Inhibiting Neddylation with MLN4924 Suppresses Growth and Delays Multicellular Development in Dictyostelium discoideum
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Media, Buffers, Chemicals, and Antibodies
2.2. Cell Proliferation Assay
2.3. Radial Bioassay of Chemotaxis
2.4. Aggregation Assay
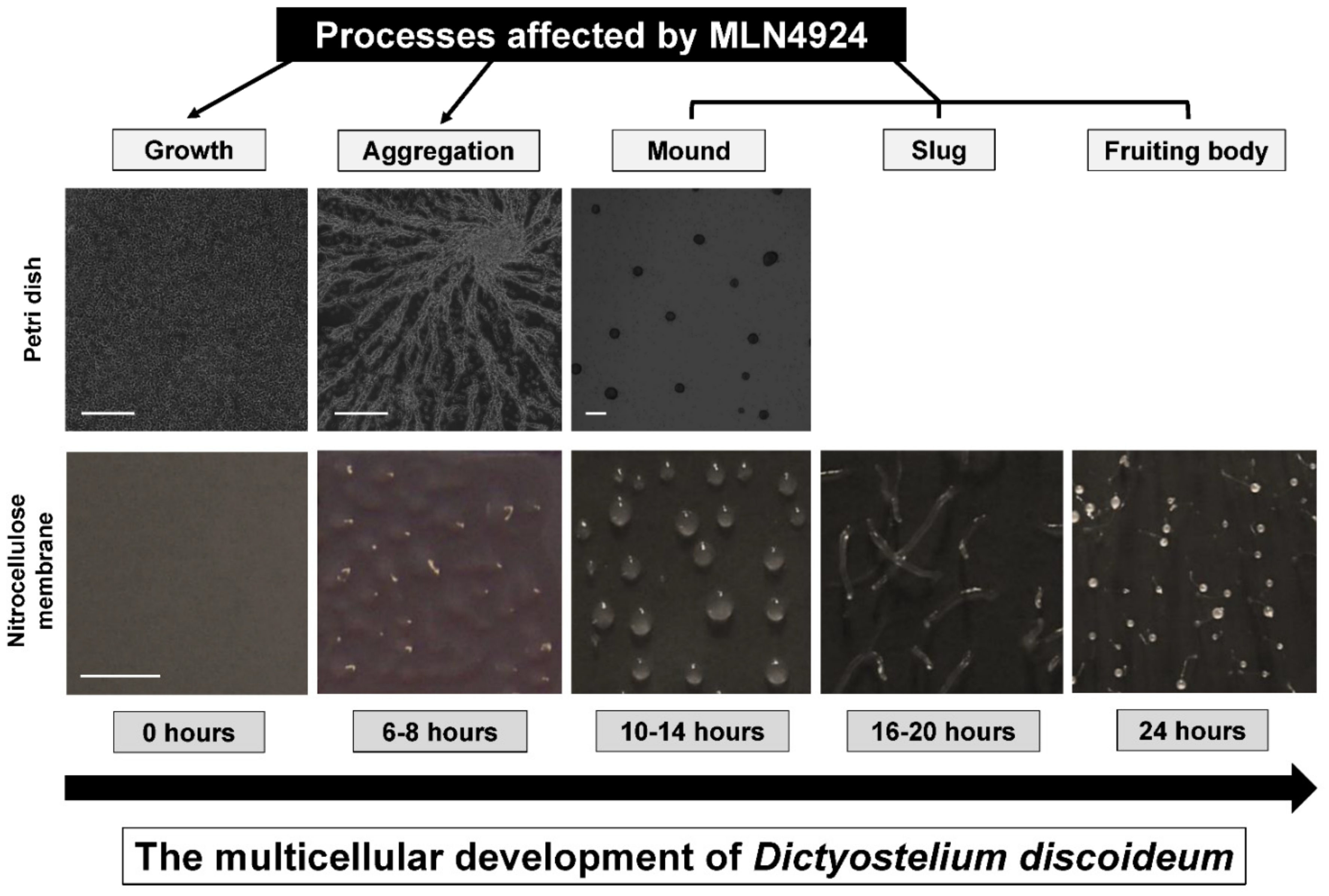
2.5. Multicellular Development Assay
2.6. SDS-PAGE and Western Blotting
2.7. Bioinformatics
3. Results
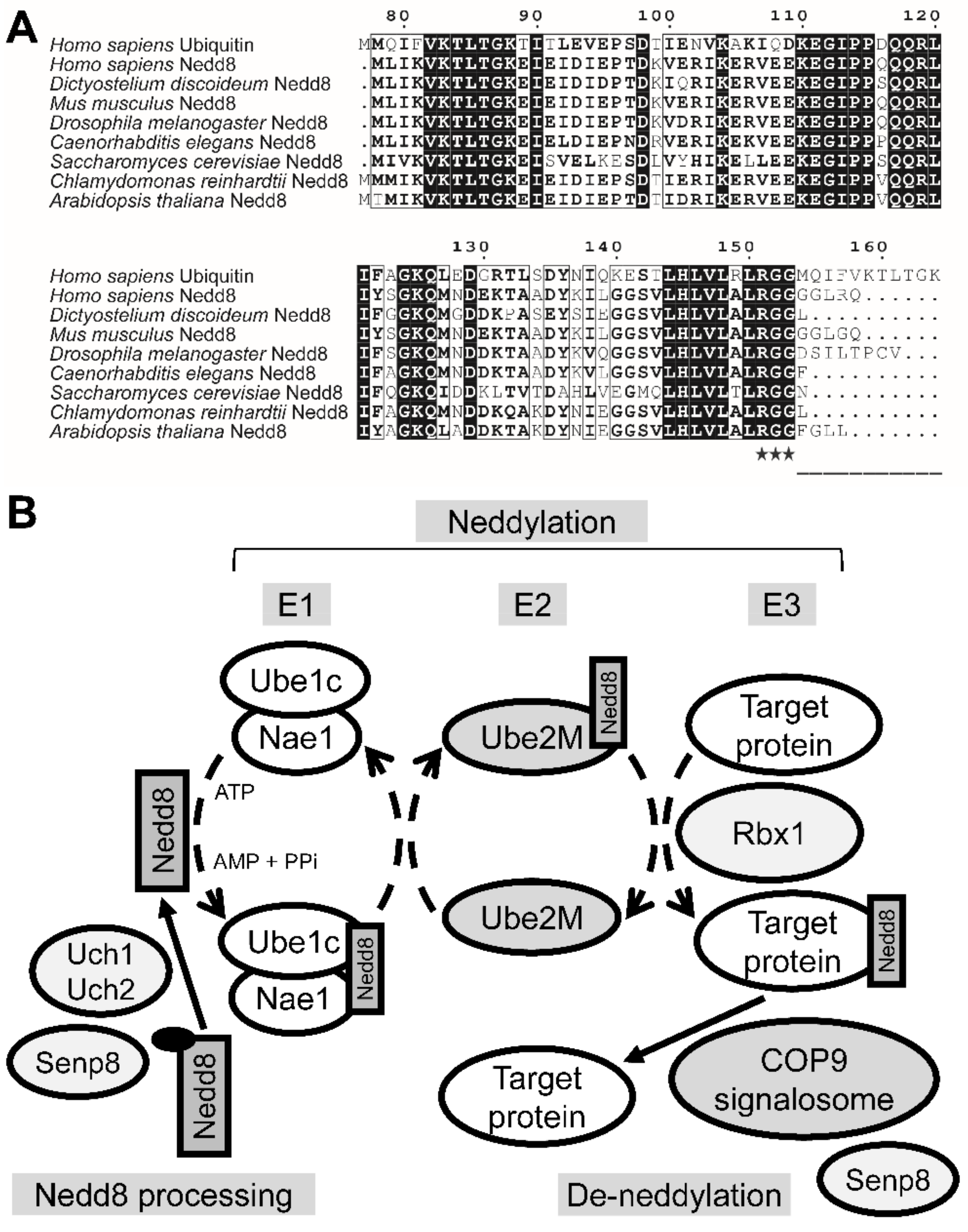
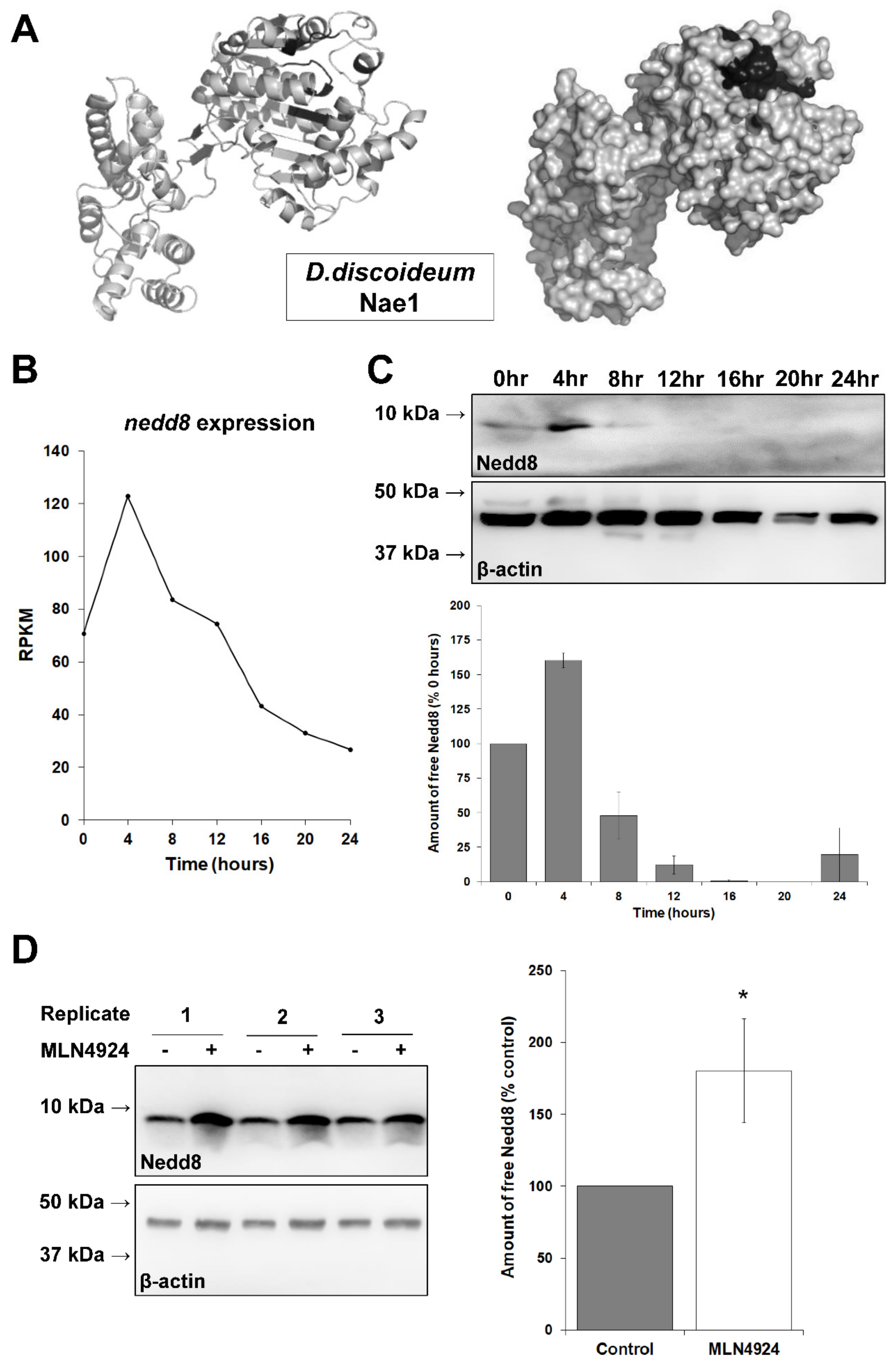
3.1. The Putative Neddylation Pathway in D. discoideum Resembles the Metazoan Neddylation Pathway
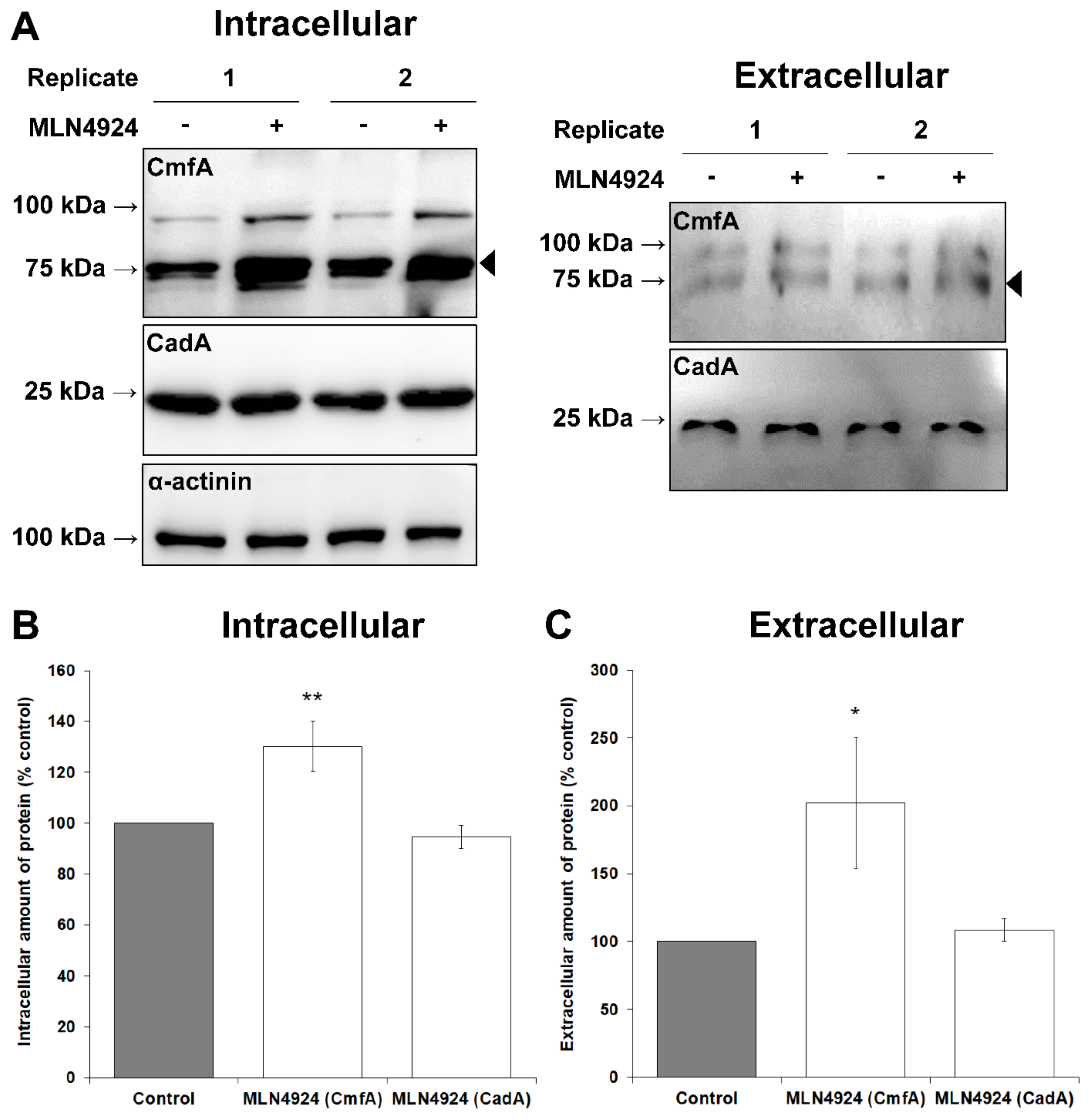
3.2. MLN4924 Inhibits Neddylation in D. discoideum
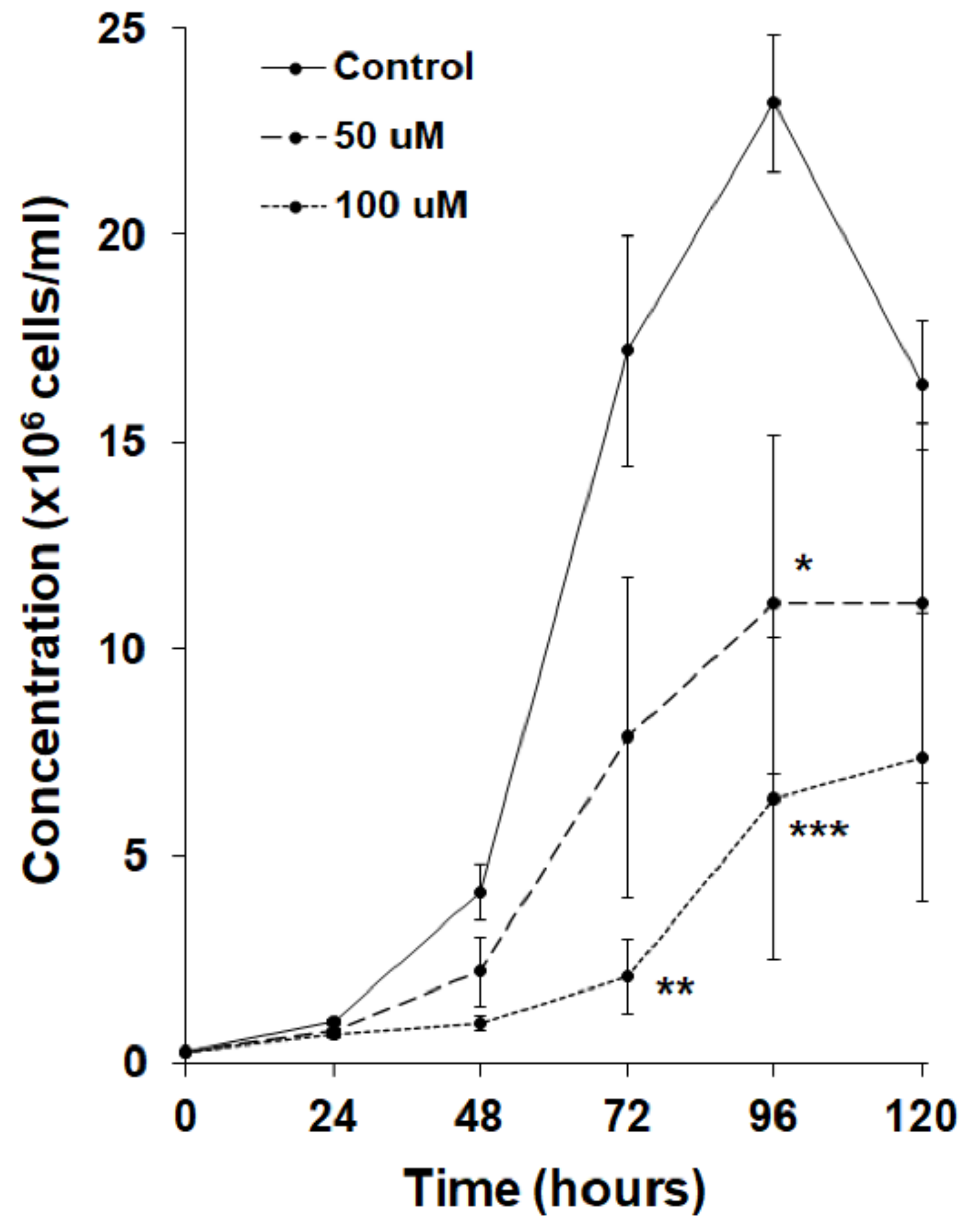
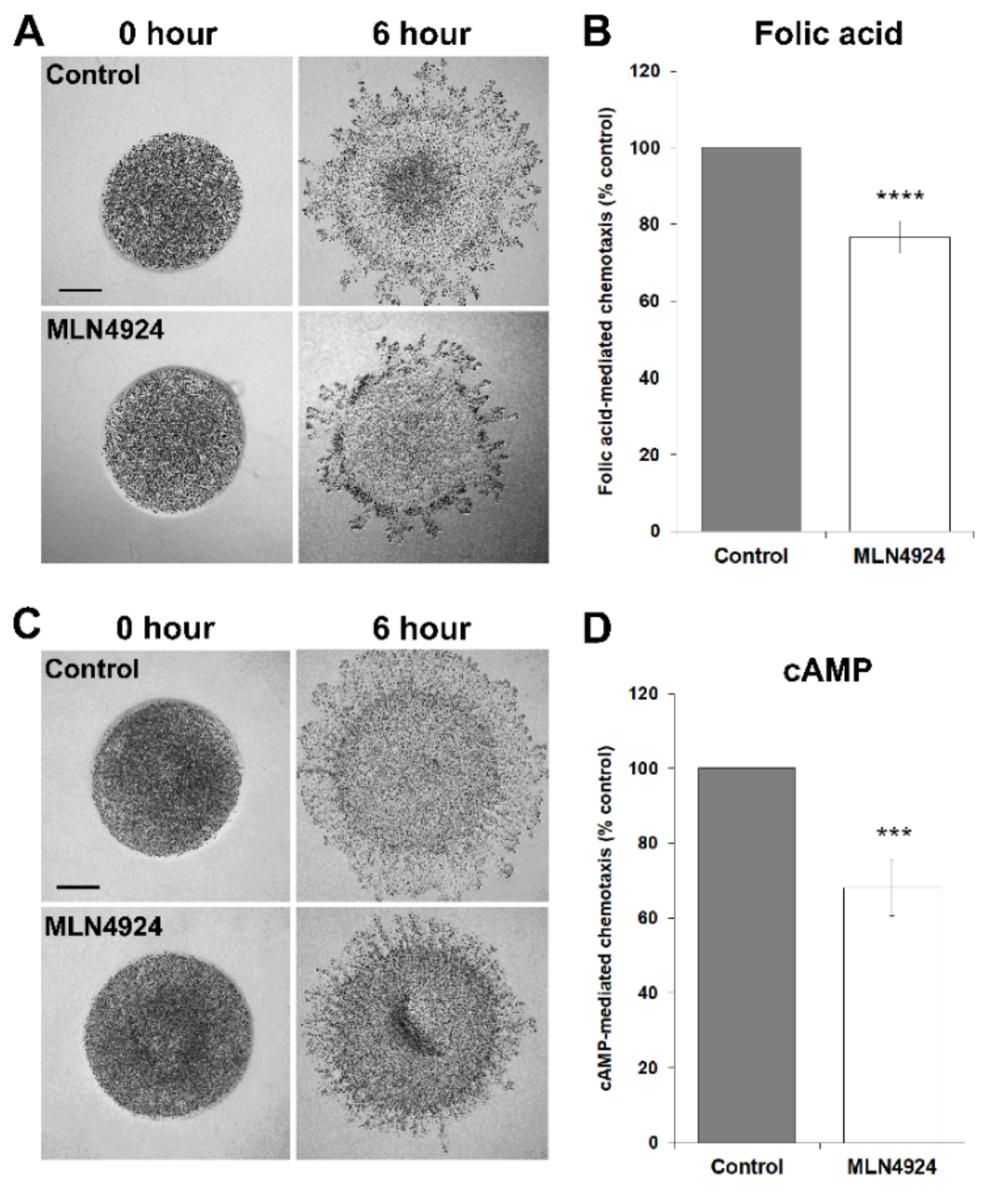
3.3. MLN4924 Inhibits Cell Proliferation and Folic Acid-Mediated Chemotaxis in D. discoideum
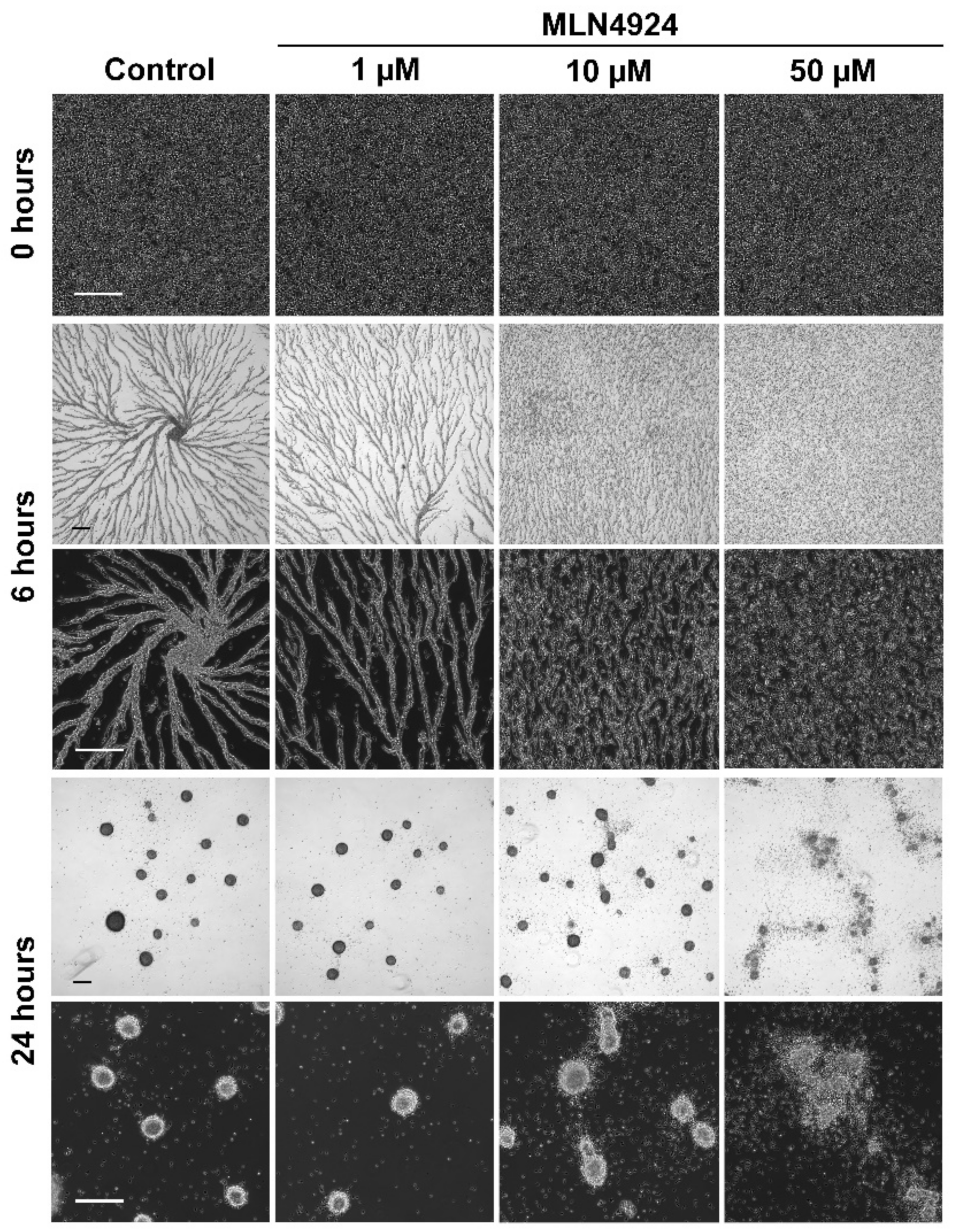
3.4. MLN4924 Treatment Inhibits cAMP-Mediated Chemotaxis and Delays the Aggregation of D. discoideum Cells
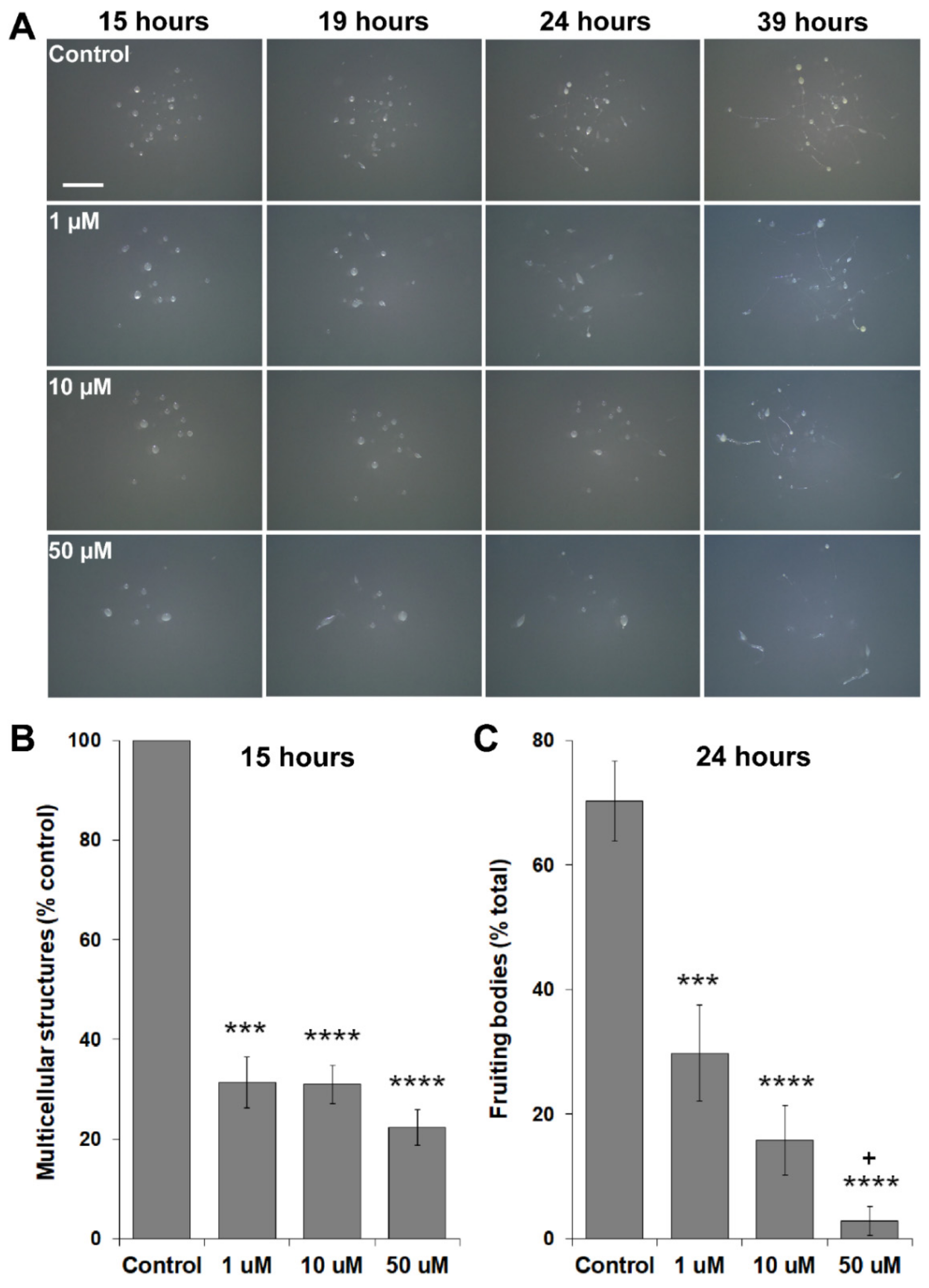
3.5. MLN4924 Treatment Inhibits Fruiting Body Formation during D. discoideum Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CadA | calcium-dependent cell adhesion molecule A |
| CAND1 | cullin-associated and neddylation dissociated 1 |
| CmfA | conditioned medium factor |
| COP9 | constitutive photomorphogenesis 9 |
| CRL | cullin–RING ligase |
| CSN | COP9 signalosome |
| CUL | cullin |
| DMSO | dimethyl sulfoxide |
| NAE1 | NEDD8 activating enzyme E1 |
| NEDD8 | neural precursor cell expressed, developmentally downregulated protein 8 |
| RBX1 | E3 ubiquitin protein ligase |
| SCF | Skp1–Cullin–F-box |
| SENP8 | sentrin-specific protease 8 |
| Ube1C | ubiquitin-activating enzyme E1C |
| Ube2M | ubiquitin-conjugating enzyme E2 M |
| UBE2M | NEDD8-conjugating enzyme Ubc12 |
| Uch1 | ubiquitin C-terminal hydrolase 1 |
| Uch2 | ubiquitin C-terminal hydrolase 2 |
| UCHL1 | ubiquitin carboxyl-terminal hydrolase isozyme L1 |
| UCHL3 | ubiquitin carboxyl-terminal hydrolase isozyme L3 |
| UCHL5 | ubiquitin carboxyl-terminal hydrolase isozyme L5 |
References
- Cappadocia, L.; Lima, C.D. Ubiquitin-like protein conjugation: Structures, chemistry, and mechanism. Chem. Rev. 2017, 118, 889–918. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 2012, 160, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Liakopoulos, D.; Doenges, G.; Matuschewski, K.; Jentsch, S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998, 17, 2208–2214. [Google Scholar] [CrossRef]
- Lammer, D.; Mathias, N.; Laplaza, J.M.; Jiang, W.; Liu, Y.; Callis, J.; Goebl, M.; Estelle, M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998, 12, 914–926. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, E.S.; Schulman, B.A.; Zheng, N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010, 20, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yang, X.; Harrell, J.M.; Ryzhikov, S.; Shim, E.-H.; Lykke-Andersen, K.; Wei, N.; Sun, H.; Kobayashi, R.; Zhang, H. CAND1 Binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 2002, 10, 1519–1526. [Google Scholar] [CrossRef]
- Dubiel, D.; Gierisch, M.E.; Huang, X.; Dubiel, W.; Naumann, M. CAND1-dependent control of cullin 1-RING Ub ligases is essential for adipogenesis. Biochim. Biophys. Acta BBA Mol. Cell Res. 2013, 1833, 1078–1084. [Google Scholar] [CrossRef]
- Liu, J.; Furukawa, M.; Matsumoto, T.; Xiong, Y. NEDD8 modification of CUL1 dissociates p120CAND1, an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 2002, 10, 1511–1518. [Google Scholar] [CrossRef]
- Girdwood, D.; Robertson, M.; Gordon, C. Constitutively active Cullin-RING-Ligases fail to rescue loss of NEDD8 conjugation in Schizosaccharomyces pombe. FEBS Lett. 2012, 586, 1522–1528. [Google Scholar] [CrossRef]
- Mergner, J.; Heinzlmeir, S.; Kuster, B.; Schwechheimer, C. DENEDDYLASE1 Deconjugates NEDD8 from non-cullin protein substrates in Arabidopsis thaliana. Plant. Cell 2015, 27, 741–753. [Google Scholar] [CrossRef][Green Version]
- Enchev, R.I.; Schulman, B.A.; Peter, M. Protein neddylation: Beyond cullin–RING ligases. Nat. Rev. Mol. Cell Biol. 2014, 16, 30–44. [Google Scholar] [CrossRef]
- Cope, G.A.; Deshaies, R.J. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 2003, 114, 663–671. [Google Scholar] [CrossRef]
- Bosu, D.R.; Kipreos, E.T. Cullin-RING ubiquitin ligases: Global regulation and activation cycles. Cell Div. 2008, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zemla, A.; Thomas, Y.; Kedziora, S.; Knebel, A.; Wood, N.T.; Rabut, G.; Kurz, T. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat. Commun. 2013, 4, 1641. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Dellaire, G. The Role of the COP9 signalosome and neddylation in DNA damage signaling and repair. Biomolecules 2015, 5, 2388–2416. [Google Scholar] [CrossRef]
- Gilberto, S.; Peter, M. Dynamic ubiquitin signaling in cell cycle regulation. J. Cell Biol. 2017, 216, 2259–2271. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Zhang, J. Diverse and pivotal roles of neddylation in metabolism and immunity. FEBS J. 2020. [Google Scholar] [CrossRef]
- Zhou, L.; Jia, L. Targeting protein neddylation for cancer therapy. In Cullin-RING Ligases and Protein Neddylation; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Berlin, Germany, 2020; Volume 1217, pp. 297–315. [Google Scholar] [CrossRef]
- He, X.; Zhu, A.; Feng, J.; Wang, X. Role of neddylation in neurological development and diseases. Biotechnol. Appl. Biochem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Osaka, F.; Saeki, M.; Katayama, S.; Aida, N.; Toh, E.A.; Kominami, K.; Toda, T.; Suzuki, T.; Chiba, T.; Tanaka, K.; et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000, 19, 3475–3484. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Omata, M.; Tanaka, K.; Chiba, T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 2001, 155, 571–580. [Google Scholar] [CrossRef]
- Pereira, R.V.; Gomes, M.D.S.; Olmo, R.P.; Souza, D.M.; Jannotti-Passos, L.K.; Baba, E.H.; Castro-Borges, W.; Guerra-Sá, R. NEDD8 conjugation in Schistosoma mansoni: Genome analysis and expression profiles. Parasitol. Int. 2013, 62, 199–207. [Google Scholar] [CrossRef][Green Version]
- Meister, C.; Thieme, K.G.; Thieme, S.; Köhler, A.M.; Schmitt, K.; Valerius, O.; Braus, G.H. COP9 signalosome interaction with UspA/Usp15 deubiquitinase controls VeA-mediated fungal multicellular development. Biomolecules 2019, 9, 238. [Google Scholar] [CrossRef]
- Mergner, J.; Kuster, B.; Schwechheimer, C. DENEDDYLASE1 protein counters automodification of neddylating enzymes to maintain NEDD8 protein homeostasis in Arabidopsis. J. Biol. Chem. 2017, 292, 3854–3865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jing, H.; Li, H.; Chen, W.; Luo, B.; Zhang, H.; Dong, Z.; Li, L.; Su, H.; Xiong, W.-C.; et al. Neddylation is critical to cortical development by regulating Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 26448–26459. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kwon, M.; Hwang, Y.; Yoon, J.; Park, S.; Kang, H.C. Stress-induced NEDDylation promotes cytosolic protein aggregation through HDAC6 in a p62-dependent manner. iScience 2021, 24, 102146. [Google Scholar] [CrossRef] [PubMed]
- Romeralo, M.; Escalante, R.; Baldauf, S.L. Evolution and diversity of Dictyostelid social amoebae. Protist 2012, 163, 327–343. [Google Scholar] [CrossRef] [PubMed]
- Mathavarajah, S.; Flores, A.; Huber, R.J. Dictyostelium discoideum: A model system for cell and developmental biology. Curr. Protoc. Essent. Lab. Tech. 2017, 15, 14.1.1–14.1.19. [Google Scholar] [CrossRef]
- Fey, P.; Dodson, R.J.; Basu, S.; Hartline, E.C.; Chisholm, R.L. DictyBase and the Dicty Stock Center (version 2.0)—A progress report. Int. J. Dev. Biol. 2019, 63, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.O.; Xu, Y.; van der Wel, H.; Walden, P.; Hartson, S.D.; West, C.M. Glycosylation of Skp1 promotes formation of Skp1–Cullin-1–F-box protein complexes in Dictyostelium. Mol. Cell. Proteom. 2015, 14, 66–80. [Google Scholar] [CrossRef]
- Rosel, D.; Kimmel, A.R. The COP9 signalosome regulates cell proliferation of Dictyostelium discoideum. Eur. J. Cell Biol. 2006, 85, 1023–1034. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Serino, G.; Callis, J.; Crosby, W.L.; Lyapina, S.; Deshaies, R.J.; Gray, W.M.; Estelle, M.; Deng, X.W. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 2001, 292, 1379–1382. [Google Scholar] [CrossRef]
- Busch, S.; Eckert, S.E.; Krappmann, S.; Braus, G.H. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2004, 49, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Pintard, L.; Kurz, T.; Glaser, S.; Willis, J.H.; Peter, M.; Bowerman, B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 2003, 13, 911–921. [Google Scholar] [CrossRef]
- Doronkin, S.; Djagaeva, I.; Beckendorf, S.K. The COP9 signalosome promotes degradation of cyclin e during early drosophila oogenesis. Dev. Cell 2003, 4, 699–710. [Google Scholar] [CrossRef]
- He, Q.; Cheng, P.; Liu, Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 2005, 19, 1518–1531. [Google Scholar] [CrossRef]
- Wee, S.; Geyer, R.K.; Toda, T.; Wolf, D.A. CSN facilitates Cullin–RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 2005, 7, 387–391. [Google Scholar] [CrossRef]
- Wu, J.-T.; Lin, H.-C.; Hu, Y.-C.; Chien, C.-T. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 2005, 7, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Cope, G.A.; Deshaies, R.J. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006, 7, 1. [Google Scholar] [CrossRef]
- Schmidt, M.W.; McQuary, P.R.; Wee, S.; Hofmann, K.; Wolf, D.A. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol. Cell 2009, 35, 586–597. [Google Scholar] [CrossRef]
- Barth, E.; Hübler, R.; Baniahmad, A.; Marz, M. The evolution of COP9 signalosome in unicellular and multicellular organisms. Genome Biol. Evol. 2016, 8, 1279–1289. [Google Scholar] [CrossRef]
- Heidel, A.J.; Lawal, H.M.; Felder, M.; Schilde, C.; Helps, N.R.; Tunggal, B.; Rivero, F.; John, U.; Schleicher, M.; Eichinger, L.; et al. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 2011, 21, 1882–1891. [Google Scholar] [CrossRef] [PubMed]
- Brownell, J.E.; Sintchak, M.D.; Gavin, J.M.; Liao, H.; Bruzzese, F.J.; Bump, N.J.; Soucy, T.A.; Milhollen, M.A.; Yang, X.; Burkhardt, A.L.; et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: The NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell 2010, 37, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Oladghaffari, M.; Islamian, J.P.; Baradaran, B.; Monfared, A.S. MLN4924 therapy as a novel approach in cancer treatment modalities. J. Chemother. 2016, 28. [Google Scholar] [CrossRef] [PubMed]
- Aubry, A.; Yu, T.; Bremner, R. Preclinical studies reveal MLN4924 is a promising new retinoblastoma therapy. Cell Death Discov. 2020, 6, 1–12. [Google Scholar] [CrossRef]
- Kim, K.; Pröbstel, A.-K.; Baumann, R.; Dyckow, J.; Landefeld, J.; Kogl, E.; Madireddy, L.; Loudermilk, R.; Eggers, E.L.; Singh, S.; et al. Cell type-specific transcriptomics identifies neddylation as a novel therapeutic target in multiple sclerosis. Brain 2020, 144, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Liang, X.; Liu, Y.; Zheng, M. Neddylation: A versatile pathway takes on chronic liver diseases. Front. Med. 2020, 7, 586881. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.; Espona-Fiedler, M.; Hamilton, C.; Holohan, C.; Crawford, N.; McIntyre, A.J.; Roberts, J.Z.; Wappett, M.; McDade, S.S.; Longley, D.B.; et al. Pevonedistat (MLN4924): Mechanism of cell death induction and therapeutic potential in colorectal cancer. Cell Death Discov. 2020, 6, 1–14. [Google Scholar] [CrossRef]
- Fey, P.; Kowal, A.S.; Gaudet, P.; Pilcher, K.E.; Chisholm, R.L. Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2007, 2, 1307–1316. [Google Scholar] [CrossRef]
- Jain, R.; Gomer, R. A developmentally regulated cell surface receptor for a density-sensing factor in Dictyostelium. J. Biol. Chem. 1994, 269, 9128–9136. [Google Scholar] [CrossRef]
- McLaren, M.D.; Mathavarajah, S.; Kim, W.D.; Yap, S.Q.; Huber, R.J. Aberrant autophagy impacts growth and multicellular development in a Dictyostelium knockout model of CLN5 disease. Front Cell Dev. Biol. 2021. under review. [Google Scholar]
- Schleicher, M.; Noegel, A.; Schwarz, T.; Wallraff, E.; Brink, M.; Faix, J.; Gerisch, G.; Isenberg, G. A Dictyostelium mutant with severe defects in alpha-actinin: Its characterization using cDNA probes and monoclonal antibodies. J. Cell Sci. 1988, 90, 59–71. [Google Scholar] [PubMed]
- Huber, R.J.; Myre, M.A.; Cotman, S.L. Loss of Cln3 function in the social amoeba Dictyostelium discoideum causes pleiotropic effects that are rescued by human CLN3. PLoS ONE 2014, 9, e110544. [Google Scholar] [CrossRef]
- Browning, D.D.; The, T.; O’Day, D.H. Comparative analysis of chemotaxis in Dictyostelium using a radial bioassay method: Protein tyrosine kinase activity is required for chemotaxis to folate but not to cAMP. Cell. Signal. 1995, 7, 481–489. [Google Scholar] [CrossRef]
- Huber, R.J.; Mathavarajah, S. Secretion and function of Cln5 during the early stages of Dictyostelium development. Biochim. Biophys. Acta BBA Mol. Cell Res. 2018, 1865, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Huber, R.J.; Myre, M.A.; Cotman, S.L. Aberrant adhesion impacts early development in a Dictyostelium model for juvenile neuronal ceroid lipofuscinosis. Cell Adhes. Migr. 2016, 11, 399–418. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nat. Cell Biol. 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Souphron, J.; Waddell, M.B.; Paydar, A.; Tokgoz-Gromley, Z.; Roussel, M.F.; Schulman, B.A. Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8’s E1. Biochemistry 2008, 47, 8961–8969. [Google Scholar] [CrossRef] [PubMed]
- Hotton, S.K.; Callis, J. Regulation of cullin RING ligases. Annu. Rev. Plant. Biol. 2008, 59, 467–489. [Google Scholar] [CrossRef] [PubMed]
- Wada, H.; Kito, K.; Caskey, L.S.; Yeh, E.T.; Kamitani, T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem. Biophys. Res. Commun. 1998, 251, 688–692. [Google Scholar] [CrossRef]
- Hakenjos, J.P.; Richter, R.; Dohmann, E.M.N.; Katsiarimpa, A.; Isono, E.; Schwechheimer, C. MLN4924 is an efficient inhibitor of NEDD8 conjugation in plants. Plant. Physiol. 2011, 156, 527–536. [Google Scholar] [CrossRef]
- Malhab, L.J.B.; Descamps, S.; Delaval, B.; Xirodimas, D.P. The use of the NEDD8 inhibitor MLN4924 (Pevonedistat) in a cyclotherapy approach to protect wild-type p53 cells from MLN4924 induced toxicity. Sci. Rep. 2016, 6, 37775. [Google Scholar] [CrossRef] [PubMed]
- Misra, M.; Kuhn, M.; Löbel, M.; An, H.; Statsyuk, A.V.; Sotriffer, C.; Schindelin, H. Dissecting the specificity of adenosyl sulfamate inhibitors targeting the ubiquitin-activating enzyme. Structure 2017, 25, 1120–1129. [Google Scholar] [CrossRef]
- Rot, G.; Parikh, A.; Curk, T.; Kuspa, A.; Shaulsky, G.; Zupan, B. DictyExpress: A Dictyostelium discoideum gene expression database with an explorative data analysis web-based interface. BMC Bioinform. 2009, 10, 265. [Google Scholar] [CrossRef]
- Echalier, A.; Pan, Y.; Birol, M.; Tavernier, N.; Pintard, L.; Hoh, F.; Ebel, C.; Galophe, N.; Claret, F.X.; Dumas, C. Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1. Proc. Natl. Acad. Sci. USA 2013, 110, 1273–1278. [Google Scholar] [CrossRef]
- Mohanty, S.; Lee, S.; Yadava, N.; Dealy, M.J.; Johnson, R.S.; Firtel, R.A. Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 2001, 15, 1435–1448. [Google Scholar] [CrossRef]
- Yuen, I.S.; Taphouse, C.; Halfant, K.A.; Gomer, R.H. Regulation and processing of a secreted protein that mediates sensing of cell density in Dictyostelium. Development 1991, 113, 1375–1385. [Google Scholar]
- Brar, S.K.; Siu, C.H. Characterization of the cell adhesion molecule gp24 in Dictyostelium discoideum. Mediation of cell-cell adhesion via a Ca2+-dependent mechanism. J. Biol. Chem. 1993, 268, 24902–24909. [Google Scholar] [CrossRef]
- Wang, B.; Kuspa, A. CulB, a putative ubiquitin ligase subunit, regulates prestalk cell differentiation and morphogenesis in Dictyostelium spp. Eukaryot. Cell 2002, 1, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Kin, K.; Forbes, G.; Cassidy, A.; Schaap, P. Cell-type specific RNA-Seq reveals novel roles and regulatory programs for terminally differentiated Dictyostelium cells. BMC Genom. 2018, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Li, H.; Sun, Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia 2011, 13, 561–569. [Google Scholar] [CrossRef]
- Chen, Y.; Du, M.; Yusuying, S.; Liu, W.; Tan, Y.; Xie, P. Nedd8-activating enzyme inhibitor MLN4924 (Pevonedistat), inhibits miR-1303 to suppress human breast cancer cell proliferation via targeting p27Kip1. Exp. Cell Res. 2020, 392, 112038. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Han, S.; Wilder-Romans, K.; Sun, G.Y.; Zhu, H.; Liu, X.; Tan, M.; Wang, G.; Feng, F.Y.; Sun, Y. Neddylation inactivation represses androgen receptor transcription and inhibits growth, survival and invasion of prostate cancer cells. Neoplasia 2020, 22, 192–202. [Google Scholar] [CrossRef]
- Liao, S.; Hu, H.; Wang, T.; Tu, X.; Li, Z. The protein neddylation pathway in Trypanosoma brucei: Functional characterization and substrate identification. J. Biol. Chem. 2017, 292, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Kuo, K.-L.; Ho, I.-L.; Shi, C.-S.; Wu, J.-T.; Lin, W.-C.; Tsai, Y.-C.; Chang, H.-C.; Chou, C.-T.; Hsu, C.-H.; Hsieh, J.-T.; et al. MLN4924, a novel protein neddylation inhibitor, suppresses proliferation and migration of human urothelial carcinoma: In vitro and in vivo studies. Cancer Lett. 2015, 363, 127–136. [Google Scholar] [CrossRef]
- Lan, H.; Tang, Z.; Jin, H.; Sun, Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci. Rep. 2016, 6, 24218. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Si, Y.; Yu, H.; Zhang, L.; Xie, P.; Jiang, W. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Sobko, A.; Ma, H.; Firtel, R.A. Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev. Cell 2002, 2, 745–756. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huber, R.J.; Kim, W.D.; Mathavarajah, S. Inhibiting Neddylation with MLN4924 Suppresses Growth and Delays Multicellular Development in Dictyostelium discoideum. Biomolecules 2021, 11, 482. https://doi.org/10.3390/biom11030482
Huber RJ, Kim WD, Mathavarajah S. Inhibiting Neddylation with MLN4924 Suppresses Growth and Delays Multicellular Development in Dictyostelium discoideum. Biomolecules. 2021; 11(3):482. https://doi.org/10.3390/biom11030482
Chicago/Turabian StyleHuber, Robert J., William D. Kim, and Sabateeshan Mathavarajah. 2021. "Inhibiting Neddylation with MLN4924 Suppresses Growth and Delays Multicellular Development in Dictyostelium discoideum" Biomolecules 11, no. 3: 482. https://doi.org/10.3390/biom11030482
APA StyleHuber, R. J., Kim, W. D., & Mathavarajah, S. (2021). Inhibiting Neddylation with MLN4924 Suppresses Growth and Delays Multicellular Development in Dictyostelium discoideum. Biomolecules, 11(3), 482. https://doi.org/10.3390/biom11030482





