. De Novo Mutation in ELOVL1 Causes Ichthyosis, Acanthosis Nigricans, Hypomyelination, Spastic Paraplegia, High Frequency Deafness and Optic Atrophy. J. Med. Genet. 2019, 56, 164–175. [Google Scholar] [CrossRef]
- Kutkowska-Kaźmierczak, A.; Rydzanicz, M.; Chlebowski, A.; Kłosowska-Kosicka, K.; Mika, A.; Gruchota, J.; Jurkiewicz, E.; Kowalewski, C.; Pollak, A.; Stradomska, T.J.; et al. Dominant ELOVL1 Mutation Causes Neurological Disorder with Ichthyotic Keratoderma, Spasticity, Hypomyelination and Dysmorphic Features. J. Med. Genet. 2018, 55, 408–414. [Google Scholar] [CrossRef]
- Miralles Fusté, J.; Shi, Y.; Wanrooij, S.; Zhu, X.; Jemt, E.; Persson, Ö.; Sabouri, N.; Gustafsson, C.M.; Falkenberg, M. In Vivo Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication. PLoS Genet. 2014, 10, e1004832. [Google Scholar] [CrossRef] [PubMed]
- Courage, C.; Jackson, C.B.; Hahn, D.; Euro, L.; Nuoffer, J.-M.; Gallati, S.; Schaller, A. SDHA Mutation with Dominant Transmission Results in Complex II Deficiency with Ocular, Cardiac, and Neurologic Involvement. Am. J. Med. Genet. A 2017, 173, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Zehavi, Y.; Saada, A.; Jabaly-Habib, H.; Dessau, M.; Shaag, A.; Elpeleg, O.; Spiegel, R. A Novel de Novo Heterozygous Pathogenic Variant in the SDHA Gene Results in Childhood Onset Bilateral Optic Atrophy and Cognitive Impairment. Metab. Brain Dis 2021. [Google Scholar] [CrossRef]
- Metodiev, M.D.; Gerber, S.; Hubert, L.; Delahodde, A.; Chretien, D.; Gérard, X.; Amati-Bonneau, P.; Giacomotto, M.-C.; Boddaert, N.; Kaminska, A.; et al. Mutations in the Tricarboxylic Acid Cycle Enzyme, Aconitase 2, Cause Either Isolated or Syndromic Optic Neuropathy with Encephalopathy and Cerebellar Atrophy. J. Med. Genet. 2014, 51, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.A.-C.; Grossmann, D.; Schimpf-Linzenbold, S.; Dayan, D.; Stingl, K.; Ben-Menachem, R.; Pines, O.; Massart, F.; Delcambre, S.; Ghelfi, J.; et al. Haploinsufficiency Due to a Novel ACO2 Deletion Causes Mitochondrial Dysfunction in Fibroblasts from a Patient with Dominant Optic Nerve Atrophy. Sci Rep. 2020, 10, 16736. [Google Scholar] [CrossRef]
- Sadat, R.; Barca, E.; Masand, R.; Donti, T.R.; Naini, A.; de Vivo, D.C.; di Mauro, S.; Hanchard, N.A.; Graham, B.H. Functional Cellular Analyses Reveal Energy Metabolism Defect and Mitochondrial DNA Depletion in a Case of Mitochondrial Aconitase Deficiency. Mol. Genet. Metab. 2016, 118, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Bosch, D.G.M.; Boonstra, F.N.; Gonzaga-Jauregui, C.; Xu, M.; de Ligt, J.; Jhangiani, S.; Wiszniewski, W.; Muzny, D.M.; Yntema, H.G.; Pfundt, R.; et al. NR2F1 Mutations Cause Optic Atrophy with Intellectual Disability. Am. J. Hum. Genet. 2014, 94, 303–309. [Google Scholar] [CrossRef]
- Tang, K.; Xie, X.; Park, J.-I.; Jamrich, M.; Tsai, S.; Tsai, M.-J. COUP-TFs Regulate Eye Development by Controlling Factors Essential for Optic Vesicle Morphogenesis. Development 2010, 137, 725–734. [Google Scholar] [CrossRef]
- Kao, C.-Y.; Xu, M.; Wang, L.; Lin, S.-C.; Lee, H.-J.; Duraine, L.; Bellen, H.J.; Goldstein, D.S.; Tsai, S.Y.; Tsai, M.-J. Elevated COUP-TFII Expression in Dopaminergic Neurons Accelerates the Progression of Parkinson’s Disease through Mitochondrial Dysfunction. PLoS Genet. 2020, 16, e1008868. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced Pluripotent Stem Cell Technology: A Decade of Progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Cowan, C.S.; Renner, M.; De Gennaro, M.; Gross-Scherf, B.; Goldblum, D.; Hou, Y.; Munz, M.; Rodrigues, T.M.; Krol, J.; Szikra, T.; et al. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell 2020, 182, 1623–1640.e34. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Jourdain, A.A.; Calvo, S.E.; Ballarano, C.A.; Doench, J.G.; Root, D.E.; Mootha, V.K. A Genome-Wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab. 2016, 24, 875–885. [Google Scholar] [CrossRef]
- Aleo, S.J.; Del Dotto, V.; Fogazza, M.; Maresca, A.; Lodi, T.; Goffrini, P.; Ghelli, A.; Rugolo, M.; Carelli, V.; Baruffini, E.; et al. Drug Repositioning as a Therapeutic Strategy for Neurodegenerations Associated with OPA1 Mutations. Hum. Mol. Genet. 2021, 29, 3631–3645. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A. The Promise and Challenge of Therapeutic Genome Editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Laha, B.; Stafford, B.K.; Huberman, A.D. Regenerating Optic Pathways from the Eye to the Brain. Science 2017, 356, 1031–1034. [Google Scholar] [CrossRef] [PubMed]
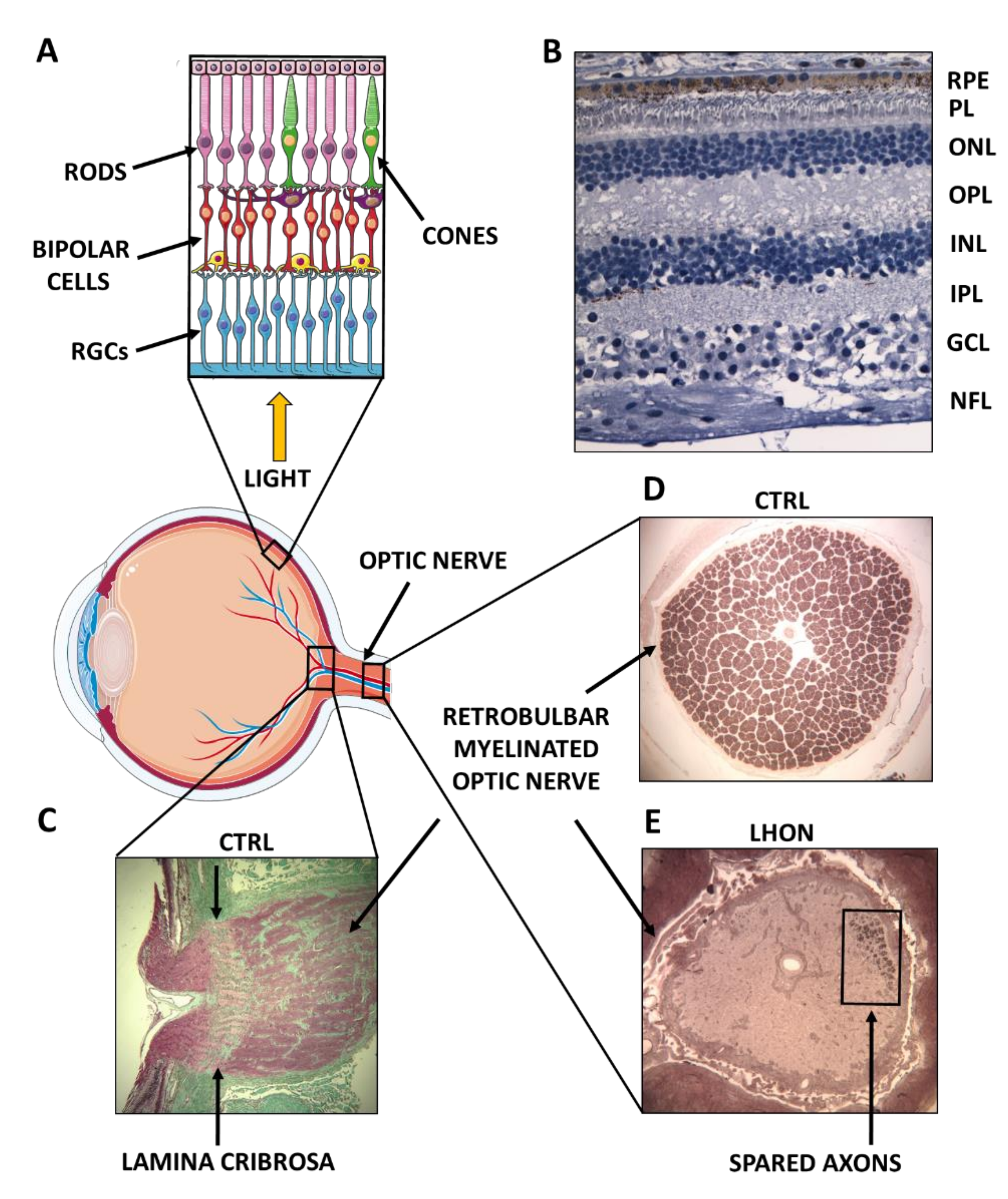
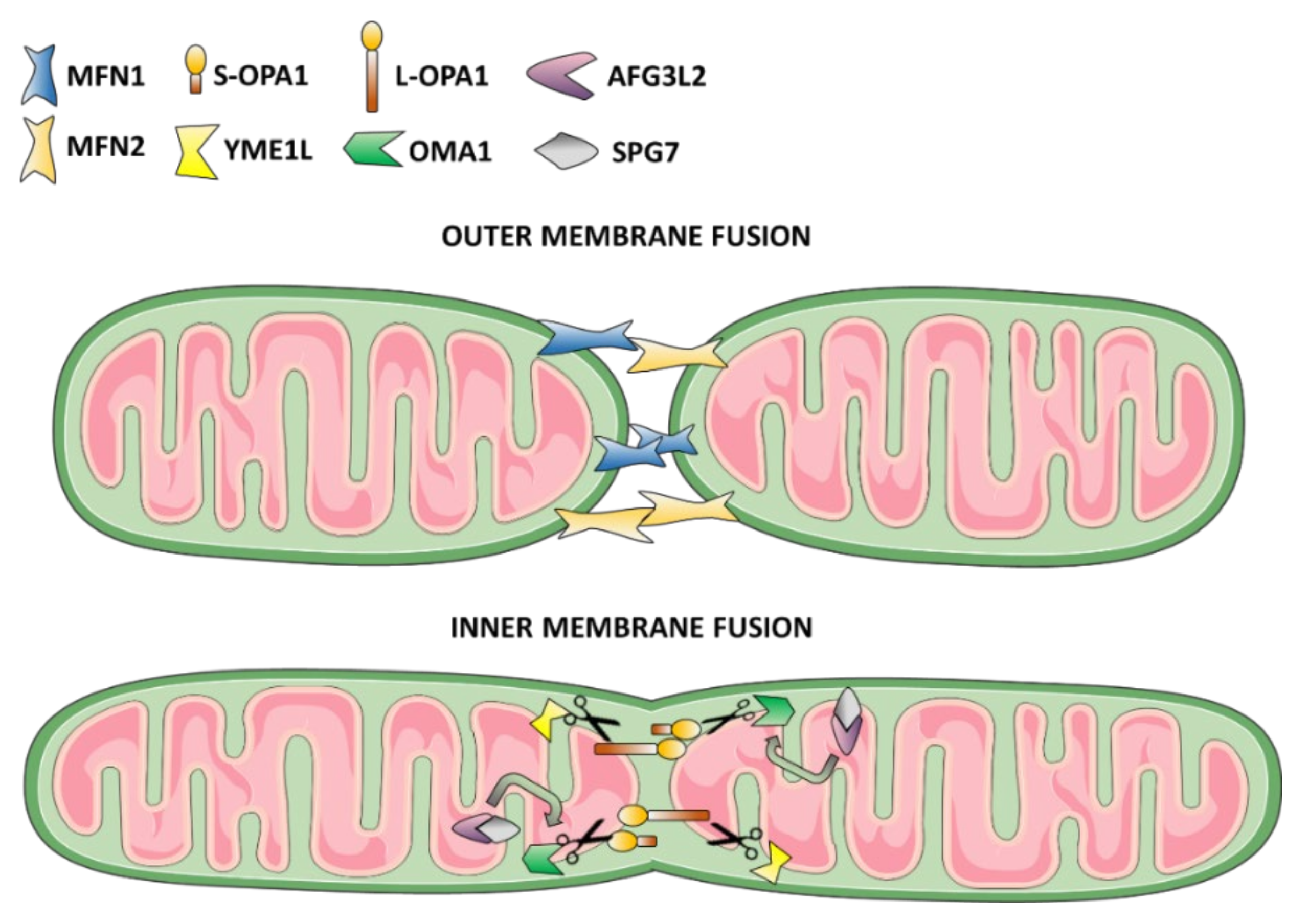
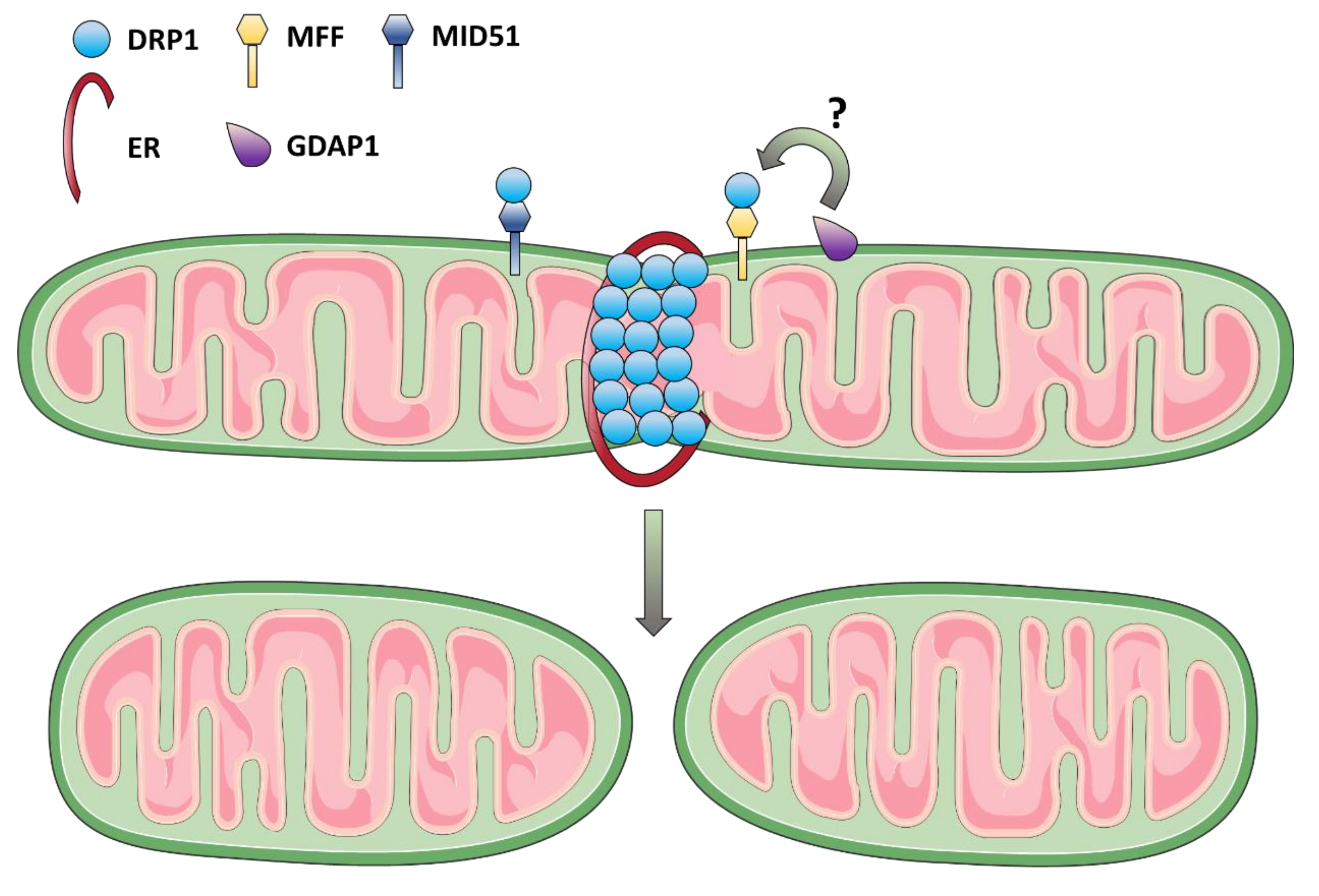
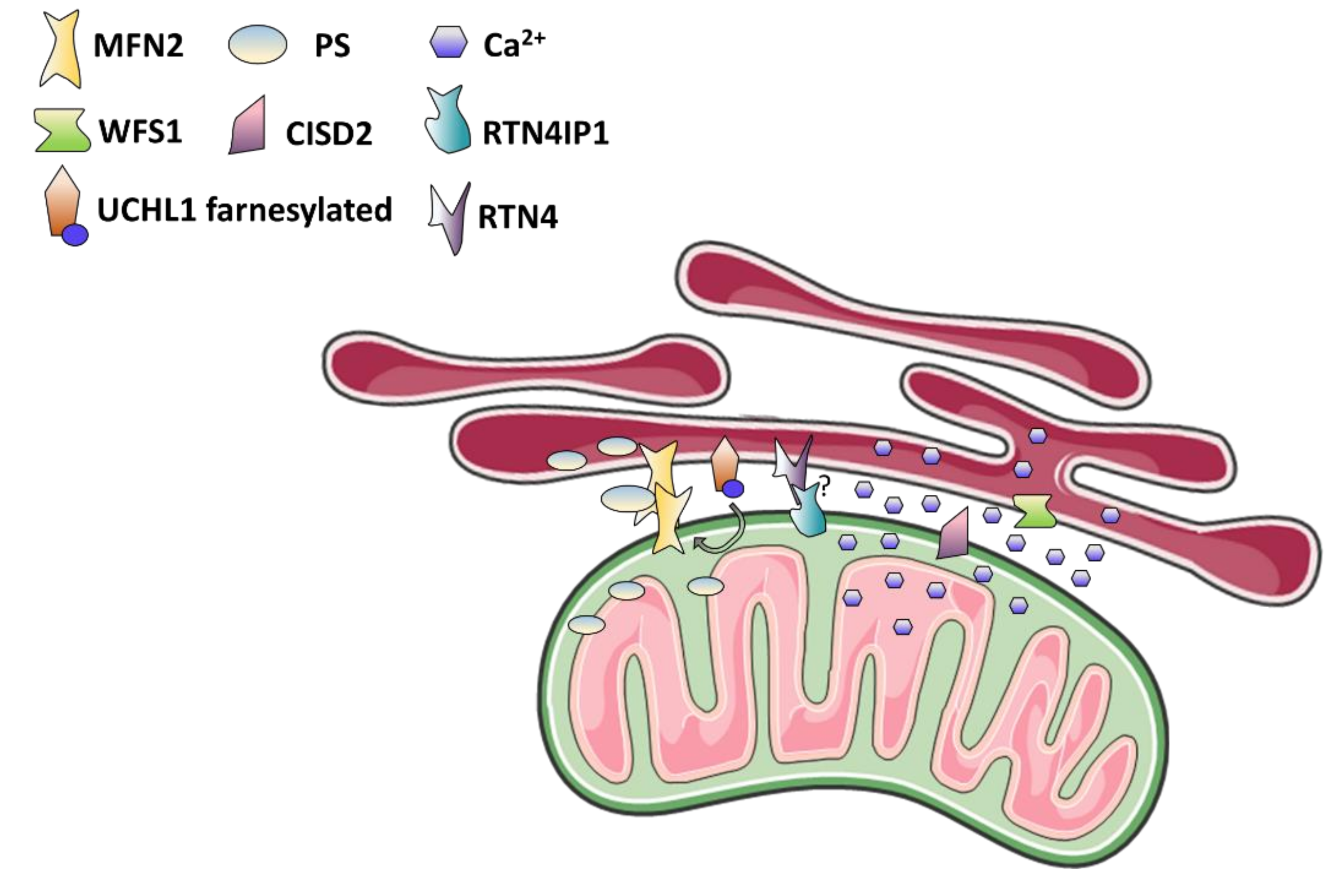
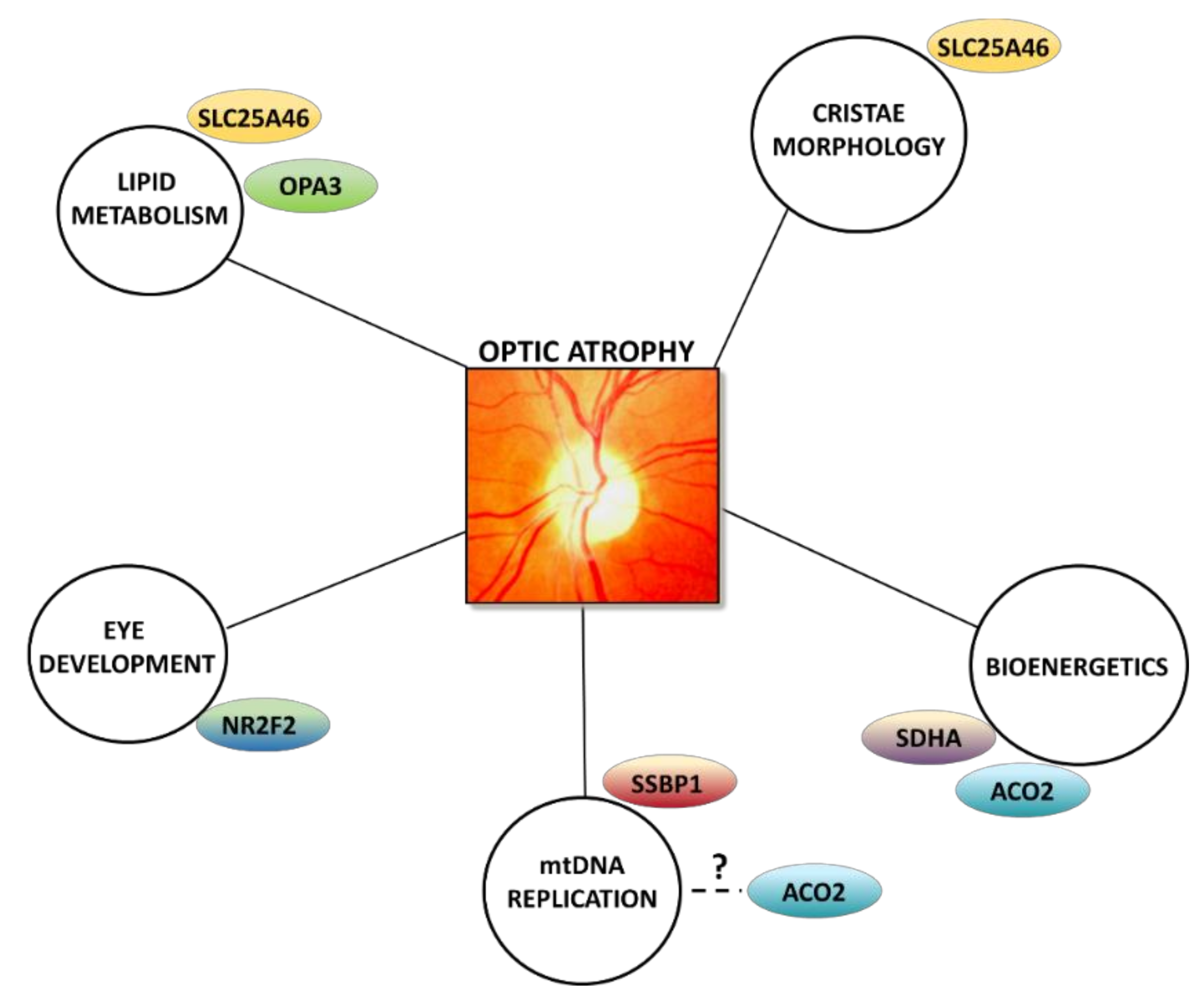
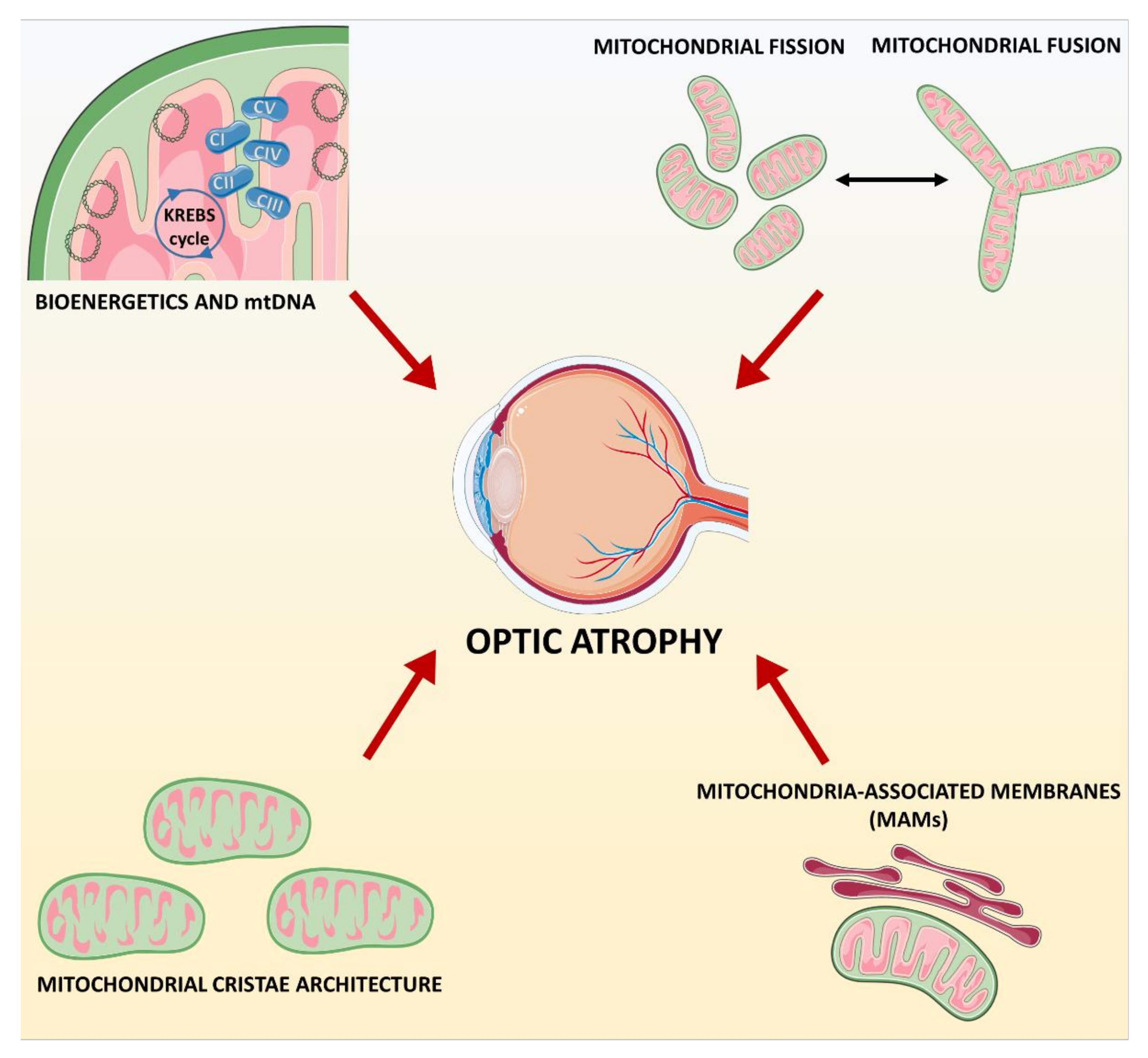
| Gene | Genome | Disease | Inheritance | Protein Function |
|---|---|---|---|---|
| MT-ND1 | mitochondrial | LHON | MI | CI subunit |
| MT-ND4 | mitochondrial | LHON | MI | CI subunit |
| MT-ND6 | mitochondrial | LHON | MI | CI subunit |
| DNAJC30 | nuclear | LHON | AR | CI turnover |
| TMEM126A | nuclear | OA and auditory neuropathy | AR | CI assembly |
| RTN4IP1 | nuclear | OA with or without ataxia, ID, and epilepsy | AR | CI assembly |
| Gene | Effect on Mitochondrial Network | Disease | Inheritance | Protein Function |
|---|---|---|---|---|
| OPA1 | Fragmented | DOA DOA-plus | AD | IMM fusion |
| YME1L | Fragmented | Mitochondrial encephalopathy with OA | AR | OPA1 processing |
| AFG3L2 | Fragmented | DOA with or without additional neurological features, CPEO | AD | OMA1 regulation |
| SPG7 | Normal or hyperfused | DOA Spastic paraparesis and OA, CPEO | AD AR | OMA1 regulation |
| MFN2 | Fragmented or hyperfused | CMT with or without OA HMSN Lipomatosis and neuropathy | AD/AR AD AR | OMM fusion; MAMs formation; PS transfer |
| DRP1 | Hyperfused | Mitochondrial encephalopathy with OA DOA | AD/AR AD | OMM fission |
| MFF | Hyperfused | Mitochondrial encephalopathy with OA | AR | DRP1 adaptor |
| MIEF1 | Fragmented | DOA | AD | DRP1 adaptor |
| GDAP1 | Fragmented or hyperfused | CMT with or without OA | AD/AR | DRP1 and FIS1 regulation? |
| Gene | Effect on MAMs Function | Disease | Inheritance | Protein Function |
|---|---|---|---|---|
| MFN2 | Reduced ER-mitochondria contacts; UPR activation; Ca2+ and lipids mishandling; altered autophagy | CMT with or without OA HMSN Lipomatosis and neuropathy | AD/AR AD AR | OMM fusion; MAMs formation; PS transfer |
| UCHL1 | Reduced ER-mitochondria contacts; Ca2+ mishandling 1 | Spastic paraplegia with OA | AR | Protein deubiquitination |
| RTN4IP1 | Unknown | OA with or without ataxia, ID, and epilepsy | AR | CI assembly; RTN4 interactor |
| WFS1 | Reduced ER-mitochondria contacts; Ca2+ mishandling | WS1 Isolated cataract, or diabetes or deafness Isolated OA WS-like | AR AD AR AD | Ca2+ homeostasis; UPR and ER stress regulation; insulin biosynthesis |
| CISD2 | Increased ER-mitochondria contact; mitochondrial Ca2+ overload; enhanced ROS | WS2 | AR | Ca2+ homeostasis; anti-oxidant activity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maresca, A.; Carelli, V. Molecular Mechanisms behind Inherited Neurodegeneration of the Optic Nerve. Biomolecules 2021, 11, 496. https://doi.org/10.3390/biom11040496
Maresca A, Carelli V. Molecular Mechanisms behind Inherited Neurodegeneration of the Optic Nerve. Biomolecules. 2021; 11(4):496. https://doi.org/10.3390/biom11040496
Chicago/Turabian StyleMaresca, Alessandra, and Valerio Carelli. 2021. "Molecular Mechanisms behind Inherited Neurodegeneration of the Optic Nerve" Biomolecules 11, no. 4: 496. https://doi.org/10.3390/biom11040496
APA StyleMaresca, A., & Carelli, V. (2021). Molecular Mechanisms behind Inherited Neurodegeneration of the Optic Nerve. Biomolecules, 11(4), 496. https://doi.org/10.3390/biom11040496






