RNA-seq Characterization of Melanoma Phenotype Switch in 3D Collagen after p38 MAPK Inhibitor Treatment
Abstract
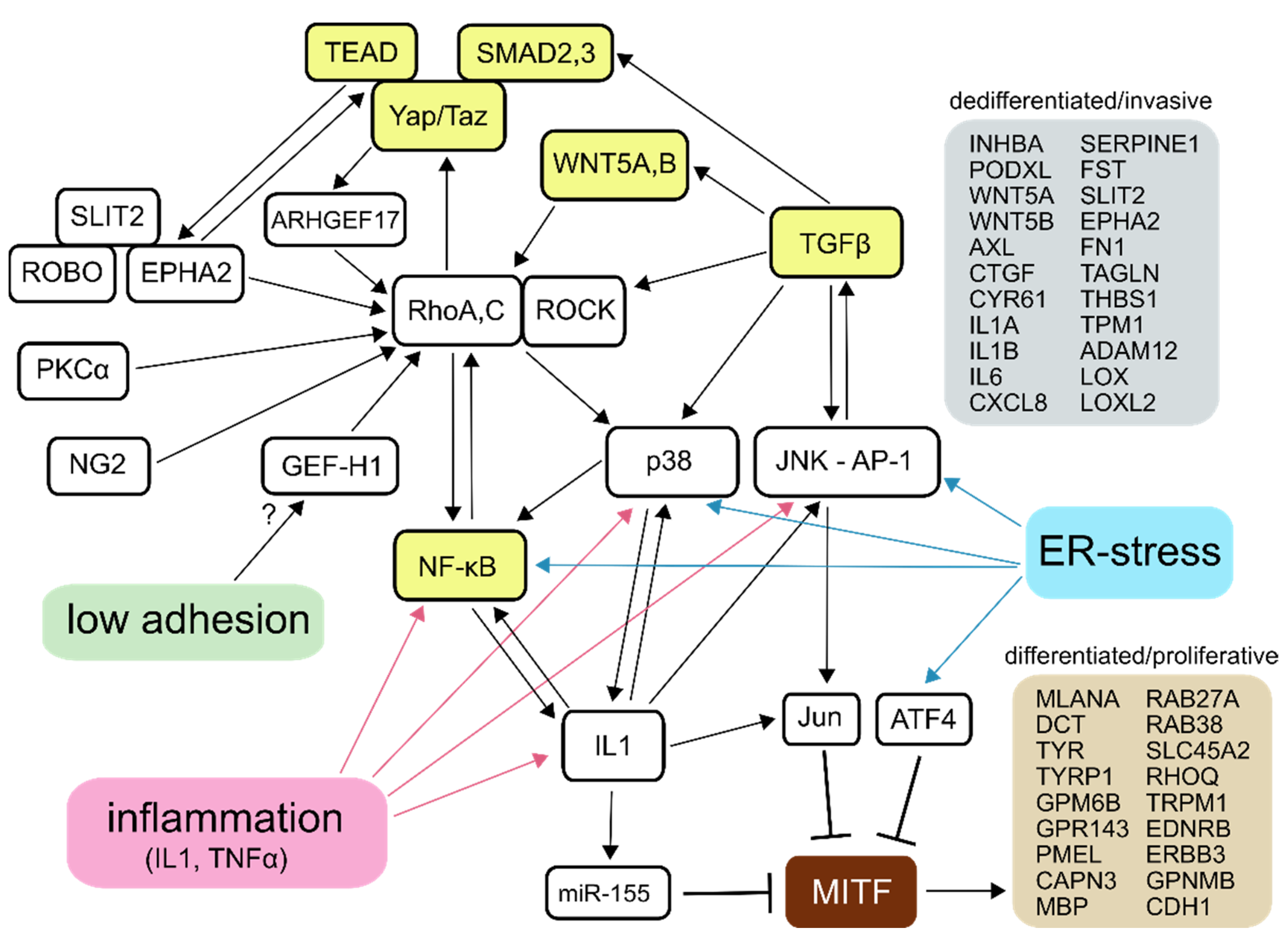
1. Introduction
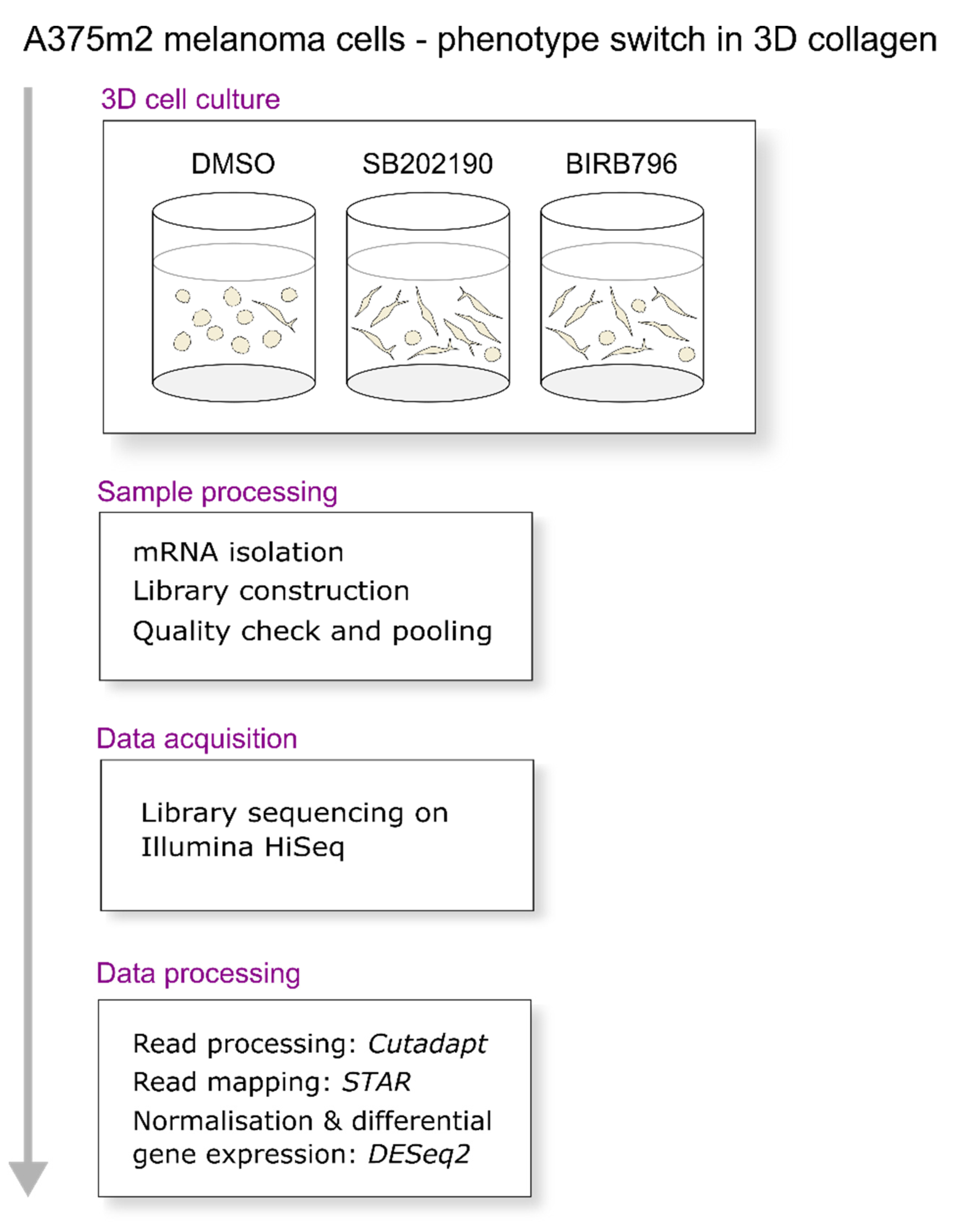
2. Materials and Methods
2.1. Cells Culture, Treatments and Morphological Analysis
2.2. RNA Extraction and Sequencing
2.3. Reverse Transcription–Quantitative Polymerase Chain Reaction (RT-qPCR)
2.4. Statistical Analysis
3. Results
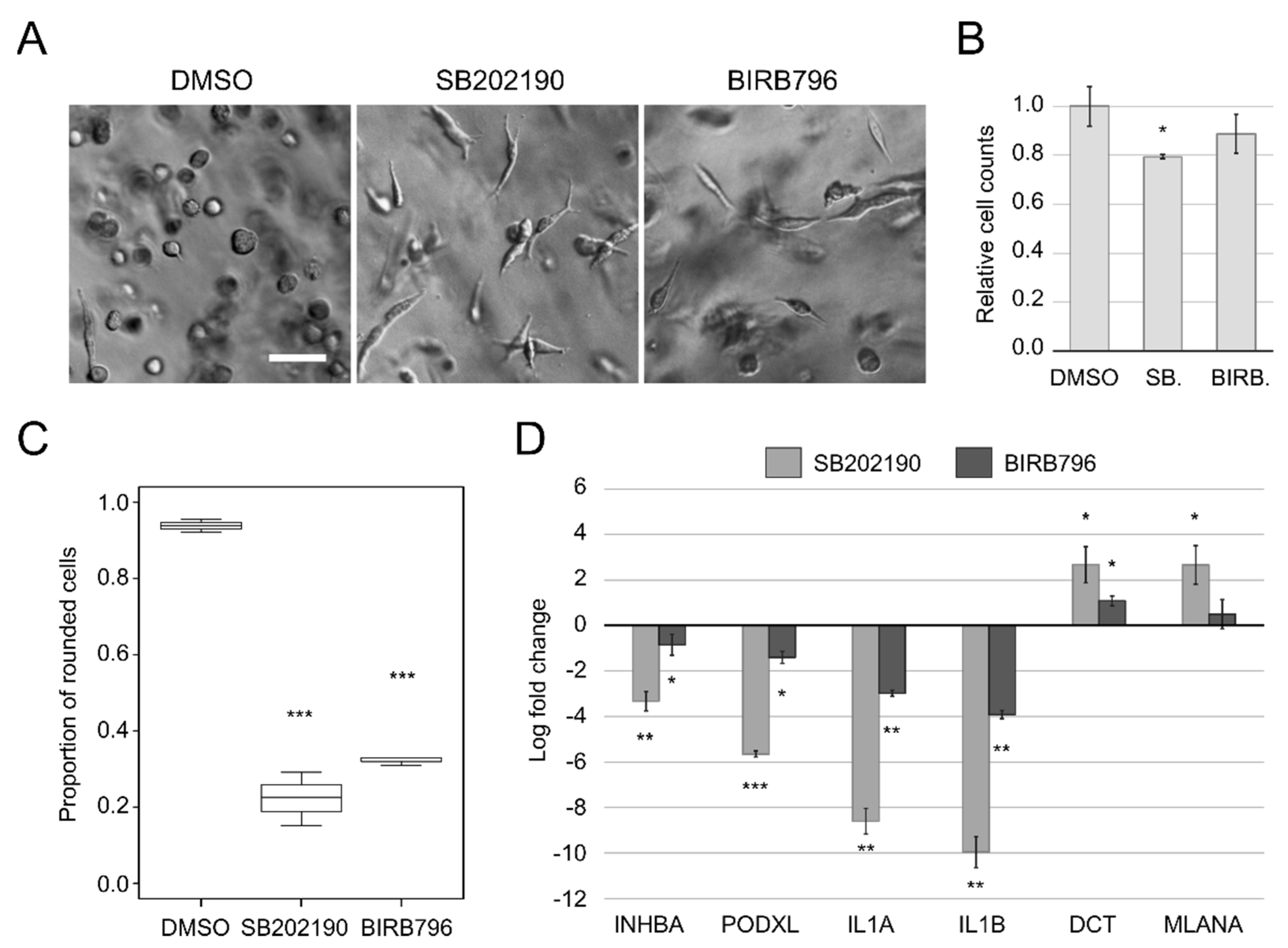
3.1. SB202190 and BIRB796 Induce a Phenotype Switch in A375M2 Cells Cultured in 3D Collagen
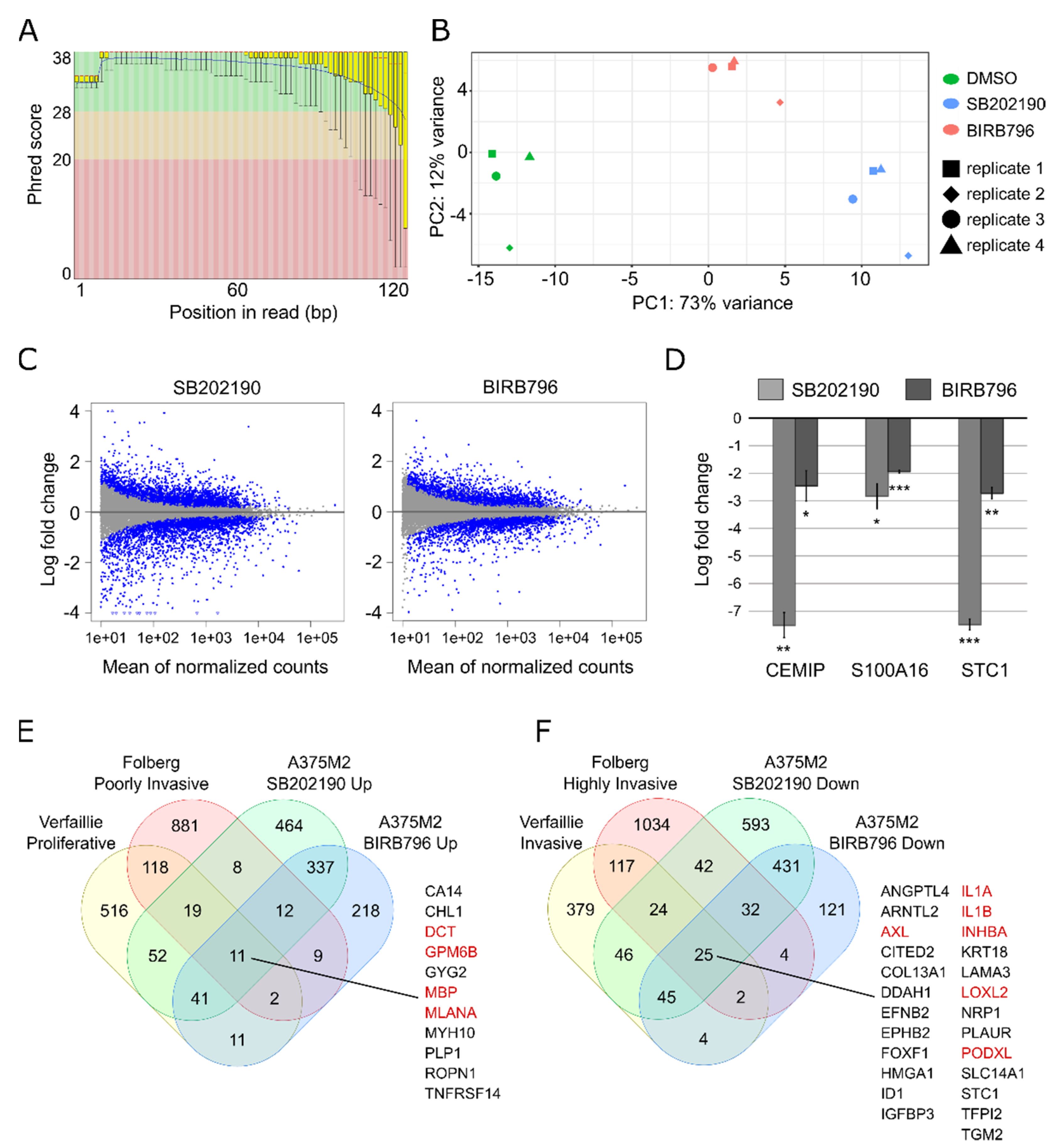
3.2. Transcriptomic Profiling of the Phenotype Switch-Associated Changes with RNA-seq
3.3. Validation and Reproducibility of the RNA-seq Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rambow, F.; Marine, J.-C.; Goding, C.R. Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef]
- Goding, C.R.; Arnheiter, H. Mitf—the First 25 Years. Genes Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.L.; Czyz, M. MITF in melanoma: Mechanisms behind its expression and activity. Cell. Mol. Life Sci. 2015, 72, 1249–1260. [Google Scholar] [CrossRef]
- Kim, J.-H.; Hong, A.-R.; Kim, Y.-H.; Yoo, H.; Kang, S.-W.; Chang, S.E.; Song, Y. JNK suppresses melanogenesis by interfering with CREB-regulated transcription coactivator 3-dependent MITF expression. Theranostics 2020, 10, 4017–4029. [Google Scholar] [CrossRef]
- Falletta, P.; Sanchez-Del-Campo, L.; Chauhan, J.; Effern, M.; Kenyon, A.; Kershaw, C.J.; Siddaway, R.; Lisle, R.J.; Freter, R.; Daniels, M.J.; et al. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 2017, 31, 18–33. [Google Scholar] [CrossRef]
- Buscà, R.; Bertolotto, C.; Abbe, P.; Englaro, W.; Ishizaki, T.; Narumiya, S.; Boquet, P.; Ortonne, J.P.; Ballotti, R. Inhibition of rho is required for CAMP-induced melanoma cell differentiation. Mol. Biol. Cell 1998, 9, 1367–1378. [Google Scholar] [CrossRef]
- Hoek, K.S.; Schlegel, N.C.; Brafford, P.; Sucker, A.; Ugurel, S.; Kumar, R.; Weber, B.L.; Nathanson, K.L.; Phillips, D.J.; Herlyn, M.; et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment. Cell Res. 2006, 19, 290–302. [Google Scholar] [CrossRef]
- Vivas-García, Y.; Falletta, P.; Liebing, J.; Louphrasitthiphol, P.; Feng, Y.; Chauhan, J.; Scott, D.A.; Glodde, N.; Chocarro-Calvo, A.; Bonham, S.; et al. Lineage-restricted regulation of SCD and fatty acid saturation by MITF controls melanoma phenotypic plasticity. Mol. Cell 2020, 77, 120–137.e9. [Google Scholar] [CrossRef]
- Misek, S.A.; Appleton, K.M.; Dexheimer, T.S.; Lisabeth, E.M.; Lo, R.S.; Larsen, S.D.; Gallo, K.A.; Neubig, R.R. Rho-mediated signaling promotes BRAF inhibitor resistance in de-differentiated melanoma cells. Oncogene 2020, 39, 1466–1483. [Google Scholar] [CrossRef] [PubMed]
- Konieczkowski, D.J.; Johannessen, C.M.; Abudayyeh, O.; Kim, J.W.; Cooper, Z.A.; Piris, A.; Frederick, D.T.; Barzily-Rokni, M.; Straussman, R.; Haq, R.; et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014, 4, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Verfaillie, A.; Imrichova, H.; Atak, Z.K.; Dewaele, M.; Rambow, F.; Hulselmans, G.; Christiaens, V.; Svetlichnyy, D.; Luciani, F.; Van Der Mooter, L.L.; et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 2015, 6, 6683. [Google Scholar] [CrossRef]
- Kholmanskikh, O.; Van Baren, N.; Brasseur, F.; Ottaviani, S.; Vanacker, J.; Arts, N.; Van Der Bruggen, P.; Coulie, P.; De Plaen, E. Interleukins 1α and 1β secreted by some melanoma cell lines strongly reduce expression of MITF-M and melanocyte differentiation antigens. Int. J. Cancer 2010, 127, 1625–1636. [Google Scholar] [CrossRef]
- Miskolczi, Z.; Smith, M.P.; Rowling, E.J.; Ferguson, J.; Barriuso, J.; Wellbrock, C. Collagen abundance controls melanoma phenotypes through lineage-specific microenvironment sensing. Oncogene 2018, 37, 3166–3182. [Google Scholar] [CrossRef]
- Strub, T.; Kobi, D.; Koludrovic, D.; Davidson, I. Research on Melanoma—A Glimpse into Current Directions and Future Trends; Murph, M., Ed.; IntechOpen: London, UK, 2011. [Google Scholar]
- Thurber, A.; Douglas, G.; Sturm, E.; Zabierowski, S.; Smit, D.; Ramakrishnan, S.; Hacker, E.; Leonard, J.; Herlyn, M.; Sturm, R. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene 2011, 30, 3036–3048. [Google Scholar] [CrossRef]
- O’Connell, M.P.; Marchbank, K.; Webster, M.R.; Valiga, A.A.; Kaur, A.; Vultur, A.; Li, L.; Herlyn, M.; Villanueva, J.; Liu, Q.; et al. Hypoxia induces phenotypic plasticity and therapy resistance in melanoma via the tyrosine kinase receptors ROR1 and ROR2. Cancer Discov. 2013, 3, 1378–1393. [Google Scholar] [CrossRef] [PubMed]
- Landsberg, J.; Kohlmeyer, J.; Renn, M.; Bald, T.; Rogava, M.; Cron, M.; Fatho, M.; Lennerz, V.; Wölfel, T.; Hölzel, M.; et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 2012, 490, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Kim, Y.J.; Robert, L.; Tsoi, J.; Comin-Anduix, B.; Berent-Maoz, B.; Cochran, A.J.; Economou, J.S.; Tumeh, P.C.; Puig-Saus, C.; et al. Immunotherapy resistance by inflammation-induced dedifferentiation. Cancer Discov. 2018, 8, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Krijgsman, O.; Tsoi, J.; Robert, L.; Hugo, W.; Song, C.; Kong, X.; Possik, P.A.; Cornelissen-Steijger, P.D.M.; Foppen, M.H.G.; et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014, 5, 5712. [Google Scholar] [CrossRef]
- Kozar, I.; Margue, C.; Rothengatter, S.; Haan, C.; Kreis, S. Many ways to resistance: How melanoma cells evade targeted therapies. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 313–322. [Google Scholar] [CrossRef]
- Arozarena, I.; Wellbrock, C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 2019, 19, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.H. Non-smad TGF-β signals. J. Cell Sci. 2005, 118, 3573–3584. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.-H. Dynamic control of TGF-β signaling and its links to the cytoskeleton. FEBS Lett. 2008, 582, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-G.; Islam, R.; Cho, J.Y.; Jeong, H.; Cap, K.-C.; Park, Y.; Hossain, A.J.; Park, J.-B. Regulation of RhoA GTPase and various transcription factors in the RhoA pathway. J. Cell. Physiol. 2018, 233, 6381–6392. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yao, E.; Zhang, K.; Jiang, X.; Croll, S.; Thompson-Peer, K.; Chuang, P.-T. YAP is essential for mechanical force production and epithelial cell proliferation during lung branching morphogenesis. eLife 2017, 6, e21130. [Google Scholar] [CrossRef]
- Edwards, D.N.; Ngwa, V.M.; Wang, S.; Shiuan, E.; Brantley-Sieders, D.M.; Kim, L.C.; Reynolds, A.B.; Chen, J. The receptor tyrosine kinase EphA2 promotes glutamine metabolism in tumors by activating the transcriptional coactivators YAP and TAZ. Sci. Signal. 2017, 10, eaan4667. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Pang, K.M.; Iida, M.; Nelson, M.S.; Liu, J.; Nam, A.; Wang, J.; Mambetsariev, I.; Pillai, R.; Mohanty, A.; et al. Activation of EPHA2-ROBO1 heterodimer by SLIT2 attenuates non-canonical signaling and proliferation in squamous cell carcinomas. iScience 2020, 23, 101692. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Kanneganti, T.-D. Function and regulation of IL-1α in inflammatory diseases and cancer. Immunol. Rev. 2018, 281, 124–137. [Google Scholar] [CrossRef]
- Paňková, D.; Jobe, N.; Kratochvílová, M.; Buccione, R.; Brábek, J.; Rösel, D. NG2-mediated Rho activation promotes amoeboid invasiveness of cancer cells. Eur. J. Cell Biol. 2012, 91, 969–977. [Google Scholar] [CrossRef]
- Vaškovičová, K.; Szabadosová, E.; Čermák, V.; Gandalovičová, A.; Kasalová, L.; Rösel, D.; Brábek, J. PKCα promotes the mesenchymal to amoeboid transition and increases cancer cell invasiveness. BMC Cancer 2015, 15, 326. [Google Scholar] [CrossRef]
- Dovas, A.; Yoneda, A.; Couchman, J.R. PKC-α-dependent activation of RhoA by syndecan-4 during focal adhension formation. J. Cell Sci. 2006, 119, 2837–2846. [Google Scholar] [CrossRef]
- Riesenberg, S.; Groetchen, A.; Siddaway, R.; Bald, T.; Reinhardt, J.; Smorra, D.; Kohlmeyer, J.; Renn, M.; Phung, B.; Aymans, P.; et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat. Commun. 2015, 6, 8755. [Google Scholar] [CrossRef]
- Arts, N.; Cané, S.; Hennequart, M.; Lamy, J.; Bommer, G.; Van Den Eynde, B.; De Plaen, E. microRNA-155, Induced by interleukin-1ß, represses the expression of microphthalmia-associated transcription factor (MITF-M) in melanoma cells. PLoS ONE 2015, 10, e0122517. [Google Scholar] [CrossRef] [PubMed]
- Soustek, M.S.; Balsa, E.; Barrow, J.J.; Jedrychowski, M.; Vogel, R.; Gygi, S.P.; Puigserver, P. Inhibition of the ER stress IRE1α inflammatory pathway protects against cell death in mitochondrial complex I mutant cells. Cell Death Dis. 2018, 9, 658. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.; Talwar, P.; Parimisetty, A.; d’Hellencourt, C.L.; Ravanan, P. A molecular web: Endoplasmic reticulum stress, inflammation, and oxidative stress. Front. Cell. Neurosci. 2014, 8, 213. [Google Scholar] [CrossRef]
- Ju, R.J.; Stehbens, S.J.; Haass, N.K. The role of melanoma cell-stroma interaction in cell motility, invasion, and metastasis. Front. Med. 2018, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Paňková, K.; Rösel, D.; Novotný, M.; Brábek, J. The molecular mechanisms of transition between mesenchymal and amoeboid invasiveness in tumor cells. Cell. Mol. Life Sci. 2009, 67, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Kosla, J.; Paňková, D.; Plachý, J.; Tolde, O.; Bicanová, K.; Dvořák, M.; Rösel, D.; Brábek, J. Metastasis of aggressive amoeboid sarcoma cells is dependent on Rho/ROCK/MLC signaling. Cell Commun. Signal. 2013, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Micuda, S.; Rösel, D.; Ryska, A.; Brábek, J. ROCK inhibitors as emerging therapeutic candidates for sarcomas. Curr. Cancer Drug Targets 2010, 10, 127–134. [Google Scholar] [CrossRef]
- Voller, J.; Zahajská, L.; Plíhalová, L.; Jeřábková, J.; Burget, D.; Pataki, A.C.; Kryštof, V.; Zatloukal, M.; Brábek, J.; Rösel, D.; et al. 6-substituted purines as ROCK inhibitors with anti-metastatic activity. Bioorg. Chem. 2019, 90, 103005. [Google Scholar] [CrossRef]
- Cantelli, G.; Orgaz, J.L.; Rodriguez-Hernandez, I.; Karagiannis, P.; Maiques, O.; Matias-Guiu, X.; Nestle, F.O.; Marti, R.M.; Karagiannis, S.N.; Sanz-Moreno, V. TGF-β-induced transcription sustains amoeboid melanoma migration and dissemination. Curr. Biol. 2015, 25, 2899–2914. [Google Scholar] [CrossRef]
- Georgouli, M.; Herraiz, C.; Crosas-Molist, E.; Fanshawe, B.; Maiques, O.; Perdrix, A.; Pandya, P.; Rodriguez-Hernandez, I.; Ilieva, K.M.; Cantelli, G.; et al. Regional activation of myosin II in cancer cells drives tumor progression via a secretory cross-talk with the immune microenvironment. Cell 2019, 176, 757–774.e23. [Google Scholar] [CrossRef] [PubMed]
- Merta, L.; Gandalovičová, A.; Čermák, V.; Dibus, M.; Gutschner, T.; Diederichs, S.; Rösel, D.; Brábek, J. Increased level of long non-coding RNA MALAT1 is a common feature of amoeboid invasion. Cancers 2020, 12, 1136. [Google Scholar] [CrossRef]
- Sanz-Moreno, V.; Gaggioli, C.; Yeo, M.; Albrengues, J.; Wallberg, F.; Viros, A.; Hooper, S.; Mitter, R.; Féral, C.C.; Cook, M.; et al. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell 2011, 20, 229–245. [Google Scholar] [CrossRef]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef]
- Karaman, M.W.; Herrgard, S.; Treiber, D.K.; Gallant, P.; Atteridge, C.E.; Campbell, B.T.; Chan, K.W.; Ciceri, P.; Davis, M.I.; Edeen, P.T.; et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008, 26, 127–132. [Google Scholar] [CrossRef]
- Bain, J.; Plater, L.; Elliott, M.; Shpiro, N.; Hastie, C.J.; Mclauchlan, H.; Klevernic, I.; Arthur, J.S.C.; Alessi, D.R.; Cohen, P. The selectivity of protein kinase inhibitors: A further update. Biochem. J. 2007, 408, 297–315. [Google Scholar] [CrossRef]
- Palušová, V.; Renzová, T.; Verlande, A.; Vaclová, T.; Medková, M.; Cetlová, L.; Sedláčková, M.; Hříbková, H.; Slaninová, I.; Krutá, M.; et al. Dual targeting of BRAF and mTOR signaling in melanoma cells with pyridinyl imidazole compounds. Cancers 2020, 12, 1516. [Google Scholar] [CrossRef]
- Yang, C.; Zhu, Z.; Tong, B.C.-K.; Iyaswamy, A.; Xie, W.-J.; Zhu, Y.; Sreenivasmurthy, S.G.; Senthilkumar, K.; Cheung, K.-H.; Song, J.-X.; et al. A stress response P38 MAP kinase inhibitor SB202190 promoted TFEB/TFE3-dependent autophagy and lysosomal biogenesis independent of P38. Redox Biol. 2020, 32, 101445. [Google Scholar] [CrossRef]
- Čermák, V.; Škarková, A.; Merta, L.; Rösel, D.; Brábek, J. Melanoma Phenotype Switch in 3D Collagen After P38 MAPK Inhibitor Treatment. Available online: https://doi.org/10.6084/m9.figshare.c.5215253 (accessed on 7 March 2021).
- Hemesath, T.J.; Steingrímsson, E.; McGill, G.; Hansen, M.J.; Vaught, J.; Hodgkinson, C.A.; Arnheiter, H.; Copeland, N.G.; Jenkins, N.A.; Fisher, D.E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994, 8, 2770–2780. [Google Scholar] [CrossRef]
- Martina, J.A.; Puertollano, R. Rag GTPases mediate amino acid–dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 2013, 200, 475–491. [Google Scholar] [CrossRef]
- Amae, S.; Fuse, N.; Yasumoto, K.I.; Sato, S.; Yajima, I.; Yamamoto, H.; Udono, T.; Durlu, Y.K.; Tamai, M.; Takahashi, K.; et al. Identification of a novel isoform of microphthalmia-associated transcription factor that is enriched in retinal pigment epithelium. Biochem. Biophys. Res. Commun. 1998, 247, 710–715. [Google Scholar] [CrossRef]
- Yasumoto, K.; Takeda, K.; Saito, H.; Watanabe, K.; Takahashi, K.; Shibahara, S. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J. 2002, 21, 2703–2714. [Google Scholar] [CrossRef]
- Čermák, V.; Gandalovičová, A.; Merta, L.; Harant, K.; Rösel, D.; Brábek, J. High-throughput transcriptomic and proteomic profiling of mesenchymal-amoeboid transition in 3D collagen. Sci. Data 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Clark, E.A.; Golub, T.R.; Lander, E.S.; Hynes, R.O. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 2000, 406, 532–535. [Google Scholar] [CrossRef]
- Kozlowski, J.M.; Fidler, I.J.; Hanna, N.; Hart, I.R. A human melanoma line heterogeneous with respect to metastatic capacity in athymic nude mice. J. Natl. Cancer Inst. 1984, 72, 913–917. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Čermák, V.; Gandalovičová, A.; Merta, L.; Rösel, D.; Brábek, J. RNA-Seq Of Human Melanoma Cell Line A375m2 Treated with SB202190 or BIRB796 against DMSO-Treated Controls. Available online: https://identifiers.org/arrayexpress:E-MTAB-9273 (accessed on 28 November 2020).
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 0034. [Google Scholar] [CrossRef]
- Perkins, J.R.; Dawes, J.M.; McMahon, S.B.; Bennett, D.L.H.; Orengo, C.; Kohl, M. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-QPCR quantification cycle (Cq) data. BMC Genom. 2012, 13, 296. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Čermák, V.; Gandalovičová, A.; Merta, L.; Fučíková, J.; Špíšek, R.; Rösel, D.; Brábek, J. RNA-seq of macrophages of amoeboid or mesenchymal migratory phenotype due to specific structure of environment. Sci. Data 2018, 5, 180198. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Folberg, R.; Arbieva, Z.; Moses, J.; Hayee, A.; Sandal, T.; Kadkol, S.H.; Lin, A.Y.; Valyi-Nagy, K.; Setty, S.; Leach, L.; et al. Tumor cell plasticity in uveal melanoma: Microenvironment directed dampening of the invasive and metastatic genotype and phenotype accompanies the generation of vasculogenic mimicry patterns. Am. J. Pathol. 2006, 169, 1376–1389. [Google Scholar] [CrossRef] [PubMed]
- Molbiotools. Multiple List Comparator. Available online: http://www.molbiotools.com/listcompare.html (accessed on 4 March 2021).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 November 2020).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Widmer, D.S.; Cheng, P.F.; Eichhoff, O.M.; Belloni, B.C.; Zipser, M.C.; Schlegel, N.C.; Javelaud, D.; Mauviel, A.; Dummer, R.; Hoek, K.S. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment. Cell Melanoma Res. 2012, 25, 343–353. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.-W.; Lee, S.-Y.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-Seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Jeffs, A.R.; Glover, A.C.; Slobbe, L.J.; Wang, L.; He, S.; Hazlett, J.A.; Awasthi, A.; Woolley, A.G.; Marshall, E.S.; Joseph, W.R.; et al. A gene expression signature of invasive potential in metastatic melanoma cells. PLoS ONE 2009, 4, e8461. [Google Scholar] [CrossRef]
- Rodriguez-Hernandez, I.; Maiques, O.; Kohlhammer, L.; Cantelli, G.; Perdrix-Rosell, A.; Monger, J.; Fanshawe, B.; Bridgeman, V.L.; Karagiannis, S.N.; Penin, R.M.; et al. WNT11-FZD7-DAAM1 signalling supports tumour initiating abilities and melanoma amoeboid invasion. Nat. Commun. 2020, 11, 1–20. [Google Scholar] [CrossRef]
- Arozarena, I.; Bischof, H.; Gilby, D.; Belloni, B.; Dummer, R.; Wellbrock, C. In melanoma, beta-catenin is a suppressor of invasion. Oncogene 2011, 30, 4531–4543. [Google Scholar] [CrossRef] [PubMed]
- Parri, M.; Taddei, M.L.; Bianchini, F.; Calorini, L.; Chiarugi, P. EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer Res. 2009, 69, 2072–2081. [Google Scholar] [CrossRef] [PubMed]
| Kinase | SB202190 | BIRB796 |
|---|---|---|
| p38alpha | xx/+++ | xx/++++ |
| p38beta | xx/++ | x/ |
| p38gamma | x/+ | |
| p38delta | x | |
| JNK2 | x/+ | xx/++ |
| JNK3 | x/++ | |
| NLK | x/++ | |
| RIPK2/RIP2 | xx/+ | |
| GAK | xx/++ | |
| CK1delta | x/++ | |
| BRAF | + | |
| GSK3beta | x | |
| CK1epsilon | + | |
| Lck | x | |
| ACVR1B | + | |
| CIT | + | |
| CDC42BPG | + | |
| EGFR | + | |
| PRKACB | + | |
| RPS6KA1 | + | |
| RPS6KA6 | + | |
| STK36 | + | |
| DDR1 | ++ | |
| TIE1 | ++ | |
| MAP4K4 | + | |
| STK10 | + | |
| SLK | + | |
| ABL1 | + | |
| DDR2 | + | |
| TIE2 | + | |
| RSK1 | x | |
| RSK2 | x | |
| BRSK2 | x |
| Gene | SB202190 | BIRB796 |
|---|---|---|
| TRPM1 | 4.00 ± 0.67 | |
| DCT | 3.38 ± 0.09 | 1.37 ± 0.09 |
| MLANA | 2.47 ± 0.18 | 0.94 ± 0.19 |
| GPM6B | 2.07 ± 0.08 | 1.00 ± 0.08 |
| PMEL | 1.58 ± 0.27 | |
| TYR | 1.39 ± 0.08 | |
| MBP | 1.17 ± 0.34 | 1.52 ± 0.34 |
| RAB27A | 0.95 ± 0.11 | |
| CAPN3 | 0.88 ± 0.13 | |
| GPNMB | 0.80 ± 0.07 | |
| IL1B | −4.48 ± 0.33 | −3.06 ± 0.29 |
| IL1A | −3.80 ± 0.18 | −1.95 ± 0.15 |
| CXCL8 | −3.60 ± 0.18 | −2.01 ± 0.15 |
| SERPINE1 | −3.07 ± 0.58 | |
| PODXL | −2.83 ± 0.16 | −1.36 ± 0.15 |
| AXL | −2.70 ± 0.41 | −1.11 ± 0.34 |
| INHBA | −2.24 ± 0.12 | −0.88 ± 0.12 |
| FN1 | −1.91 ± 0.09 | −1.23 ± 0.09 |
| LOXL2 | −1.81 ± 0.10 | −0.78 ± 0.10 |
| FST | −1.61 ± 0.11 | −1.13 ± 0.11 |
| ADAM12 | −1.34 ± 0.10 | −0.60 ± 0.10 |
| WNT5B | −0.82 ± 0.35 | |
| WNT5A | −0.80 ± 0.11 | |
| THBS1 | −0.73 ± 0.11 |
| Database | Data | SB202190 | BIRB796 |
|---|---|---|---|
| NCI-60 cancer cell line panel vs. upregulated genes | UACC257 | 1.83 × 10−10 | 1.33 × 10−4 |
| SKMEL5 | 1.83 × 10−10 | 7.06 × 10−2 | |
| SKMEL28 | 1.46 × 10−6 | 2.49 × 10−3 | |
| MALME 3M | 4.46 × 10−5 | 1.85 × 10−2 | |
| M14 | 5.64 × 10−3 | 1.07 × 10−1 | |
| GO—Biological Process vs. downregulated genes | extracellular matrix organization (GO:0030198) | 3.44 × 10−20 | 2.27 × 10−11 |
| regulation of cell proliferation (GO:0042127) | 2.79 × 10−11 | 2.09 × 10−5 | |
| regulation of apoptotic process (GO:0042981) | 4.35 × 10−11 | 1.38 × 10−9 | |
| regulation of cell migration (GO:0030334) | 6.14 × 10−11 | 4.86 × 10−7 | |
| regulation of angiogenesis (GO:0045765) | 8.39 × 10−9 | 3.34 × 10−5 | |
| negative regulation of programmed cell death (GO:0043069) | 9.99 × 10−9 | 3.34 × 10−5 | |
| cellular response to cytokine stimulus (GO:0071345) | 1.86 × 10−8 | 2.11 × 10−4 | |
| positive regulation of angiogenesis (GO:0045766) | 3.98 × 10−8 | 6.02 × 10−5 | |
| regulation of signal transduction (GO:0009966) | 3.98 × 10−8 | 6.02 × 10−5 | |
| negative regulation of apoptotic process (GO:0043066) | 7.51 × 10−8 | 2.06 × 10−5 | |
| positive regulation of cell migration (GO:0030335) | 7.51 × 10−8 | 7.08 × 10−5 | |
| regulation of MAPK cascade (GO:0043408) | 2.39 × 10−7 | 6.17 × 10−5 | |
| TRRUST vs. downregulated genes | NFKB1 human | 8.76 × 10−9 | 1.85 × 10−6 |
| RELA human | 3.55 × 10−8 | 1.08 × 10−5 | |
| NFKB1 mouse | 7.76 × 10−8 | 1.01 × 10−11 | |
| VHL human | 5.70 × 10−7 | 5.01 × 10−4 | |
| STAT3 mouse | 7.85 × 10−7 | 1.08 × 10−5 | |
| SP1 mouse | 8.33 × 10−7 | 1.07 × 10−11 | |
| EGR1 mouse | 2.54 × 10−6 | 3.75 × 10−6 | |
| ETS1 human | 2.72 × 10−6 | 4.92 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čermák, V.; Škarková, A.; Merta, L.; Kolomazníková, V.; Palušová, V.; Uldrijan, S.; Rösel, D.; Brábek, J. RNA-seq Characterization of Melanoma Phenotype Switch in 3D Collagen after p38 MAPK Inhibitor Treatment. Biomolecules 2021, 11, 449. https://doi.org/10.3390/biom11030449
Čermák V, Škarková A, Merta L, Kolomazníková V, Palušová V, Uldrijan S, Rösel D, Brábek J. RNA-seq Characterization of Melanoma Phenotype Switch in 3D Collagen after p38 MAPK Inhibitor Treatment. Biomolecules. 2021; 11(3):449. https://doi.org/10.3390/biom11030449
Chicago/Turabian StyleČermák, Vladimír, Aneta Škarková, Ladislav Merta, Veronika Kolomazníková, Veronika Palušová, Stjepan Uldrijan, Daniel Rösel, and Jan Brábek. 2021. "RNA-seq Characterization of Melanoma Phenotype Switch in 3D Collagen after p38 MAPK Inhibitor Treatment" Biomolecules 11, no. 3: 449. https://doi.org/10.3390/biom11030449
APA StyleČermák, V., Škarková, A., Merta, L., Kolomazníková, V., Palušová, V., Uldrijan, S., Rösel, D., & Brábek, J. (2021). RNA-seq Characterization of Melanoma Phenotype Switch in 3D Collagen after p38 MAPK Inhibitor Treatment. Biomolecules, 11(3), 449. https://doi.org/10.3390/biom11030449







