The Interplay of Microtubules with Mitochondria–ER Contact Sites (MERCs) in Glioblastoma
Abstract
1. Introduction
2. Cytoskeleton and Glial Cells: A Focus on Microtubule and Microfilaments
3. Cytoskeletal Proteins: Key Players of Glioblastoma Cell Invasive Properties
4. Mitochondria Dynamics and Glial Cells
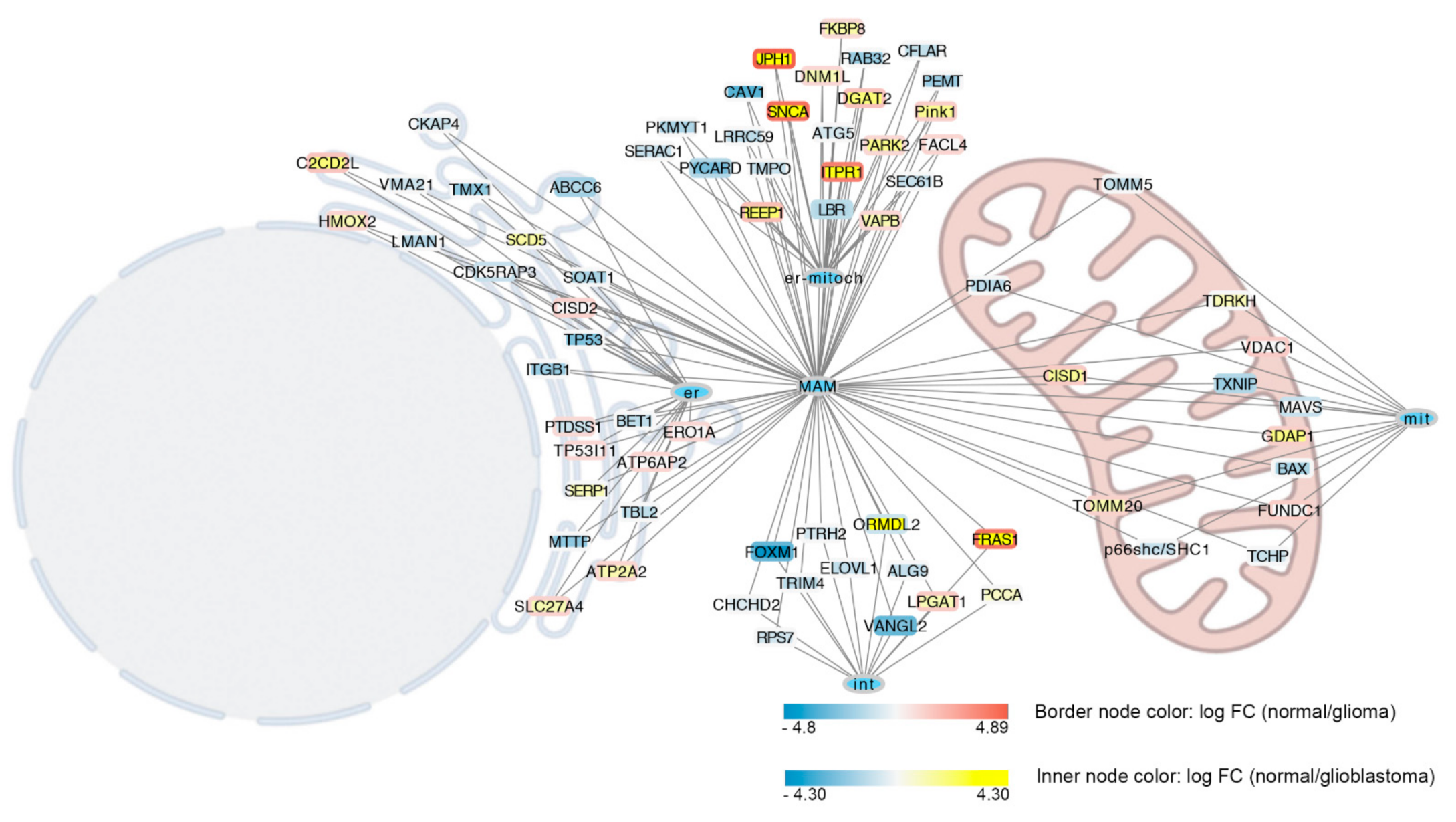
5. MERCs in Glioblastoma
6. MERCs and Microtubules Crosstalk in Glioblastoma
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pisapia, D.J. The Updated World Health Organization Glioma Classification: Cellular and Molecular Origins of Adult Infiltrating Gliomas. Arch. Pathol. Lab. Med. 2017, 141, 1633–1645. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The Epidemiology of Glioma in Adults: A State of the Science Review. Neuro-Oncology 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Perry, A.; Wesseling, P. Histologic Classification of Gliomas. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 134, pp. 71–95. ISBN 9780128029978. [Google Scholar]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Hayden, E.C. Genomics Boosts Brain-Cancer Work. Nature 2010, 463, 278. [Google Scholar] [CrossRef]
- Kuehn, B.M. Genomics Illuminates a Deadly Brain Cancer. JAMA-J. Am. Med. Assoc. 2010, 303, 925–927. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849. [Google Scholar] [CrossRef]
- Fisher, J.L.; Schwartzbaum, J.A.; Wrensch, M.; Wiemels, J.L. Epidemiology of Brain Tumors. Neurol. Clin. 2007, 25, 867–890. [Google Scholar] [CrossRef]
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef]
- Katz, A.M.; Amankulor, N.M.; Pitter, K.; Helmy, K.; Squatrito, M.; Holland, E.C. Astrocyte-Specific Expression Patterns Associated with the PDGF-Induced Glioma Microenvironment. PLoS ONE 2012, 7, e32453. [Google Scholar] [CrossRef]
- Chakravarti, A.; Delaney, M.A.; Loeffler, J.S.; Muzikansky, A.; Dyson, N.J.; Noll, E.; Black, P.M.L. Prognostic and Pathologic Significance of Quantitative Protein Expression Profiling in Human Gliomas. Clin. Cancer Res. 2001, 7, 2387–2395. [Google Scholar]
- Bachoo, R.M.; Maher, E.A.; Ligon, K.L.; Sharpless, N.E.; Chan, S.S.; You, M.J.; Tang, Y.; DeFrances, J.; Stover, E.; Weissleder, R.; et al. Epidermal Growth Factor Receptor and Ink4a/Arf: Governing Terminal Differentiation and Transformation Stem Cell to Astrocyte Axis. Cancer Cell 2002, 1, 269–277. [Google Scholar] [CrossRef]
- Guan, X.; Hasan, M.N.; Maniar, S.; Jia, W.; Sun, D. Reactive Astrocytes in Glioblastoma Multiforme. Mol. Neurobiol. 2018, 55, 6927–6938. [Google Scholar] [CrossRef]
- Ishizaki, T.; Morishima, Y.; Okamoto, M.; Furuyashiki, T.; Kato, T.; Narumiya, S. Coordination of Microtubules and the Actin Cytoskeleton by the Rho Effector MDia1. Nat. Cell Biol. 2001, 3, 8–14. [Google Scholar] [CrossRef]
- Bunse, L.; Pusch, S.; Bunse, T.; Sahm, F.; Sanghvi, K.; Friedrich, M.; Alansary, D.; Sonner, J.K.; Green, E.; Deumelandt, K.; et al. Suppression of Antitumor T Cell Immunity by the Oncometabolite (R)-2-Hydroxyglutarate. Nat. Med. 2018, 24, 1192–1203. [Google Scholar] [CrossRef]
- Bassoy, E.Y.; Kasahara, A.; Chiusolo, V.; Jacquemin, G.; Boydell, E.; Zamorano, S.; Riccadonna, C.; Pellegatta, S.; Hulo, N.; Dutoit, V.; et al. ER–Mitochondria Contacts Control Surface Glycan Expression and Sensitivity to Killer Lymphocytes in Glioma Stem-like Cells. EMBO J. 2017, 36, 1493–1512. [Google Scholar] [CrossRef]
- Lee-Chang, C.; Rashidi, A.; Miska, J.; Zhang, P.; Pituch, K.C.; Hou, D.; Xiao, T.; Fischietti, M.; Kang, S.J.; Appin, C.L.; et al. Myeloid-Derived Suppressive Cells Promote B Cell-Mediated Immunosuppression via Transfer of PD-L1 in Glioblastoma. Cancer Immunol. Res. 2019, 7, 1928–1943. [Google Scholar] [CrossRef]
- Yilmaz, M.; Christofori, G. EMT, the Cytoskeleton, and Cancer Cell Invasion. Cancer Metastasis Rev. 2009, 28, 15–33. [Google Scholar] [CrossRef]
- Yeung, C.Y.C.; Taylor, S.H.; Garva, R.; Holmes, D.F.; Zeef, L.A.; Soininen, R.; Boot-Handford, R.P.; Kadler, K.E. Arhgap28 Is a RhoGAP That Inactivates RhoA and Downregulates Stress Fibers. PLoS ONE 2014, 9, e107036. [Google Scholar] [CrossRef]
- Fortin Ensign, S.P.; Mathews, I.T.; Symons, M.H.; Berens, M.E.; Tran, N.L. Implications of Rho GTPase Signaling in Glioma Cell Invasion and Tumor Progression. Front. Oncol. 2013, 3, 241. [Google Scholar] [CrossRef]
- McLendon, R.; Friedman, A.; Bigner, D.; van Meir, E.G.; Brat, D.J.; Mastrogianakis, G.M.; Olson, J.J.; Mikkelsen, T.; Lehman, N.; Aldape, K.; et al. Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Liang, B.C.; Grootveld, M. The Importance of Mitochondria in the Tumourigenic Phenotype: Gliomas as the Paradigm (Review). Int. J. Mol. Med. 2011, 27, 159–171. [Google Scholar] [CrossRef][Green Version]
- Arismendi-Morillo, G. Electron Microscopy Morphology of the Mitochondrial Network in Gliomas and Their Vascular Microenvironment. Biochim. Biophys. Acta-Bioenerg. 2011, 1807, 602–608. [Google Scholar] [CrossRef]
- Giacomello, M.; Pellegrini, L. The Coming of Age of the Mitochondria-ER Contact: A Matter of Thickness. Cell Death Differ. 2016, 23, 1417–1427. [Google Scholar] [CrossRef]
- Gurel, P.S.; Hatch, A.L.; Higgs, H.N. Connecting the Cytoskeleton to the Endoplasmic Reticulum and Golgi. Curr. Biol. 2014, 24, 660–672. [Google Scholar] [CrossRef]
- Friedman, J.R.; Webster, B.M.; Mastronarde, D.N.; Verhey, K.J.; Voeltz, G.K. ER Sliding Dynamics and ER-Mitochondrial Contacts Occur on Acetylated Microtubules. J. Cell Biol. 2010, 190, 363–375. [Google Scholar] [CrossRef]
- Almeida, C.G.; Yamada, A.; Tenza, D.; Louvard, D.; Raposo, G.; Coudrier, E. Myosin 1b Promotes the Formation of Post-Golgi Carriers by Regulating Actin Assembly and Membrane Remodelling at the Trans-Golgi Network. Nat. Cell Biol. 2011, 13, 779–789. [Google Scholar] [CrossRef]
- Nogales, E.; Wang, H.W. Structural Intermediates in Microtubule Assembly and Disassembly: How and Why? Curr. Opin. Cell Biol. 2006, 18, 179–184. [Google Scholar] [CrossRef]
- Akhmanova, A.; Steinmetz, M.O. Tracking the Ends: A Dynamic Protein Network Controls the Fate of Microtubule Tips. Nat. Rev. Mol. Cell Biol. 2008, 9, 309–322. [Google Scholar] [CrossRef]
- Desai, A.; Mitchison, T.J. Microtubule Polymerization Dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef]
- Dumontet, C.; Jordan, M.A. Microtubule-Binding Agents: A Dynamic Field of Cancer Therapeutics. Nat. Rev. Drug Discov. 2010, 9, 790–803. [Google Scholar] [CrossRef]
- Liu, M.; Nadar, V.C.; Kozielski, F.; Kozlowska, M.; Yu, W.; Baas, P.W. Kinesin-12, a Mitotic Microtubule-Associated Motor Protein, Impacts Axonal Growth, Navigation, and Branching. J. Neurosci. 2010, 30, 14896–14906. [Google Scholar] [CrossRef]
- Chen, Y.; Sheng, Z.H. Kinesin-1-Syntaphilin Coupling Mediates Activity-Dependent Regulation of Axonal Mitochondrial Transport. J. Cell Biol. 2013, 202, 351–364. [Google Scholar] [CrossRef]
- Brickley, K.; Stephenson, F.A. Trafficking Kinesin Protein (TRAK)-Mediated Transport of Mitochondria in Axons of Hippocampal Neurons. J. Biol. Chem. 2011, 286, 18079–18092. [Google Scholar] [CrossRef]
- Wloga, D.; Joachimiak, E.; Louka, P.; Gaertig, J. Posttranslational Modifications of Tubulin and Cilia. Cold Spring Harb. Perspect. Biol. 2017, 9, a028159. [Google Scholar] [CrossRef]
- Caron, J.M. Posttranslational Modification of Tubulin by Palmitoylation: I. Mol. Biol. Cell. 1997, 8, 637–645. [Google Scholar] [CrossRef][Green Version]
- Cardanho-Ramos, C.; Faria-Pereira, A.; Morais, V.A. Orchestrating Mitochondria in Neurons: Cytoskeleton as the Conductor. Cytoskeleton 2020, 77, 65–75. [Google Scholar] [CrossRef]
- Kapitein, L.C.; Hoogenraad, C.C. Building the Neuronal Microtubule Cytoskeleton. Neuron 2015, 87, 492–506. [Google Scholar] [CrossRef]
- Nulty, J.; Alsaffar, M.; Barry, D. Radial Glial Cells Organize the Central Nervous System via Microtubule Dependant Processes. Brain Res. 2015, 1625, 171–179. [Google Scholar] [CrossRef]
- Bossing, T.; Barros, C.S.; Fischer, B.; Russell, S.; Shepherd, D. Disruption of Microtubule Integrity Initiates Mitosis during CNS Repair. Dev. Cell 2012, 23, 433–440. [Google Scholar] [CrossRef]
- Ren, W.; Zhao, W.; Cao, L.; Huang, J. Involvement of the Actin Machinery in Programmed Cell Death. Front. Cell Dev. Biol. 2021, 8, 1889. [Google Scholar] [CrossRef]
- Gentile, J.E.; Carrizales, M.G.; Koleske, A.J. Control of Synapse Structure and Function by Actin and Its Regulators. Cells 2022, 11, 603. [Google Scholar] [CrossRef]
- Illescas, M.; Peñas, A.; Arenas, J.; Martín, M.A.; Ugalde, C. Regulation of Mitochondrial Function by the Actin Cytoskeleton. Front. Cell Dev. Biol. 2021, 9, 795838. [Google Scholar] [CrossRef]
- Tokuraku, K.; Kuragano, M.; Uyeda, T.Q.P. Long-Range and Directional Allostery of Actin Filaments Plays Important Roles in Various Cellular Activities. Int. J. Mol. Sci. 2020, 21, 3209. [Google Scholar] [CrossRef]
- Yang, T.D.; Park, K.; Park, J.S.; Lee, J.H.; Choi, E.; Lee, J.; Choi, W.; Choi, Y.; Lee, K.J. Two Distinct Actin Waves Correlated with Turns-and-Runs of Crawling Microglia. PLoS ONE 2019, 14, e0220810. [Google Scholar] [CrossRef]
- Das, R.; Chinnathambi, S. Actin-Mediated Microglial Chemotaxis via G-Protein Coupled Purinergic Receptor in Alzheimer’s Disease. Neuroscience 2020, 448, 325–336. [Google Scholar] [CrossRef]
- Uhlemann, R.; Gertz, K.; Boehmerle, W.; Schwarz, T.; Nolte, C.; Freyer, D.; Kettenmann, H.; Endres, M.; Kronenberg, G. Actin Dynamics Shape Microglia Effector Functions. Brain Struct. Funct. 2016, 221, 2717–2734. [Google Scholar] [CrossRef]
- Kreplak, L.; Bär, H.; Leterrier, J.F.; Herrmann, H.; Aebi, U. Exploring the Mechanical Behavior of Single Intermediate Filaments. J. Mol. Biol. 2005, 354, 569–577. [Google Scholar] [CrossRef]
- Fan, C.; Shi, X.; Zhao, K.; Wang, L.; Shi, K.; Liu, Y.-J.; Li, H.; Ji, B.; Jiu, Y. Cell Migration Orchestrates Migrasome Formation by Shaping Retraction Fibers. J. Cell Biol. 2022, 221, e202109168. [Google Scholar] [CrossRef]
- Velez-Delvalle, C.; Marsch-Moreno, M.; Castro-Muñozledo, F.; Galván-Mendoza, I.J.; Kuri-Harcuch, W. Epithelial Cell Migration Requires the Interaction between the Vimentin and Keratin Intermediate Filaments. Sci. Rep. 2016, 6, 24389. [Google Scholar] [CrossRef]
- Maurer, M.; Lammerding, J. The Driving Force: Nuclear Mechanotransduction in Cellular Function, Fate, and Disease. Annu. Rev. Biomed. Eng. 2019, 21, 443–468. [Google Scholar] [CrossRef]
- Katsetos, C.D.; Anni, H.; Dráber, P. Mitochondrial Dysfunction in Gliomas. Semin. Pediatric Neurol. 2013, 20, 216–227. [Google Scholar] [CrossRef]
- Dráberová, E.; Lukáš, Z.; Ivanyi, D.; Viklický, V.; Dráber, P. Expression Class III β-Tubulin in Normal and Neoplastic Human Tissues. Histochem. Cell Biol. 1998, 109, 231–239. [Google Scholar] [CrossRef]
- Suzuki, S.O.; Kitai, R.; Llena, J.; Lee, S.C.; Goldman, J.E.; Shafit-Zagardo, B. MAP-2e, a Novel MAP-2 Isoform, Is Expressed in Gliomas and Delineates Tumor Architecture and Patterns of Infiltration. J. Neuropathol. Exp. Neurol. 2002, 61, 403–412. [Google Scholar] [CrossRef]
- Lim, E.J.; Suh, Y.; Yoo, K.C.; Lee, J.H.; Kim, I.G.; Kim, M.J.; Chang, J.H.; Kang, S.G.; Lee, S.J. Tumor-Associated Mesenchymal Stem-like Cells Provide Extracellular Signaling Cue for Invasiveness of Glioblastoma Cells. Oncotarget 2017, 8, 1438–1448. [Google Scholar] [CrossRef]
- Suzuki, S.O.; McKenney, R.J.; Mawatari, S.Y.; Mizuguchi, M.; Mikami, A.; Iwaki, T.; Goldman, J.E.; Canoll, P.; Vallee, R.B. Expression Patterns of LIS1, Dynein and Their Interaction Partners Dynactin, NudE, NudEL and NudC in Human Gliomas Suggest Roles in Invasion and Proliferation. Acta Neuropathol. 2007, 113, 591–599. [Google Scholar] [CrossRef]
- Xie, Z.; Janczyk, P.; Zhang, Y.; Liu, A.; Shi, X.; Singh, S.; Facemire, L.; Kubow, K.; Li, Z.; Jia, Y.; et al. A Cytoskeleton Regulator AVIL Drives Tumorigenesis in Glioblastoma. Nat. Commun. 2020, 11, 3457. [Google Scholar] [CrossRef]
- Kobielak, A.; Pasolli, H.A.; Fuchs, E. Mammalian Formin-1 Participates in Adherens Junctions and Polymerization of Linear Actin Cables. Nat. Cell Biol. 2004, 6, 21–30. [Google Scholar] [CrossRef]
- Monzo, P.; Crestani, M.; Chong, Y.K.; Ghisleni, A.; Hennig, K.; Li, Q.; Kakogiannos, N.; Giannotta, M.; Richichi, C.; Dini, T.; et al. Adaptive Mechanoproperties Mediated by the Formin FMN1 Characterize Glioblastoma Fitness for Invasion. Dev. Cell 2021, 56, 2841–2855.e8. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, H.; Hu, J.; Fu, H.; Qu, Y.; Yang, Y.; Cai, Q.; Efimov, A.; Wu, M.; Yen, T.; et al. Nestin Is Required for Spindle Assembly and Cell Cycle Progression in Glioblastoma Cells. Mol. Cancer Res. 2021, 19, 1651–1665. [Google Scholar] [CrossRef]
- Gatti, G.; Vilardo, L.; Musa, C.; di Pietro, C.; Bonaventura, F.; Scavizzi, F.; Torcinaro, A.; Bucci, B.; Saporito, R.; Arisi, I.; et al. Role of Lamin A/C as Candidate Biomarker of Aggressiveness and Tumorigenicity in Glioblastoma Multiforme. Biomedicines 2021, 9, 1343. [Google Scholar] [CrossRef]
- Mottaghitalab, F.; Lanjanian, H.; Masoudi-Nejad, A. Revealing Transcriptional and Post-Transcriptional Regulatory Mechanisms of γ-Glutamyl Transferase and Keratin Isoforms as Novel Cooperative Biomarkers in Low-Grade Glioma and Glioblastoma Multiforme. Genomics 2021, 113, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.M.; Judy, K.D.; Dunphy, I.; Jenkins, W.T.; Hwang, W.-T.; Nelson, P.T.; Lustig, R.A.; Jenkins, K.; Magarelli, D.P.; Hahn, S.M.; et al. Hypoxia Is Important in the Biology and Aggression of Human Glial Brain Tumors. Clin. Cancer Res. 2004, 10, 8177–8184. [Google Scholar] [CrossRef] [PubMed]
- Theos, A.C.; Martina, A.; Hurbain, I.; Peden, A.A.; Sviderskaya, E.V.; Stewart, A.; Robinson, M.S.; Bennett, D.C.; Cutler, D.F.; Bonifacino, J.S.; et al. Functions of Adaptor Protein (AP)-3 and AP-1 in Tyrosinase Sorting from Endosomes to Melanosomes. Mol. Biol. Cell 2005, 16, 5356–5372. [Google Scholar] [CrossRef] [PubMed]
- van Spronsen, M.; Mikhaylova, M.; Lipka, J.; Schlager, M.A.; van den Heuvel, D.J.; Kuijpers, M.; Wulf, P.S.; Keijzer, N.; Demmers, J.; Kapitein, L.C.; et al. TRAK/Milton Motor-Adaptor Proteins Steer Mitochondrial Trafficking to Axons and Dendrites. Neuron 2013, 77, 485–502. [Google Scholar] [CrossRef]
- Glater, E.E.; Megeath, L.J.; Stowers, R.S.; Schwarz, T.L. Axonal Transport of Mitochondria Requires Milton to Recruit Kinesin Heavy Chain and Is Light Chain Independent. J. Cell Biol. 2006, 173, 545–557. [Google Scholar] [CrossRef]
- Smirnova, E.; Griparic, L.; Shurland, D.-L.; van der Bliek, A.M. Drp1 Is Required for Mitochondrial Division in Mammalian Cells. Mol. Biol. Cell 2001, 12, 2245–2256. [Google Scholar] [CrossRef]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The Cell Biology of Mitochondrial Membrane Dynamics. Nat. Rev. Mol. Cell Biol. 2020, 21, 204–224. [Google Scholar] [CrossRef]
- Xie, Q.; Wu, Q.; Horbinski, C.M.; Flavahan, W.A.; Yang, K.; Zhou, W.; Dombrowski, S.M.; Huang, Z.; Fang, X.; Shi, Y.; et al. Mitochondrial Control by DRP1 in Brain Tumor Initiating Cells. Nat. Neurosci. 2015, 18, 501–510. [Google Scholar] [CrossRef]
- De Mario, A.; Tosatto, A.; Hill, J.M.; Kriston-Vizi, J.; Ketteler, R.; Vecellio Reane, D.; Cortopassi, G.; Szabadkai, G.; Rizzuto, R.; Mammucari, C. Identification and Functional Validation of FDA-Approved Positive and Negative Modulators of the Mitochondrial Calcium Uniporter. Cell Rep. 2021, 35, 109275. [Google Scholar] [CrossRef]
- Iranmanesh, Y.; Jiang, B.; Favour, O.C.; Dou, Z.; Wu, J.; Li, J.; Sun, C. Mitochondria’s Role in the Maintenance of Cancer Stem Cells in Glioblastoma. Front. Oncol. 2021, 11, 101. [Google Scholar] [CrossRef]
- Caino, M.C.; Ghosh, J.C.; Chae, Y.C.; Vaira, V.; Rivadeneira, D.B.; Faversani, A.; Rampini, P.; Kossenkov, A.V.; Aird, K.M.; Zhang, R.; et al. PI3K Therapy Reprograms Mitochondrial Trafficking to Fuel Tumor Cell Invasion. Proc. Natl. Acad. Sci. USA 2015, 112, 8638–8643. [Google Scholar] [CrossRef] [PubMed]
- Altieri, D.C. Mitochondrial Dynamics and Metastasis. Cell. Mol. Life Sci. 2019, 76, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Templeton, K.; Ramos, M.; Rose, J.; Le, B.; Zhou, Q.; Cressman, A.; Ferreyra, S.; Lebrilla, C.B.; Fierro, F.A. Mesenchymal Stromal Cells Regulate Sialylations of N-Glycans, Affecting Cell Migration and Survival. Int. J. Mol. Sci. 2021, 22, 6868. [Google Scholar] [CrossRef] [PubMed]
- De Mario, A.; Scarlatti, C.; Costiniti, V.; Primerano, S.; Lopreiato, R.; Calì, T.; Brini, M.; Giacomello, M.; Carafoli, E. Calcium Handling by Endoplasmic Reticulum and Mitochondria in a Cell Model of Huntington’s Disease. PLoS Curr. 2016, 8. [Google Scholar] [CrossRef]
- De Mario, A.; Quintana-Cabrera, R.; Martinvalet, D.; Giacomello, M. (Neuro)Degenerated Mitochondria-ER Contacts. Biochem. Biophys. Res. Commun. 2017, 483, 1096–1109. [Google Scholar] [CrossRef]
- Vance, J.E. Phospholipid Synthesis and Transport in Mammalian Cells. Traffic 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Cui, Z.; Vance, J.E.; Chen, M.H.; Voelker, D.R.; Vance, D.E. Cloning and Expression of a Novel Phosphatidylethanolamine N-Methyltransferase. A Specific Biochemical and Cytological Marker for a Unique Membrane Fraction in Rat Liver. J. Biol. Chem. 1993, 268, 16655–16663. [Google Scholar] [CrossRef]
- Galmes, R.; Houcine, A.; Vliet, A.R.; Agostinis, P.; Jackson, C.L.; Giordano, F. ORP5/ORP8 Localize to Endoplasmic Reticulum–Mitochondria Contacts and Are Involved in Mitochondrial Function. EMBO Rep. 2016, 17, 800–810. [Google Scholar] [CrossRef]
- Stone, S.J.; Levin, M.C.; Zhou, P.; Han, J.; Walther, T.C.; Farese, R.V. The Endoplasmic Reticulum Enzyme DGAT2 Is Found in Mitochondria-Associated Membranes and Has a Mitochondrial Targeting Signal That Promotes Its Association with Mitochondria. J. Biol. Chem. 2009, 284, 5352–5361. [Google Scholar] [CrossRef]
- Prasad, M.; Kaur, J.; Pawlak, K.J.; Bose, M.; Whittal, R.M.; Bose, H.S. Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Regulates Steroidogenic Activity via Steroidogenic Acute Regulatory Protein (StAR)-Voltage-Dependent Anion Channel 2 (VDAC2) Interaction. J. Biol. Chem. 2015, 290, 2604–2616. [Google Scholar] [CrossRef]
- Lewin, T.M.; Kim, J.H.; Granger, D.A.; Vance, J.E.; Coleman, R.A. Acyl-CoA Synthetase Isoforms 1, 4, and 5 Are Present in Different Subcellular Membranes in Rat Liver and Can Be Inhibited Independently. J. Biol. Chem. 2001, 276, 24674–24679. [Google Scholar] [CrossRef] [PubMed]
- Giacomello, M.; Drago, I.; Bortolozzi, M.; Scorzeto, M.; Gianelle, A.; Pizzo, P.; Pozzan, T. Ca2+ Hot Spots on the Mitochondrial Surface Are Generated by Ca2+ Mobilization from Stores, but Not by Activation of Store-Operated Ca2+ Channels. Mol. Cell 2010, 38, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, A.P.; Dal Bello, F.; Knedlik, T.; Volpin, F.; Shin, S.H.; Giacomello, M. Chemical modulation of Mitochondria-Endoplasmic Reticulum Contact Sites. Cells 2020, 9, 1637. [Google Scholar] [CrossRef] [PubMed]
- De Mario, A.; Peggion, C.; Massimino, M.L.; Viviani, F.; Castellani, A.; Giacomello, M.; Lim, D.; Bertoli, A.; Sorgato, M.C. The Prion Protein Regulates Glutamate-Mediated Ca2+ Entry and Mitochondrial Ca2+ Accumulation in Neurons. J. Cell Sci. 2017, 130, 2736–2746. [Google Scholar] [CrossRef]
- Csordás, G.; Várnai, P.; Golenár, T.; Roy, S.; Purkins, G.; Schneider, T.G.; Balla, T.; Hajnóczky, G. Imaging Interorganelle Contacts and Local Calcium Dynamics at the ER-Mitochondrial Interface. Mol. Cell 2010, 39, 121–132. [Google Scholar] [CrossRef]
- Szabadkai, G.; Bianchi, K.; Várnai, P.; de Stefani, D.; Wieckowski, M.R.; Cavagna, D.; Nagy, A.I.; Balla, T.; Rizzuto, R. Chaperone-Mediated Coupling of Endoplasmic Reticulum and Mitochondrial Ca2+ Channels. J. Cell Biol. 2006, 175, 901–911. [Google Scholar] [CrossRef]
- Vandecaetsbeek, I.; Vangheluwe, P.; Raeymaekers, L.; Wuytack, F.; Vanoevelen, J. The Ca2+ Pumps of the Endoplasmic Reticulum and Golgi Apparatus. Cold Spring Harb. Perspect. Biol. 2011, 3, a004184. [Google Scholar] [CrossRef]
- Das, S.; Fransson, Å.; Aspenstrom, P. Bidirectional Ca2+-Dependent Control of Mitochondrial Dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. USA 2008, 105, 20728–20733. [Google Scholar] [CrossRef]
- Berenguer-Escuder, C.; Grossmann, D.; Massart, F.; Antony, P.; Burbulla, L.F.; Glaab, E.; Imhoff, S.; Trinh, J.; Seibler, P.; Grünewald, A.; et al. Variants in Miro1 Cause Alterations of ER- Mitochondria Contact Sites in Fibroblasts from Parkinson’s Disease Patients. J. Clin. Med. 2019, 8, 2226. [Google Scholar] [CrossRef]
- Grossmann, D.; Berenguer-Escuder, C.; Bellet, M.E.; Scheibner, D.; Bohler, J.; Massart, F.; Rapaport, D.; Skupin, A.; Fouquier D’Hérouël, A.; Sharma, M.; et al. Mutations in RHOT1 Disrupt Endoplasmic Reticulum-Mitochondria Contact Sites Interfering with Calcium Homeostasis and Mitochondrial Dynamics in Parkinson’s Disease. Antioxid. Redox Signal. 2019, 31, 1213–1234. [Google Scholar] [CrossRef]
- Liu, S.; Sawada, T.; Lee, S.; Yu, W.; Silverio, G.; Alapatt, P.; Millan, I.; Shen, A.; Saxton, W.; Kanao, T.; et al. Parkinson’s Disease-Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria. PLoS Genet. 2012, 8, e1002537. [Google Scholar] [CrossRef] [PubMed]
- Birsa, N.; Norkett, R.; Wauer, T.; Mevissen, T.E.T.; Wu, H.C.; Foltynie, T.; Bhatia, K.; Hirst, W.D.; Komander, D.; Plun-Favreau, H.; et al. Lysine 27 Ubiquitination of the Mitochondrial Transport Protein Miro Is Dependent on Serine 65 of the Parkin Ubiquitin Ligase. J. Biol. Chem. 2014, 289, 14569–14582. [Google Scholar] [CrossRef]
- Giacomello, M.; Drago, I.; Pizzo, P.; Pozzan, T. Mitochondrial Ca2+ as a Key Regulator of Cell Life and Death. Cell Death Differ. 2007, 14, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Boehning, D.; Patterson, R.L.; Sedaghat, L.; Glebova, N.O.; Kurosaki, T.; Snyder, S.H. Cytochrome c Binds to Inositol (1,4,5) Trisphosphate Receptors, Amplifying Calcium-Dependent Apoptosis. Nat. Cell Biol. 2003, 5, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Pinton, P.; Giorgi, C.; Pandolfi, P.P. The Role of PML in the Control of Apoptotic Cell Fate: A New Key Player at ER-Mitochondria Sites. Cell Death Differ. 2011, 18, 1450–1456. [Google Scholar] [CrossRef]
- Molina, J.R.; Hayashi, Y.; Stephens, C.; Georgescu, M.M. Invasive Glioblastoma Cells Acquire Stemness and Increased Akt Activation. Neoplasia 2010, 12, 453–463. [Google Scholar] [CrossRef]
- Arismendi-Morillo, G.J.; Castellano-Ramirez, A.V. Ultrastructural Mitochondrial Pathology in Human Astrocytic Tumors: Potentials Implications pro-Therapeutics Strategies. J. Electron. Microsc. 2008, 57, 33–39. [Google Scholar] [CrossRef]
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Matsutani, T.; Masui, K.; Poulou, M.; Popescu, R.; della Donna, L.; Evers, P.; Dekmezian, C.; et al. Metabolic State of Glioma Stem Cells and Nontumorigenic Cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16062–16067. [Google Scholar] [CrossRef]
- Arismendi-Morillo, G.; Castellano-Ramírez, A.; Seyfried, T.N. Functional and Therapeutic Implications of Mitochondrial Network and Mitochondria-Associated Membranes: The Glioma’s Case. Glioma-Contemp. Diagn. Ther. Approaches 2019. [Google Scholar] [CrossRef]
- Moser, J.J.; Fritzler, M.J.; Rattner, J.B. Ultrastructural Characterization of Primary Cilia in Pathologically Characterized Human Glioblastoma Multiforme (GBM) Tumors. BMC Clin. Pathol. 2014, 14, 40. [Google Scholar] [CrossRef]
- Moser, J.J.; Fritzler, M.J.; Rattner, J.B. Primary Ciliogenesis Defects Are Associated with Human Astrocytoma/Glioblastoma Cells. BMC Cancer 2009, 9, 448. [Google Scholar] [CrossRef] [PubMed]
- Carré, M.; André, N.; Carles, G.; Borghi, H.; Brichese, L.; Briand, C.; Braguer, D.; Medicine, U.F.R.; La, U.O. Tubulin Is an Inherent Component of Mitochondrial Membranes That Interacts with the Voltage-Dependent Anion Channel. J. Biol. Chem. 2002, 277, 33664–33669. [Google Scholar] [CrossRef] [PubMed]
- Puurand, M.; Tepp, K.; Timohhina, N.; Aid, J.; Shevchuk, I.; Chekulayev, V.; Kaambre, T. Tubulin ΒII and ΒIII Isoforms as the Regulators of VDAC Channel Permeability in Health and Disease. Cells 2019, 8, 239. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, E.N.; Sheldon, K.L.; Dehart, D.N.; Patnaik, J.; Manevich, Y.; Townsend, D.M.; Bezrukov, S.M.; Rostovtseva, T.K.; Lemasters, J.J. Voltage-Dependent Anion Channels Modulate Mitochondrial Metabolism in Cancer Cells: Regulation by Free Tubulin and Erastin. J. Biol. Chem. 2013, 288, 11920–11929. [Google Scholar] [CrossRef] [PubMed]
- Strickland, M.; Stoll, E.A. Metabolic Reprogramming in Glioma. Front. Cell Dev. Biol. 2017, 5, 43. [Google Scholar] [CrossRef]
- Mucignat-Caretta, C.; Cavaggioni, A.; Redaelli, M.; Malatesta, M.; Zancanaro, C.; Caretta, A. Selective Distribution of Protein Kinase A Regulatory Subunit Rllα in Rodent Gliomas. Neuro-Oncology 2008, 10, 958–967. [Google Scholar] [CrossRef]
- Zhao, P.; Li, Q.; Shi, Z.; Li, C.; Wang, L.; Liu, X.; Jiang, C.; Qian, X.; You, Y.; Liu, N.; et al. GSK-3β Regulates Tumor Growth and Angiogenesis in Human Glioma Cells Theseauthorshavecontributedequallytothiswork. Oncotarget 2021, 6, 31901–31915. [Google Scholar] [CrossRef]
- Collet, B.; Guitton, N.; Saïkali, S.; Avril, T.; Pineau, C.; Hamlat, A.; Mosser, J.; Quillien, V. Differential Analysis of Glioblastoma Multiforme Proteome by a 2D-DIGE Approach. Proteome Sci. 2011, 9, 16. [Google Scholar] [CrossRef]
- Cui, Y.; Li, J.; Weng, L.; Wirbisky, S.E.; Freeman, J.L.; Liu, J.; Liu, Q.; Yuan, X.; Irudayaraj, J. Regulatory Landscape and Clinical Implication of MBD3 in Human Malignant Glioma. Oncotarget 2016, 7, 81698–81714. [Google Scholar] [CrossRef]
- Emig, D.; Salomonis, N.; Baumbach, J.; Lengauer, T.; Conklin, B.R.; Albrecht, M. AltAnalyze and DomainGraph: Analyzing and Visualizing Exon Expression Data. Nucleic Acids Res. 2010, 38, 755–762. [Google Scholar] [CrossRef]
- Alessio, E.; Buson, L.; Chemello, F.; Peggion, C.; Grespi, F.; Martini, P.; Massimino, M.L.; Pacchioni, B.; Millino, C.; Romualdi, C.; et al. Single Cell Analysis Reveals the Involvement of the Long Non-Coding RNA Pvt1 in the Modulation of Muscle Atrophy and Mitochondrial Network. Nucleic Acids Res. 2019, 47, 1653–1670. [Google Scholar] [CrossRef]
- Pagliari, M.; Munari, F.; Toffoletto, M.; Lonardi, S.; Chemello, F.; Codolo, G.; Millino, C.; Della Bella, C.; Pacchioni, B.; Vermi, W.; et al. Helicobacter Pylori Affects the Antigen Presentation Activity of Macrophages Modulating the Expression of the Immune Receptor CD300E through MiR-4270. Front. Immunol. 2017, 8, 1288. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Springer: Berlin/Heidelberg, Germany, 2010; pp. 267–277. ISBN 9781441957122. [Google Scholar]
- Mi, H.; Thomas, P. PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. Revigo Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, A.; Ericsson, M.; Bachi, T.; Griffiths, G.; Hauri, H.P. Characterization of a Novel 63 KDa Membrane Protein. Implications for the Organization of the ER-to-Golgi Pathway. J. Cell Sci. 1993, 104, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Sada, R.; Osugi, Y.; Matsumoto, S.; Matsuda, T.; Hayashi-Nishino, M.; Nagai, T.; Harada, A.; Kikuchi, A. Palmitoylated CKAP4 Regulates Mitochondrial Functions through an Interaction with VDAC2 at ER-Mitochondria Contact Sites. J. Cell Sci. 2020, 133, jcs249045. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, A.; Fumoto, K.; Kimura, H. The Dickkopf1-Cytoskeleton-Associated Protein 4 Axis Creates a Novel Signalling Pathway and May Represent a Molecular Target for Cancer Therapy. Br. J. Pharmacol. 2017, 174, 4651–4665. [Google Scholar] [CrossRef]
- Noda, C.; Kimura, H.; Arasaki, K.; Matsushita, M.; Yamamoto, A.; Wakana, Y.; Inoue, H.; Tagaya, M. Valosin-Containing Protein-Interacting Membrane Protein (VIMP) Links the Endoplasmic Reticulum with Microtubules in Concert with Cytoskeleton-Linking Membrane Protein (CLIMP)-63. J. Biol. Chem. 2014, 289, 24304–24313. [Google Scholar] [CrossRef]
- Kuznetsov, A.V.; Javadov, S.; Grimm, M.; Margreiter, R.; Ausserlechner, M.J.; Hagenbuchner, J. Crosstalk between Mitochondria and Cytoskeleton in Cardiac Cells. Cells 2020, 9, 222. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grespi, F.; Vianello, C.; Cagnin, S.; Giacomello, M.; De Mario, A. The Interplay of Microtubules with Mitochondria–ER Contact Sites (MERCs) in Glioblastoma. Biomolecules 2022, 12, 567. https://doi.org/10.3390/biom12040567
Grespi F, Vianello C, Cagnin S, Giacomello M, De Mario A. The Interplay of Microtubules with Mitochondria–ER Contact Sites (MERCs) in Glioblastoma. Biomolecules. 2022; 12(4):567. https://doi.org/10.3390/biom12040567
Chicago/Turabian StyleGrespi, Francesca, Caterina Vianello, Stefano Cagnin, Marta Giacomello, and Agnese De Mario. 2022. "The Interplay of Microtubules with Mitochondria–ER Contact Sites (MERCs) in Glioblastoma" Biomolecules 12, no. 4: 567. https://doi.org/10.3390/biom12040567
APA StyleGrespi, F., Vianello, C., Cagnin, S., Giacomello, M., & De Mario, A. (2022). The Interplay of Microtubules with Mitochondria–ER Contact Sites (MERCs) in Glioblastoma. Biomolecules, 12(4), 567. https://doi.org/10.3390/biom12040567






