Lipidomic Signatures for Colorectal Cancer Diagnosis and Progression Using UPLC-QTOF-ESI+MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Compliance with Ethical Standards
2.2. Blood Collection and Processing
2.3. HPLC–ESI(+)-QTOF-MS Analysis of Blood Serum
2.4. Statistical Analysis
3. Results
3.1. Multivariate Analysis
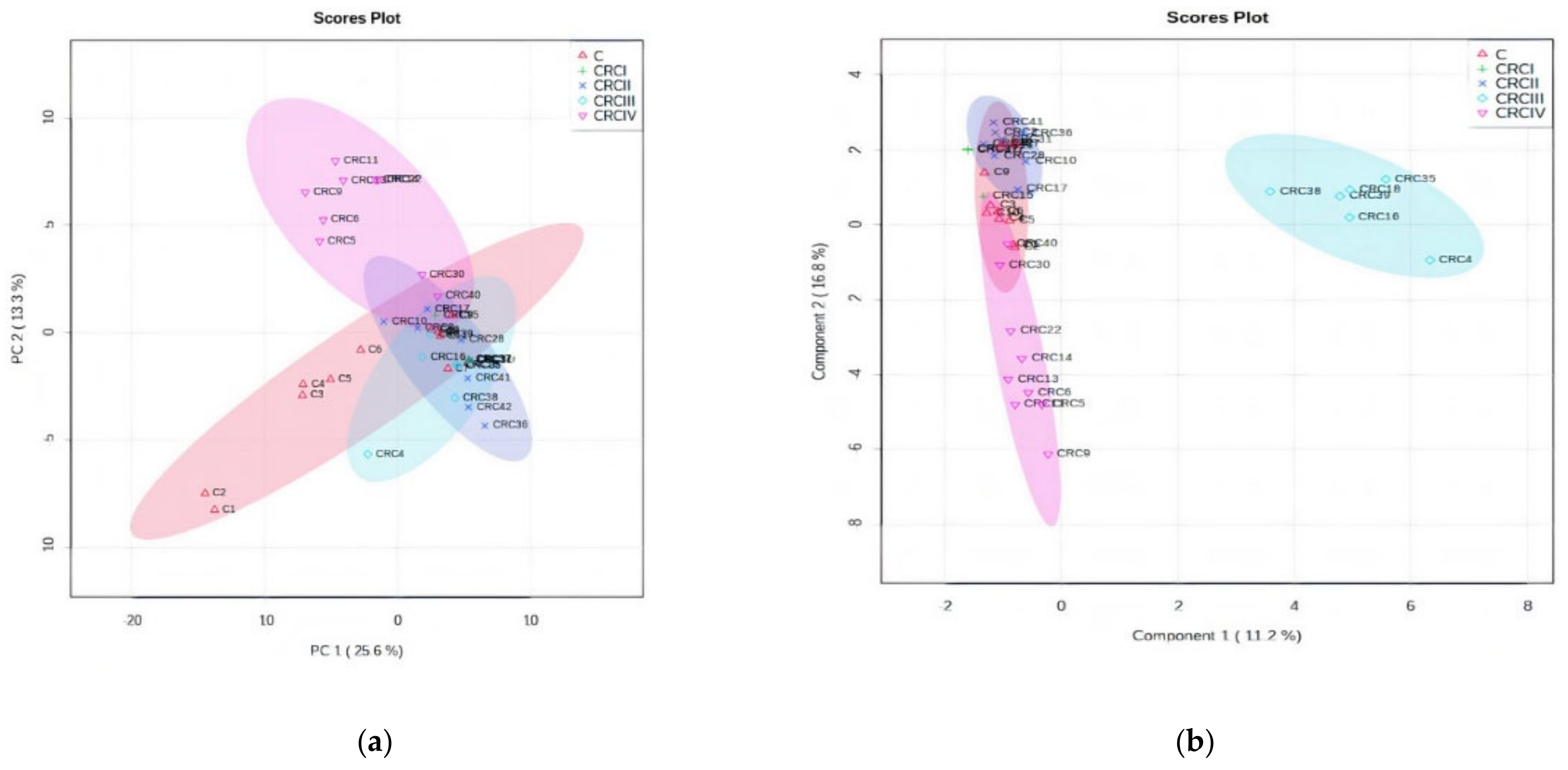
3.1.1. PCA and PLSDA Analysis
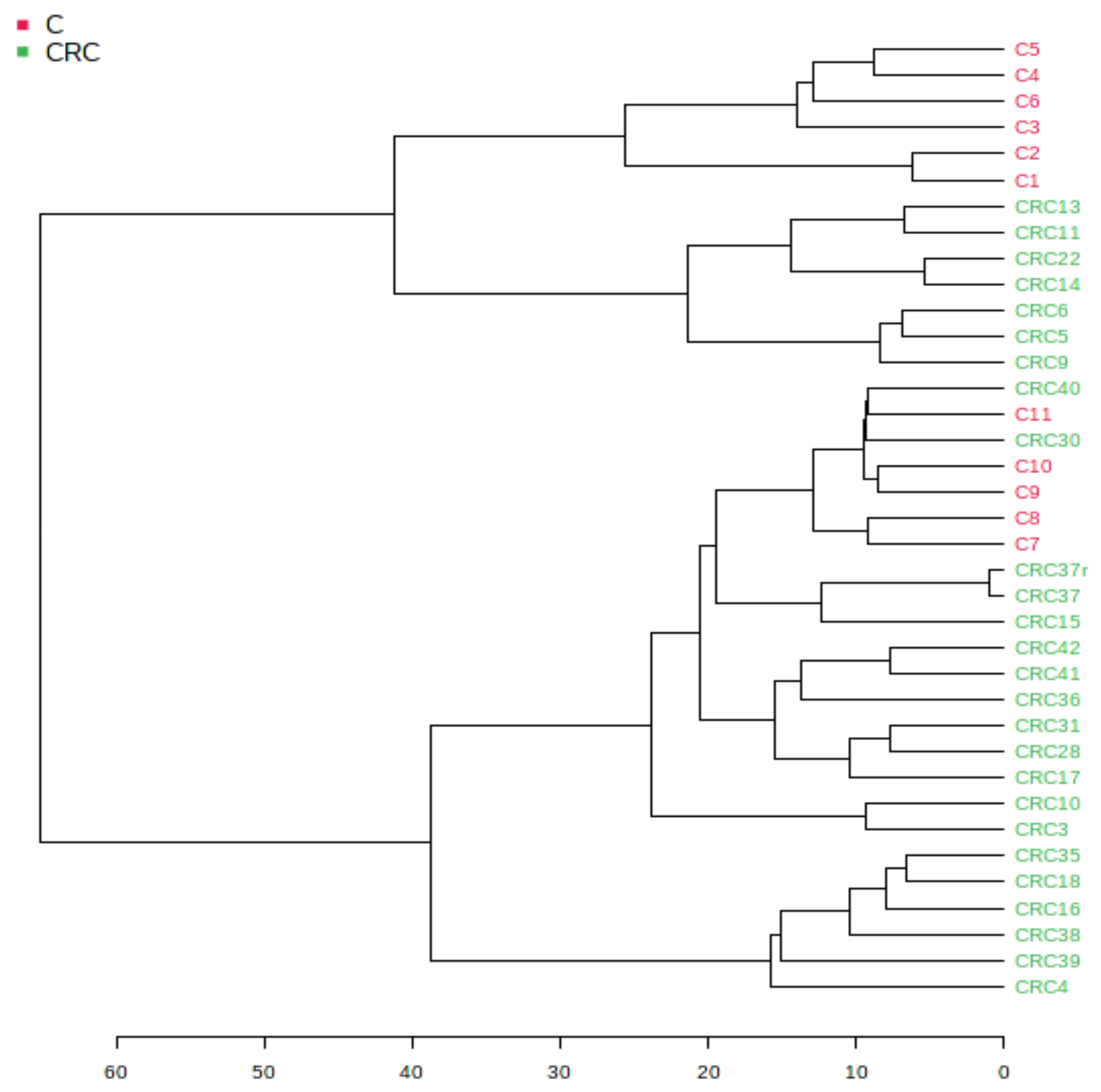
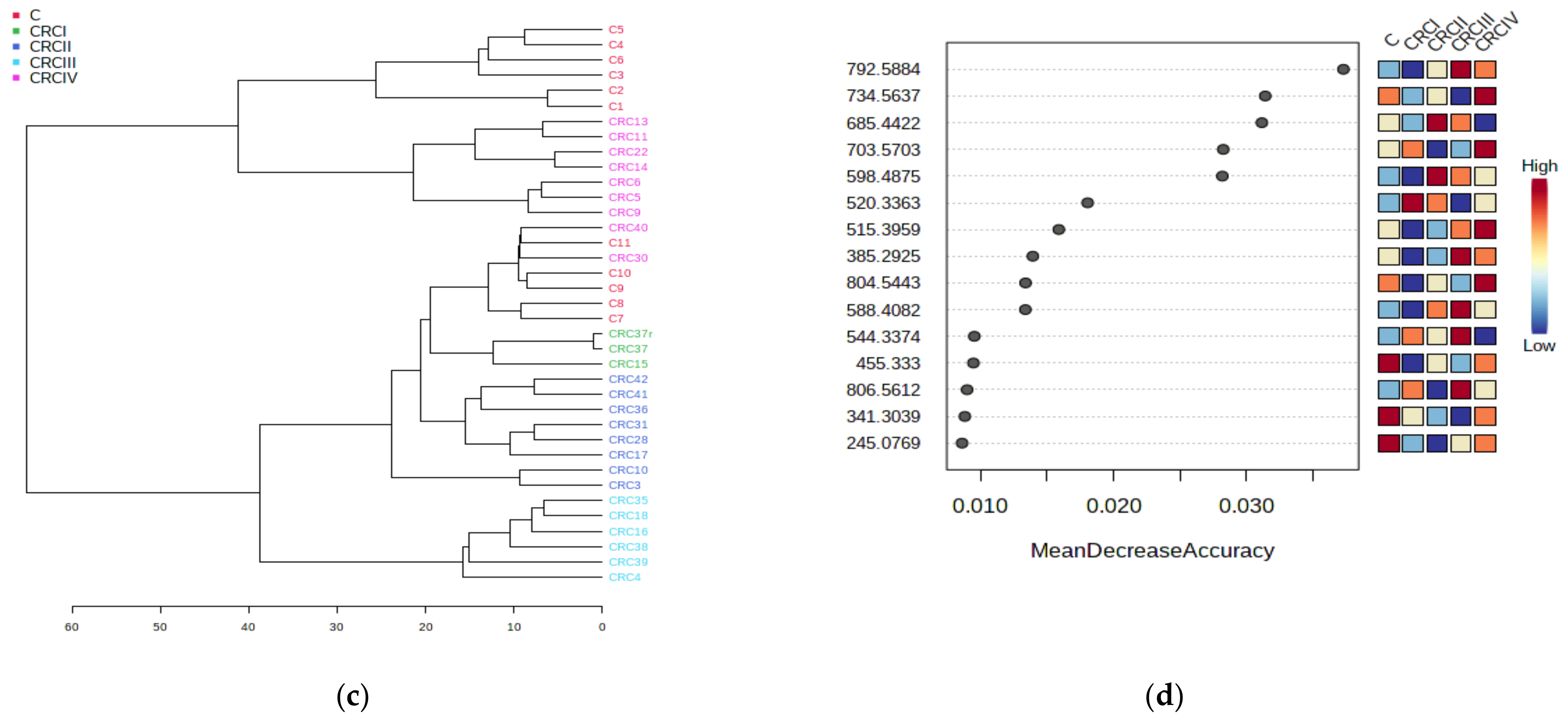
3.1.2. Euclidean Dendrogram and Correlation Heatmaps
3.1.3. The Random Forest Algorithm and Its Predictive Value
3.1.4. Biomarker Analysis
3.2. Univariate Analysis ANOVA: Discrimination of CRC Stages
3.2.1. One-Way ANOVA to Identify Biomarkers for CRC Progression (Stages I to IV)
3.2.2. Statistical Analysis Based on MS Peak Intensity Values for CRC Subgroups
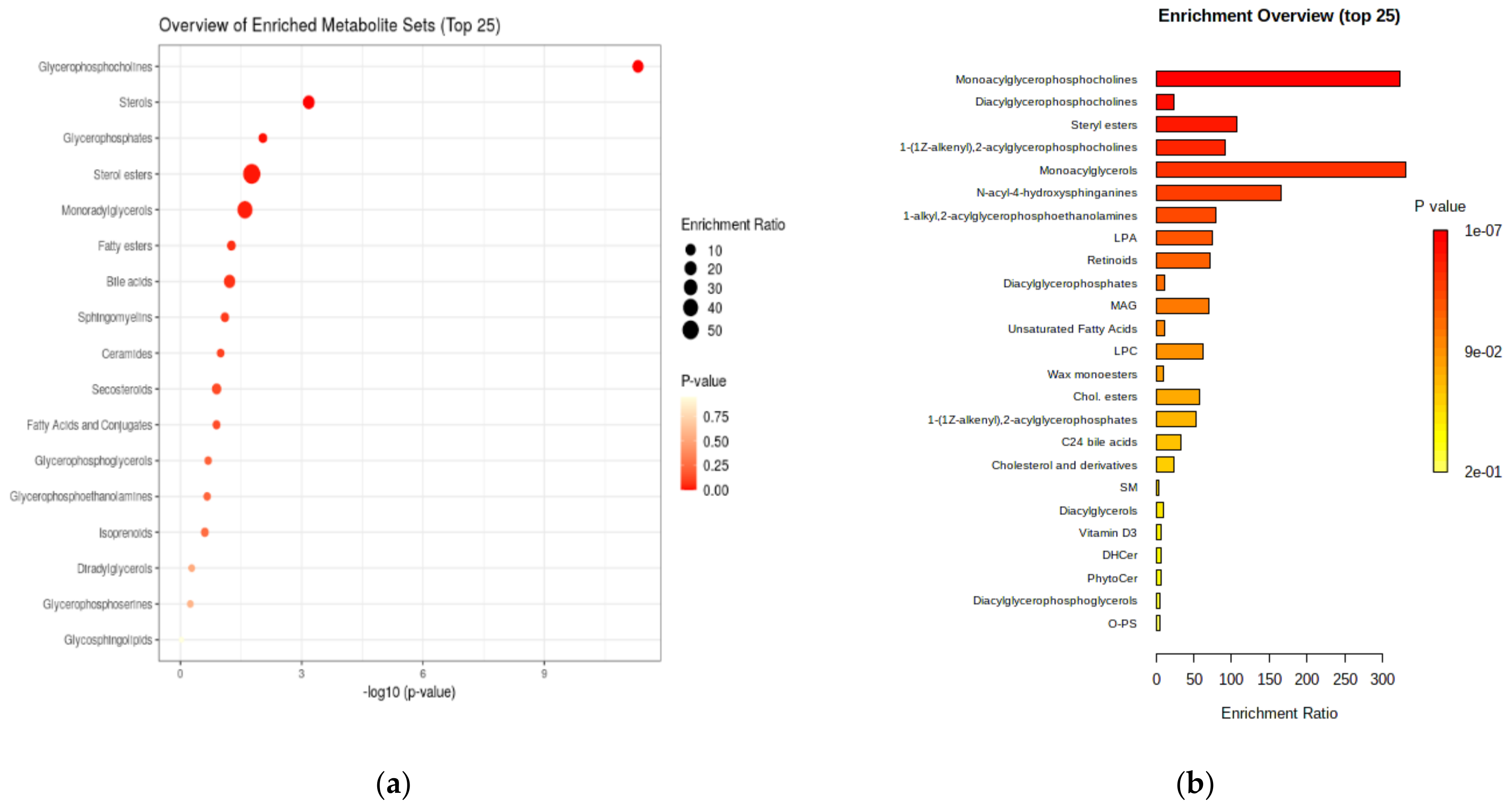
3.2.3. Enrichment and Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Cer | ceramide |
| CRC | colorectal cancer |
| DG | diacylglycerol |
| FFA | free fatty acid |
| GlcCer | glucosylceramide |
| HETE | hydroxyeicosatetraenoic acid |
| HODE | 13-hydroxyoctadecadienoic acid |
| LA | linoleic acid |
| LPA | lysophosphatidic acid |
| LPC | lysophosphatidylcholine |
| MG | monoacylglycerol |
| MUFA | monounsaturated fatty acid |
| PA | phosphatidic acid |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| PL | phospholipid |
| PUFA | polyunsaturated fatty acid |
| SM | sphingomyelin |
| SPL | sphingolipid |
| TG | triacylglycerol. |
References
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A. Colorectal cancer statistics. CA Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Cancer Facts & Figures. 2019. Available online: https://www.cancer.org/research/cancer-facts-statistics.html (accessed on 24 January 2021).
- American Cancer Society. Colorectal Cancer Facts & Figures 2020–2022; American Cancer Society: Atlanta, GA, USA, 2020. [Google Scholar]
- Beggs, A.D.; Dilworth, M.P. Surgery in the era of the ’omics revolution. Br. J. Surg. 2015, 102, e29–e40. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98. [Google Scholar] [CrossRef]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Abu Zaid, M.; Chiorean, E.G.; Raftery, D. Targeted serum metabolite profiling and sequential metabolite ratio analysis for colorectal cancer progression monitoring. Anal. Bioanal. Chem. 2015, 407, 7857–7863. [Google Scholar] [CrossRef]
- Johnson, C.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell. Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, D.J.; Hoeferlin, L.A.; Chalfant, C.E. Lipidomics in translational research and the clinical significance of lipid-based biomarkers. Transl. Res. 2017, 189, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Muc-Wierzgoń, M.; Nowakowska-Zajdel, E.; Dzięgielewska-Gęsiak, S.; Kokot, T.; Klakla, K.; Fatyga, E.; Grochowska-Niedworok, E.; Waniczek, D.; Wierzgoń, J. Specific metabolic biomarkers as risk and prognostic factors in colorectal cancer. World Gastroenterol. 2014, 20, 9759–9774. [Google Scholar] [CrossRef]
- Djukovic, D.; Zhang, J.; Raftery, D. Colorectal cancer detection using targeted LC-MS metabolic profiling. In Colorectal Cancer. Methods in Molecular Biology; Beaulieu, J.F., Ed.; Humana Press: New York, NY, USA, 2018; Volume 1765, pp. 229–240. [Google Scholar] [CrossRef]
- Triebl, A.; Trötzmüller, M.; Hartler, J.; Stojakovic, T.; Köfeler, H.C. Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J. Chromatogr. B 2017, 1053, 72–80. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, H.; Liu, R.; Gao, R.; Yang, X.; Jin, W.; Zhang, Y.; Bi, K.; Li, Q. A novel strategy for targeted lipidomics based on LC-tandem-MS parameters prediction, quantification, and multiple statistical data mining: Evaluation of lysophosphatidylcholines as potential cancer biomarker. Anal. Chem. 2019, 91, 3389–3396. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, C.; Gui, Q.; Chen, Y.; Yang, Y. Ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry-based metabolic profiling of human serum prior to and following radical resection of colorectal carcinoma. Mol. Med. Rep. 2015, 12, 6879–6886. [Google Scholar] [CrossRef][Green Version]
- Kondo, Y.; Nishiumi, S.; Shinohara, M.; Hatano, N.; Ikeda, A.; Yoshie, T.; Kobayashi, T.; Shiomi, Y.; Irino, Y.; Takenawa, T.; et al. Serum fatty acid profiling of colorectal cancer by gas chromatography/mass spectrometry. Biomark. Med. 2011, 5, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Kawana, S.; Unno, Y.; Sakai, T.; Okamoto, K.; Yoshida, M. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget 2017, 8, 17115–17126. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics analysis in serum from patients with colorectal polyp and colorectal cancer by 1H-NMR spectrometry. Dis. Markers 2019. [Google Scholar] [CrossRef]
- Li, F.; Qin, X.; Chen, H.; Qiu, L.; Guo, Y.; Liu, H.; Li, Z.L. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 24–34. [Google Scholar] [CrossRef]
- Xu, X.D.; Shao, S.X.; Jiang, H.P.; Cao, Y.W.; Wang, Y.H.; Yang, X.C.; Wang, Y.L.; Wang, X.S.; Ni, H.T. Warburg effect or reverse Warburg effect? A review of cancer metabolism. Oncol. Res. Treat. 2015, 38, 117–122. [Google Scholar] [CrossRef]
- Chekulayev, V.; Mado, K.; Shevchuk, I.; Koit, A.; Kaldma, A.; Klepinin, A.; Ounpuua, L.; Heckc, K.; Truud, L.; Plankene, A.; et al. Metabolic remodeling in human colorectal cancer and surrounding tissues: Alterations in regulation of mitochondrial respiration and metabolic fluxes. Biochem. Biophys. Rep. 2015, 4, 111–125. [Google Scholar] [CrossRef]
- Hao, Y.; Samuels, Y.; Li, Q.; Krokowski, D.; Guan, B.-J.; Wang, C.; Wang, Z. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat. Commun. 2016, 7, 11971. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, Y.Y.; Harris, J.W.; Mitov, M.I.; Kim, J.T.; Butterfield, D.A.; Lee, E.Y.; Weiss, H.L.; Evers, B.M. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget 2015, 6, 18891–18904. [Google Scholar] [CrossRef] [PubMed]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and Cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29–38. [Google Scholar] [CrossRef]
- Shen, S.; Yang, L.; Li, L.; Bai, Y.; Cai, C.; Liu, H.C. A plasma lipidomics strategy reveals perturbed lipid metabolic pathways and potential lipid biomarkers of human colorectal cancer. J. Chromatogr. B 2017, 1068–1069, 41–48. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, B.; Zhang, J.; Li, J.; Yang, Q.; Zhong, Q.; Zhan, S.; Liu, H.; Cai, C. Serum polyunsaturated fatty acid metabolites as useful tool for screening potential biomarker of colorectal cancer. Prostaglandins Leukot. Essent. Fat Acids 2017, 120, 25–31. [Google Scholar] [CrossRef]
- Yan, F.; Zhao, H.; Zeng, Y. Lipidomics: A promising cancer biomarker. Clin. Trans. Med. 2018, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Charkoftaki, G.; Thompson, D.C.; Golla, J.P.; Garcia-Milian, R.; Lam, T.K.T.; Engel, J.; Vasiliou, V. Integrated multi-omics approach reveals a role of ALDH1A1 in lipid metabolism in human colon cancer cells. Chem. Biol. Interact. 2019, 304, 88–96. [Google Scholar] [CrossRef]
- Wang, Y.; Hinz, S.; Uckermann, O.; Hönscheid, P.; Von Schönfels, W.; Burmeister, G.; Hendricks, A. Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. BBA Mol. Cell Biol. Lipids 2020, 1865, 158579. [Google Scholar] [CrossRef]
- Jung, J.H.; Taniguchi, K.; Lee, H.M.; Lee, M.Y.; Bandu, R.; Komura, K.; Lee, K.Y.; Akao, Y.; Kim, K.P. Comparative lipidomics of 5-Fluorouracil–sensitive and –resistant colorectal cancer cells reveals altered sphingomyelin and ceramide controlled by acid sphingomyelinase (SMPD1). Sci. Rep. 2020, 10, 6124. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Cai, G.; Zhou, B.; Li, D.; Zhao, A.; Xie, G.; Li, H.A.; Cai, S.; Xie, D.; Huang, C.; et al. Distinct metabolic signature of human colorectal cancer with prognostic potential. Clin. Cancer Res. 2014, 20, 2136–2147. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Yoshida, M. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef] [PubMed]
- Perttula, K.; Schiffman, C.; Edmands, W.M.B.; Petrick, L.; Grigoryan, H.; Cai, X.; Rappaport, S.M. Untargeted lipidomic features associated with colorectal cancer in a prospective cohort. BMC Cancer 2018, 18, 996. [Google Scholar] [CrossRef] [PubMed]
- Kühn, T.; Floegel, A.; Sookthai, D.; Johnson, T.; Rolle-Kampczyk, U.; Otto, W.; Kaaks, R. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Xiang, Y.B.; Rothman, N.; Yu, D.; Li, H.L.; Yang, G.; Zheng, W. Prospective study of blood metabolites associated with colorectal cancer risk. Int. J. Cancer 2018, 143, 527–534. [Google Scholar] [CrossRef]
- Răchieriu, C.; Eniu, D.T.; Moiş, E.; Graur, F.; Socaciu, C.; Socaciu, M.A.; Alhajjar, N. Lipidomics: Advanced analytical technology to identify biomarkers of colorectal cancer. Studia Univ. Babes Bolyai Chem. 2020, 65, 203–215. [Google Scholar] [CrossRef]
- Răchieriu, C.; Graur, F.; Moiş, E.; Socaciu, C.; Eniu, D.T.; Alhajjar, N. Metabolomic profile of colorectal cancer patients and its clinical implications. Rom. Biotechnol. Lett. 2020, 25, 2045–2054. [Google Scholar] [CrossRef]
- Zhao, Z.; Xiao, Y.; Elson, P.; Tan, H.; Plummer, S.J.; Berk, M.; Aung, P.P.; Lavery, I.C.; Achkar, J.P.; Li, L.; et al. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J. Clin. Oncol. 2007, 25, 2696–2701. [Google Scholar] [CrossRef]
- Li, S.; Guo, B.; Song, J.W.; Deng, X.L.; Cong, Y.; Li, P.; Zhao, K.; Liu, L.; Xiao, G.; Xu, F.; et al. Plasma choline-containing phospholipids: Potential biomarkers for colorectal cancer progression. Metabolomics 2013, 9, 202–212. [Google Scholar] [CrossRef]
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Jia, W. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef] [PubMed]
- Kurabe, N.; Hayasaka, T.; Ogawa, M.; Masaki, N.; Ide, Y.; Waki, M.; Nakamura, T.; Kurachi, K.; Kahyo, T.; Shinmura, K.; et al. Accumulated phosphatidylcholine (16:0/16:1) in human colorectal cancer; possible involvement of LPCAT4. Cancer Sci. 2013, 104, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Geijsen, J.M.R.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.J.B.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer 2019, 145, 1221–1231. [Google Scholar] [CrossRef]
- Zuo, X.; Shureiqi, I. Eicosanoid profiling in colon cancer: Emergence of a pattern. Prostaglandins Other Lipid Mediat. 2013, 104–105, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Shureiqi, I.; Chen, D.; Day, R.S.; Zuo, X.; Hochman, F.L.; Ross, W.A.; Lippman, S.M. Profiling lipoxygenase metabolism in specific steps of colorectal tumorigenesis. Cancer Prev. Res. 2010, 3, 829–838. [Google Scholar] [CrossRef]
- Crotti, S.; Agnoletto, E.; Cancemi, G.; Di Marco, V.; Traldi, P.; Pucciarelli, S.; Agostini, M. Altered plasma levels of decanoic acid in colorectal cancer as a new diagnostic biomarker. Anal. Bioanal. Chem. 2016, 408, 6321–6328. [Google Scholar] [CrossRef]
- Uchiyama, K.; Yagi, N.; Mizushima, K.; Higashimura, Y.; Hirai, Y.; Okayama, T.; Naito, Y. Serum metabolomics analysis for early detection of colorectal cancer. J. Gastroenterol. 2017, 52, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Farshidfar, F.; Kopciuk, K.A.; Hilsden, R.; Mcgregor, S.E.; Mazurak, V.C.; Buie, W.D. A quantitative multimodal metabolomic assay for colorectal cancer. BMC Cancer 2018, 18, 26. [Google Scholar] [CrossRef]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.; Hilsden, S.E.; Mcgregor, F.; Buie, W.D.; Bathe, O.F. A validated metabolomic signature for colorectal cancer: Exploration of the clinical value of metabolomics. Br. J. Cancer 2016, 115, 848–857. [Google Scholar] [CrossRef]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.; Buie, W.D.; Maclean, A.; Dixon, E.; Sutherland, F.R.; Molckovsky, A.; Vogel, H.J.; Bathe, O.F. Serum metabolomic profile as a means to distinguish stage of colorectal cancer. Genome Med. 2012, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Butler, L.M.; Yuan, J.M.; Huang, J.; Su, J.J.; Wang, R.; Koh, W.-P.; Ong, C.-N. Plasma fatty acids and risk of colon and rectal cancers in the Singapore Chinese Health Study. Precis. Oncol. 2017, 1, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, L.; Ye, X.; Chen, L.; Zhang, L.; Gao, Y.; Kang, J.X.; Cai, C. Characteristics of fatty acid distribution is associated with colorectal cancer prognosis. Prostaglandins Leukot. Essent. Fat Acids 2013, 88, 355–360. [Google Scholar] [CrossRef]
- Liu, T.; Tan, Z.; Yu, J.; Peng, F.; Guo, J.; Meng, W.; Chen, Y.; Rao, T.; Liu, Z.; Peng, J. A conjunctive lipidomic approach reveals plasma ethanolamine plasmalogens and fatty acids as early diagnostic biomarkers for colorectal cancer patient. Expert Rev. Proteom. 2020, 17, 233–242. [Google Scholar] [CrossRef]
- Guo, S.; Wang, Y.; Zhou, D.; Li, Z. Significantly increased monounsaturated lipids relative to polyunsaturated lipids in six types of cancer microenvironment are observed by mass spectrometry imaging. Sci. Rep. 2014, 4, 5959. [Google Scholar] [CrossRef]
- Mika, A.; Kobiela, J.; Czumaj, A.; Chmielewski, M.; Stepnowski, P.; Sledzinski, T. Hyper-elongation in colorectal cancer tissue-cerotic acid is a potential novel serum metabolic marker of colorectal malignancies. Cell. Physiol. Biochem. 2017, 41, 722–730. [Google Scholar] [CrossRef]
- Mika, A.; Pakiet, A.; Czumaj, A.; Kaczynski, Z.; Liakh, I.; Kobiela, J.; Perdyan, A.; Adrych, K.; Makarewicz, W.; Sledzinski, T. Decreased triacylglycerol content and elevated contents of cell membrane lipids in colorectal cancer tissue: A lipidomic study. J. Clin. Med. 2020, 9, 1095. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Peng, F.; Yu, J. LC-MS-based lipid profile in colorectal cancer patients: TAGs are the main disturbed lipid markers of colorectal cancer progression. Anal. Bioanal. Chem. 2019, 411, 5079–5088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, Y.; Zhao, W.; Deng, K.; Wang, Z.; Yang, C.; Ma, L.; Openkova, M.S.; Hou, Y.; Li, K. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: A systematic review. Oncotarget 2017, 8, 35460–35472. [Google Scholar] [CrossRef] [PubMed]

| Biospecimen | CRC | Tumor Site | Co-Morbidities |
|---|---|---|---|
| Number of participants | 25 | - | - |
| Male Age (mean ± SD) Female Age (mean ± SD) | 64.12 ± 13.94 69.11 ± 9.34 | - | - |
| Male/female Nr | 16/9 | - | - |
| Body mass index Male/female | 32.5 ± 4.7 25.5 ± 6.7 | - | - |
| pT2NoMoLoVo, Stage I | 2 (8%) | Rectosigmoid/rectum | Obesity |
| pT3NoMoLoVo, Stage-IIA | 8 (32%) | Left/right colon | Obesity, IHD, Aortic stenosis, Hemorrhoids |
| pT3/4aNoMoLoVo, Stage-IIIB | 5 (20%) | Rectosigmoid/rectum/colon | NIDDM, obesity, IHD |
| pT4aNoMoLoVo, Stage-IIIC | 1 (4%) | Rectosigmoid | Obesity, HTN, NIDDM |
| pT3/4aNoMoLoVo, TNM Stage-IV | 9 (36%) | Sigmoid/rectum/colon and metastasis | Obesity, HTN, NIDDM, Hemorrhoids |
| m/z | MDA | CRC vs. C | m/z | MDA | CRC vs. C |
|---|---|---|---|---|---|
| 723.5055 | 0.011848 | D | 377.1835 | 0.004234 | D |
| 792.5884 | 0.010493 | I | 381.2972 | 0.004234 | D |
| 598.4875 | 0.007986 | I | 579.2966 | 0.004096 | D |
| 524.37 | 0.007733 | D | 359.3152 | 0.003745 | D |
| 341.3039 | 0.007204 | D | 804.5443 | 0.003715 | D |
| 391.2841 | 0.006768 | D | 529.3726 | 0.002683 | D |
| 455.333 | 0.006587 | D | 707.486 | 0.002601 | D |
| 520.3363 | 0.006484 | I | 611.3532 | 0.002516 | D |
| 679.4944 | 0.00603 | D | 703.5703 | 0.002481 | D |
| 588.4082 | 0.005828 | I | 758.5642 | 0.00238 | D |
| 751.5213 | 0.004866 | I | 830.5572 | 0.002307 | D |
| 794.5973 | 0.004584 | D | 685.4422 | 0.002176 | D |
| 722.5123 | 0.004329 | D | 473.3446 | 0.002169 | D |
| 808.5757 | 0.004291 | D | 683.43 | 0.002123 | D |
| 628.46 | 0.004263 | D | 782.5624 | 0.002123 | D |
| m/z | Tentative Identification | AUC | p-Value | Log2FC | CRC vs. C |
|---|---|---|---|---|---|
| 598.4875 | Cer(t18:0/19:0) | 0.94056 | 2.0072 × 10−4 | −1.0491 | I |
| 792.5884 | PC(P-18:0/20:5) | 0.88811 | 0.00741 | −2.199 | I |
| 760.578 | PC(18:1/16:0) | 0.88462 | 3.498 × 10−4 | −0.84091 | I |
| 533.2813 | Linoleyl stearate | 0.84615 | 9.6101 × 10−4 | −0.32704 | I |
| 642.5126 | GlcCer(d14:2/16:0) | 0.83566 | 0.0051776 | −0.50237 | I |
| 509.4034 | Stearyl palmitate | 0.83217 | 0.0015194 | −0.30749 | I |
| 758.5642 | PC(18:1(11Z)/16:1(9Z)) | 0.82867 | 0.010393 | −0.8229 | I |
| 675.54 | 20:3 Cholesterol ester | 0.81818 | 0.099092 | −0.68236 | I |
| 551.3605 | Retinol oleate | 0.81119 | 0.0018414 | −0.92325 | I |
| 520.3363 | PC(18:2(9Z,12Z)/0:0) | 0.8042 | 0.032773 | −1.7684 | I |
| 732.5489 | PC(16:0/16:1) | 0.7972 | 0.063087 | −0.74595 | I |
| 341.3039 | 9-Hexadecenoylcholine | 0.78671 | 0.0049616 | 0.84315 | D |
| 485.3469 | PA(22:5(7Z,10Z,13Z,16Z,19Z)/0:0) | 0.78322 | 0.026174 | −0.42322 | I |
| 515.3959 | PA (24:4/0:0) | 0.78322 | 0.010955 | −0.8075 | I |
| 588.4082 | Cer(d18:3/20:1) | 0.77273 | 0.010821 | −1.0561 | I |
| 716.5108 | PE(18:2/16:0) | 0.76923 | 0.10133 | −0.33135 | I |
| 808.5757 | PC(18:0/20:5(5Z,8Z,11Z,14Z,17Z)) | 0.76573 | 0.011384 | 0.35853 | I |
| 663.4599 | PG(14:1/14:1) | 0.76224 | 0.053647 | −1.1222 | I |
| 679.4944 | 20:1 Cholesterol ester | 0.76224 | 0.43243 | 0.85535 | D |
| 359.3152 | Tetracosapentaenoic acid (24:5n-3) | 0.75874 | 0.019955 | 0.79973 | D |
| 597.4554 | DG(16:0/18:0/0:0) | 0.75874 | 0.0029561 | −0.45245 | I |
| 814.5707 | PC(18:0/20:2(5Z,11Z)) | 0.75175 | 0.068745 | −1.2218 | I |
| 355.2819 | MG(18:2(9Z,12Z)/0:0/0:0)[rac] | 0.75175 | 0.042076 | −0.39775 | I |
| 498.3996 | Cer(d18:0/13:0) | 0.75175 | 0.053514 | −0.16722 | I |
| 703.5703 | 22:3Cholesterol ester | 0.75175 | 0.017635 | −0.60197 | I |
| m/z | Tentative Identification | MDA | m/z | Tentative Identification | MDA |
|---|---|---|---|---|---|
| 792.5884 | PC(P-18:0/20:5) | 0.037295 | 828.5433 | PC(22:6/18:3) | 0.007843 |
| 734.5637 | PE(O-18:0/18:0) | 0.031412 | 732.5489 | PE(O-18:0/18:1(9Z)) | 0.007805 |
| 685.4422 | PA(P-18:0/18:2) | 0.03116 | 732.5489 | PE(O-18:0/18:1(9Z)) | 0.007805 |
| 703.5703 | CE(22:3) | 0.028253 | 524.37 | PC(18:0/0:0) | 0.007498 |
| 598.4875 | Cer(t18:0/19:0) | 0.028186 | 429.3186 | Cholesteryl acetate | 0.007431 |
| 520.3363 | PC(18:2(9Z,12Z)/0:0) | 0.018047 | 701.4414 | PA(18:2/18:0) | 0.007245 |
| 515.3959 | PA (24:4/0:0) | 0.015878 | 760.578 | PC(18:1/16:0) | 0.006782 |
| 385.2925 | 22-dehydrocholesterol | 0.013922 | 780.5458 | PC(18:2/18:3) | 0.006763 |
| 804.5443 | PC(18:2/20:5) | 0.013375 | 512.4243 | Cer(d16:0/16:0) | 0.00673 |
| 588.4082 | Cer(d18:3/20:1) | 0.013362 | 794.5973 | PC(P-18:0/20:4 | 0.005945 |
| 544.3374 | PC(20:4/0:0) | 0.009519 | 267.2647 | Norlinoleic acid | 0.004452 |
| 455.333 | Vitamin D3 butyrate | 0.009452 | 533.2813 | Stearyl palmitate | 0.004352 |
| 806.5612 | PC(18:1/20:5) | 0.008977 | 808.5757 | PC(18:0/20:5 (5Z,8Z,11Z,14Z,17Z)) | 0.00432 |
| 341.3039 | 9-Hexadecenoylcholine | 0.008805 | 642.5126 | GlcCer(d14:2/16:0) | 0.004228 |
| 245.0769 | Uridine | 0.008599 | 723.5055 | PG(16:0/16:0) | 0.004146 |
| 828.5433 | PC(22:6/18:3 | 0.007843 | 707.486 | CE(22:1) | 0.004095 |
| m/z | CRCIV/C | CRCIV/I | CRCIV/III | CRCIII/C | Tentative Identification | PubChem |
|---|---|---|---|---|---|---|
| 267.265 | 0.987 | 1.534 * | 1.393 * | 0.708 | Norlinoleic acid | 13932174 |
| 341.304 | 0.715 | 1.079 | 1.635 * | 0.437 | 9-Hexadecenoylcholine | 22155839 |
| 355.282 | 0.957 | 0.715 | 0.556 | 1.721 * | MG(18:2(9Z,12Z)/0:0/0:0)[rac] | 5283469 |
| 359.315 | 0.755 | 3.134 | 1.260 | 0.599 | Tetracosapentaenoic acid (24:5n-3) | 52921801 |
| 385.293 | 1.114 | 0.976 | 0.232 | 4.808 * | 22-Dehydrocholesterol | 5283661 |
| 391.284 | 0.826 | 0.814 | 0.660 | 1.253 | 12-Ketolithocholic acid | 3080612 |
| 455.333 | 0.813 | 0.632 | 0.756 | 1.075 | Vitamin D3 butyrate | 14260146 |
| 485.347 | 1.393 * | 0.933 | 0.829 | 1.680 * | PA(22:5(7Z,10Z,13Z,16Z,19Z)/0:0) | 25099711 |
| 498.400 | 1.122 | 0.700 | 1.267 | 0.885 | Cer(d18:0/13:0) | 52931113 |
| 509.403 | 1.303 * | 0.908 | 0.986 | 1.322 * | Stearyl palmitate | 75778 |
| 515.396 | 2.401 * | 3.506 * | 0.768 | 3.124 * | PA (24:4/0:0) | 138233301 |
| 520.336 | 2.183 * | 0.087 | 3.290 * | 0.664 | PC(18:2(9Z,12Z)/0:0) | 11005824 |
| 522.354 | 1.440 | 0.654 | 1.296 | 1.111 | PC(18:1(9Z)/0:0) | 16081932 |
| 524.370 | 0.341 | 0.293 | 0.509 | 0.670 | PC(18:0/0:0) | 497299 |
| 533.281 | 1.420 * | 1.346 * | 1.061 | 1.338 | Stearyl palmitate | 75778 |
| 544.337 | 0.243 | 0.104 | 0.072 | 3.376 * | PC(20:4(5Z,8Z,11Z,14Z)/0:0) | 24779476 |
| 551.361 | 1.787 * | 0.792 | 1.067 | 1.675 * | Retinol oleate | 11699609 |
| 588.408 | 1.227 | 3.575 * | 0.198 | 6.202 * | Cer(d18:3/20:1) | 70678688 |
| 597.455 | 1.295 | 0.785 | 0.857 | 1.512 * | DG(16:0/18:0/0:0) | 9543688 |
| 598.488 | 1.365 * | 1.022 | 0.567 | 2.405 * | Cer(t18:0/19:0) | 5322154 |
| 628.460 | 0.407 | 0.319 | 0.377 | 1.080 | Cer(t18:0/20:0(2OH)) | 70678864 |
| 642.513 | 1.142 | 0.829 | 0.489 | 2.337 * | GlcCer(d14:2(4E,6E)/16:0) | 70699233 |
| 663.460 | 1.107 | 0.424 | 0.522 | 2.122 | PA(16:0/17:0) | 52929500 |
| 675.540 | 1.532 * | 0.897 | 1.225 | 1.251 | SM(d16:1/16:0) | 52931133 |
| 679.494 | 0.401 | 1.231 | 1.184 | 0.339 | 20:1 Cholesterol ester | 16061337 |
| 685.442 | 0.704 | 0.522 | 0.200 | 3.523* | PA(P-18:0/18:2(9Z,12Z)) | 52929695 |
| 701.441 | 1.637 * | 1.541 * | 2.614 * | 0.626 | PA(18:2(9Z,12Z)/18:0) | 52929468 |
| 701.530 | 1.006 | 2.091 * | 5.920 * | 0.170 | SM(d18:1/16:1) | 52931145 |
| 703.570 | 1.851 * | 0.648 | 1.400 | 1.322 * | 22:3 Cholesterol ester | 70699301 |
| 707.486 | 0.367 | 0.292 | 0.368 | 0.997 | 22:1 Cholesterol ester | 16219158 |
| 716.511 | 0.981 | 0.607 | 0.732 | 1.341 * | PC(P-16:0/16:1(9Z)) | 52923882 |
| 722.512 | 1.042 | 1.157 | 0.881 | 1.184 | PS(O-16:0/16:0) | 52926171 |
| 723.506 | 0.569 | 0.973 | 0.676 | 0.841 | PG(16:0/16:0) | 446440 |
| 732.549 | 1.949 * | 2.911 * | 0.994 | 1.961 * | PE(O-18:0/18:1(9Z)) | 52924982 |
| 734.564 | 2.722 * | 3.062 * | 4.165 * | 0.654 | PE(O-18:0/18:0) | 9547051 |
| 751.521 | 1.567 * | 0.790 | 0.971 | 1.613 * | PG(18:0/16:0) | 52927153 |
| 758.564 | 1.156 | 0.544 | 0.512 | 2.258 * | PC(18:1(11Z)/16:1(9Z)) | 53478719 |
| 760.578 | 2.368 * | 1.818 * | 1.314 * | 1.802 * | PC(18:1(11Z)/16:0) | 53478717 |
| 792.588 | 3.593 * | 5.039 * | 0.280 | 4.850 * | PC(P-18:0/20:5(5Z,8Z,11Z,14Z,17Z)) | 52923964 |
| 794.597 | 0.740 | 0.428 | 0.557 | 1.329 * | PC(P-18:0/20:4(5Z,8Z,11Z,14Z)) | 24779390 |
| 804.544 | 1.052 | 5.931 * | 3.524 * | 0.299 | PC(18:2(9Z,12Z)/20:5(5Z,8Z,11Z,14Z,17Z)) | 52922747 |
| 806.561 | 1.303 * | 0.480 | 0.412 | 3.160 * | PC(18:1(9Z)/20:5(5Z,8Z,11Z,14Z,17Z)) | 24778949 |
| 808.576 | 0.698 | 0.672 | 0.799 | 0.873 | PC(18:0/20:5(5Z,8Z,11Z,14Z,17Z)) | 24778860 |
| 814.571 | 1.030 | 0.236 | 0.818 | 1.260 | PC(18:0/20:2(5Z,11Z)) | 24778848 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Răchieriu, C.; Eniu, D.T.; Moiş, E.; Graur, F.; Socaciu, C.; Socaciu, M.A.; Hajjar, N.A. Lipidomic Signatures for Colorectal Cancer Diagnosis and Progression Using UPLC-QTOF-ESI+MS. Biomolecules 2021, 11, 417. https://doi.org/10.3390/biom11030417
Răchieriu C, Eniu DT, Moiş E, Graur F, Socaciu C, Socaciu MA, Hajjar NA. Lipidomic Signatures for Colorectal Cancer Diagnosis and Progression Using UPLC-QTOF-ESI+MS. Biomolecules. 2021; 11(3):417. https://doi.org/10.3390/biom11030417
Chicago/Turabian StyleRăchieriu, Claudiu, Dan Tudor Eniu, Emil Moiş, Florin Graur, Carmen Socaciu, Mihai Adrian Socaciu, and Nadim Al Hajjar. 2021. "Lipidomic Signatures for Colorectal Cancer Diagnosis and Progression Using UPLC-QTOF-ESI+MS" Biomolecules 11, no. 3: 417. https://doi.org/10.3390/biom11030417
APA StyleRăchieriu, C., Eniu, D. T., Moiş, E., Graur, F., Socaciu, C., Socaciu, M. A., & Hajjar, N. A. (2021). Lipidomic Signatures for Colorectal Cancer Diagnosis and Progression Using UPLC-QTOF-ESI+MS. Biomolecules, 11(3), 417. https://doi.org/10.3390/biom11030417









