Abstract
Metabolomics coupled with bioinformatics may identify relevant biomolecules such as putative biomarkers of specific metabolic pathways related to colorectal diagnosis, classification and prognosis. This study performed an integrated metabolomic profiling of blood serum from 25 colorectal cancer (CRC) cases previously classified (Stage I to IV) compared with 16 controls (disease-free, non-CRC patients), using high-performance liquid chromatography and mass spectrometry (UPLC-QTOF-ESI+ MS). More than 400 metabolites were separated and identified, then all data were processed by the advanced Metaboanalyst 5.0 online software, using multi- and univariate analysis, including specificity/sensitivity relationships (area under the curve (AUC) values), enrichment and pathway analysis, identifying the specific pathways affected by cancer progression in the different stages. Several sub-classes of lipids including phosphatidylglycerols (phosphatidylcholines (PCs), phosphatidylethanolamines (PEs) and PAs), fatty acids and sterol esters as well as ceramides confirmed the “lipogenic phenotype” specific to CRC development, namely the upregulated lipogenesis associated with tumor progression. Both multivariate and univariate bioinformatics confirmed the relevance of some putative lipid biomarkers to be responsible for the altered metabolic pathways in colorectal cancer.
1. Introduction
Colorectal cancer (CRC) is an important public health issue, among the three leading causes of cancer-related mortality in both men and women, according to recent cancer statistics [1,2,3,4], particularly in Western countries but also in developing countries, and is strongly related to lifestyle, stress, food diet and habits.
The early detection and endoscopic resection of adenomatous polyps (premalignant conditions) and screening colonoscopy significantly improve the survival rate, being considered a gold standard for the detection of colorectal neoplasms, beside sigmoidoscopy, colon capsule endoscopy and magnetic resonance colonography. The biopsy specimens of colorectal mucosa and colonic lesions are also useful diagnosis procedures; however, all these techniques are invasive. This is the reason why scientists are more and more keen on using non-invasive techniques with good predictive value and high sensitivity such as metabolomics. As reviewed recently, the management of colorectal cancer was changed radically, using omics technologies for finding diagnosis, stratification and prognosis biomarkers, as well as for treatment monitoring [5].
Metabolomics-based procedures using biofluids (especially blood serum or plasma) assure a systematic screening or fingerprinting of small metabolites (with less than 2000 Da) related to the metabolic signature and pathway alterations in different stages of CRC. Metabolomics and metabonomics offer a qualitative untargeted signature (fingerprint) or a targeted methodology with quantitative evaluation of putative biomarkers [6,7,8,9,10,11].
Metabolomic investigations include mainly gas chromatography or high-performance liquid chromatography coupled with mass spectrometry (GC-MS, HPLC-MS) and magnetic resonance (NMR). By far the most applied technique is based on LC-MS, as it has superior detection and identification capability [12,13,14,15]. Recently, the serum fatty acid profiling of colorectal cancer was reported, using either gas chromatography – mass spectrometry [16,17], NMR [18] or Fourier transform ion cyclotron resonance mass spectrometry [19].
Different metabolic alterations are associated with colorectal cancer (CRC), since cancer cells are able to generate energy even in a nutrient-deficient environment and prefer glycolysis against oxidative phosphorylation as demonstrated for years (the Warburg effect). However, recently this paradigm shifted towards a “reversed Warburg effect”: some cancer cell types, including CRC cells, may synthesize ATP by mitochondrial phosphorylation [20], realizing metabolic remodeling and alterations of mitochondrial respiration [21], opening new research directions for the identification of molecular therapeutic targets, such as fatty acid (FA) synthesis and oxidation.
The metabolization of exogenous glutamine represents another dependence of cancer cell, with many oncogenic mutations affecting glutamine metabolism [22]. Meanwhile, alterations of lipid metabolism in CRC lead to structural changes in cell membranes and disruption of energy homeostasis, cell signaling, gene expression and protein distribution, affecting a number of cell functions, such as proliferation, differentiation, apoptosis, autophagy, necrosis, and drug and chemotherapy resistance [23].
The growing interest related to the role of lipids and their metabolism in cancer development has been presented in previous reviews [1,24,25]. The lipid metabolic pathways are affected in CRC cells and include FA synthesis, desaturation, elongation and mitochondrial oxidation. A plasma lipidomic signature reveals perturbed lipid metabolic pathways and potential lipid biomarkers of human colorectal cancer [26]. The complex lipid metabolic changes may be explained by the high proliferation rate of the CRC cells, with high energetic needs and changes in the serum levels of phospholipid components derived from cell membrane degradation, accompanied by inflammation and changes in the arachidonic acid metabolites in serum or tissue. Recently, an integrated multi-omics approach and lipidomic-based characterization of the lipid metabolism in colorectal cancer were reported [27,28,29,30].
The explanation for increased levels of choline-related metabolites in tumors is probably the result of the accelerated lipid membrane metabolism involved in ATP generation, due to rapid cell proliferation. Glucose changes were consecutive to glycolysis and upregulated in CRC, while increases in 3-hydroxybutyrate, an end metabolite of fatty acids, suggested that the upregulation of fatty acid β-oxidation needed as energy support for cancer cell proliferation [31]. Increased oxidative stress is usually associated with increased oxidation of fatty acids, which may result in an accumulation of 3-hydroxybutyrate [32]. The predictive value of lipid biomarkers is very important, the main predictive factor being considered the stage of diagnosis [4], which explains the importance of CRC screening and early diagnosis.
Different prediction models for CRC patients compared with controls included metabolites such as 2-hydroxybutyrate, aspartic acid, kynurenine and cysteamine [33], or pyruvic, fumaric and glycolic acids; palmitoleic acid; ornithine; lysine tryptophan and 3-hydroxyisovaleric acid [17]. When serum fingerprints from CRC patients were compared prior to surgery and one month after, the potential biomarkers belonged to lipid classes (phosphatidylcholines (PCs), lysophosphatidylcholines (LPCs) and diacylglycerols (DGs)), without a significant difference between the pre-operative and post-operative status [15]. For the assessment of the CRC recurrence rate and survival of the patients after surgical intervention or chemotherapy, 15 metabolites, including four lipids (glycerol, myristate, palmitoleate and 2-aminobutyrate), were selected as potential biomarkers [32].
Recently, a relevant serum MS study for lipophilic metabolites performed under the European Prospective Investigation into Cancer and Nutrition (EPIC), reported nine metabolites to be related to CRC etiology and were recommended for further CRC prospective studies [34]. It was concluded that changes in plasma lipid composition preceded the appearance of neoplasia and that tumor changes can induce a global change in LPC metabolism [35]. Another prospective study found 35 metabolites associated with CRC risk, including 12 glycerophospholipids with an important role in the risk of developing colorectal cancer [36].
Our previous reports focused on the selection of some lipidomic biomarkers to diagnose CRC, based on literature surveys [37,38]. In this context, this experimental study aimed at the identification of specific blood serum biomarkers from patients diagnosed with CRC in four progression stages. The UPLC-QTOF-ESI+ MS results, combined with a succession of multivariate and univariate statistical models, including ANOVA, partial least squares discriminant analysis (PLSDA), cluster analysis, random forest and pathway analysis, showed the predictive value of specific biomolecules to be considered as putative CRC biomarkers.
2. Materials and Methods
2.1. Patients and Compliance with Ethical Standards
The protocol of this study was approved by the Ethics Committee of the Cluj-Napoca “Iuliu Hatieganu” University of Medicine and Pharmacy, including collection of the details about samples and the individual, and the written consent of all subjects, before entering them in the study. The CRC patient group (25 patients) included 16 men and 9 women operated on in the Surgery Department, Regional Institute of Gastroenterology and Hepatology “Octavian Fodor” Cluj-Napoca, Romania, between May and December 2018, with confirmed CRC, either before surgery (suspicion through colonoscopy) or post-surgery (having preoperative radiological suspicion of CRC). The clinical and pathological features as well the stage of CRC tumor were established according to histological evaluation and pTNM classification, as presented in Table 1.

Table 1.
Clinical pathological features of the colorectal cancer (CRC) patients included in this study.
The control group included 7 males (53.1 ± 6.7 years) and 4 females (56.33 ± 8.98 years) considered to be CRC-free, with a negative colonoscopy for cancer or adenoma during the last 12 months. The controls were operated on in the same hospital for other benign diseases (inguinal or incisional hernia, benign skin lesions, cholelithiasis).
Co-morbidities like morbid obesity, insulin-dependent diabetes mellitus, liver cirrhosis, adenomatous polyps and a history of inflammatory bowel disease were excluded in both groups. The blood samples were collected similarly from all participants, in hospital, prior to surgery, in the morning, after a minimum of 12 h fasting.
In parallel, data about other clinical and preclinical parameters of the patients and controls were registered but not included in this report.
2.2. Blood Collection and Processing
Blood serum samples were collected according to standardized procedures in accordance with the ethical standards of the institutional and national research ethical committee and with the 1964 Helsinki Declaration and its later amendments for ethical standards.
The blood was collected in vacutainer tubes without anticoagulant, kept at room temperature for 30 min to allow clotting and centrifuged for 10 min at 3000 rpm (4 °C) to separate clear serum. After separation, the blood serum was stored at −80 °C. To a volume of 0.2 mL serum, 0.8 mL of a mixture of methanol and acetonitrile (1:1) was added to precipitate proteins. The mixture was vortexed for 1 min, kept at −20 °C overnight and then vortexed again for 1 min. After mixing, the vials were centrifuged at 12,500 rpm for 10 min and the supernatant was collected and filtered through PTFE filters of 0.25 µm.
2.3. HPLC–ESI(+)-QTOF-MS Analysis of Blood Serum
Aliquots of 3 µL of serum were subjected to ultrahigh-pressure chromatography on a Thermo Scientific HPLC UltiMate 3000 (Waltham, MA, USA) system equipped with a quaternary pump system DionexUltiMate 3000 (UHPLC) (ThermoFischer, Waltham, MA, USA), a DionexUltimate 3000 photodiode array detector, a column oven and autosampler. Serum metabolites were separated using a Thermo Scientific C18 reverse-phase column (Acquity, UPLC C18 BEH, Waters Corporation, Milford, MA, USA) (5 µm, 2.1 × 75 mm) at 25 °C and a flow rate of 0.3 mL/min. The mobile phase was represented by a gradient of Eluent A (water containing 0.1% formic acid) and Eluent B (methanol:acetonitrile, 1:1, containing 0.1% formic acid). The gradient system consisted of 99% A (Minute 0), 70% A (Minute 1), 40% A (Minute 2), 20% A (Minute 6) and 100% B (Minute 9–10), followed by 5 min with 99% A. The total running time was 15 min. The mass spectrometry was performed on a Bruker Daltonics MaXis Impact QTOF (Bremen, Germany) instrument, operating in positive ion mode (ESI+). The mass range was set between 50 and 1000 m/z. For measurements, the nebulizing gas pressure was set at 2.8 bar, the drying gas flow at 12 L/min and the drying gas temperature at 300 °C. Before each chromatographic run, a calibrant solution of sodium formate was injected. The control of the instrument, the acquisition and data processing were done using Chromeleon, TofControl 3.2, Hystar 3.2 and Data Analysis 4.2 (Bruker Daltonics, Bremen, Germany).
2.4. Statistical Analysis
The Bruker software attached to the instrument, Data Analysis 4.2, was used to process the acquired data. First, from the total ion chromatogram, using specific algorithms base peak chromatograms were obtained, and the Find Molecular Features (FMF) algorithm generated an advanced bucket matrix. The matrix released by Data Analysis contained the retention time, the peak areas and intensities and the signal/noise (S/N) ratio for each component, together with its m/z value. Generally, the number of separated compounds ranged between 600 and 800.
In this first step, a matrix for all samples was obtained and stored in an Excel file. In order to eliminate the small signals with S/N values under 10, an initial filtration (1) was made and then a second matrix containing m/z values and peak intensities was saved and filtered in a second step eliminating the small intensities (2). Generally, the number of peaks remained at 180–220. Only metabolites which were detected in more than 80% of the samples were included in the statistical analysis, so to make an adequate alignment of the peak m/z values, the online software from bioinformatica.isa.cnr.it/NEAPOLIS was applied. The aligned matrix (3) allowed the calculation of mean Intensity values and standard deviations for the control group and for subgroups corresponding to CRC Stages I–IV. The aligned matrix was then converted to a csv file and introduced in the specialized online software Metaboanalyst 5.0.
After successive alignment and normalization of the matrix data, the multivariate analysis consisted of the representation of fold change, volcano plot, principal component analysis (PCA), partial least squares discriminant analysis (PLSDA) and random forest, finding correlations between samples and between variables (m/z values), as well as building the heatmap which represents the correlation between variables and samples. Finally, using the biomarker analysis, the receiver operating curves (ROCs) were obtained and the values of the areas under the ROC curves (AUCs) were obtained and the molecules identified were ranked according to their sensitivity/specificity. The enrichment analysis and the MS to pathways algorithm allowed the identification of specific alterations of metabolic pathways induced in CRC.
The identification of molecules which can be considered potential biomarkers was made using the 2 most relevant databases, LIPID MAPS Lipidomics Gateway and the Human Metabolome Database.
3. Results
3.1. Multivariate Analysis
3.1.1. PCA and PLSDA Analysis
By unsupervised PCA, the co-variance for the first five components was evaluated. The explained variance in serum groups (CRC and C) was 25.6% (PC1) and 13.3% (PC2), covered by a total variance of 38.9% (Figure 1a). The discrimination between CRC and C groups was better represented by PLSDA (covariance of 35.1%) (Figure 1b).
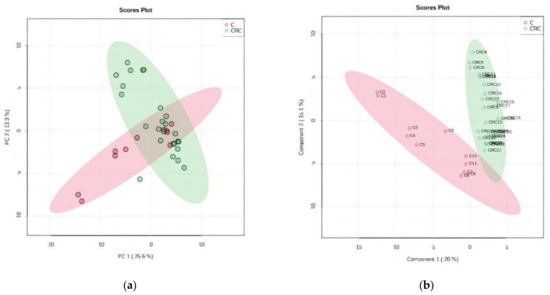
Figure 1.
(a) Principal component score plot showing the homogeneity and C and CRC groups. (b) Partial least squares discriminant analysis (PLSDA) plot with sample identification, showing the discrimination between C and CRC groups.
According to the PCA and PLSDA plots, the C group was less homogeneous than the CRC group, two or three subgroups being visible in this group. This can be explained by the diverse co-morbidities of the patients from this non-CRC group.
The cross-validation algorithm showed a high accuracy (close to 1), high R2 and significantly high Q2 values, its performance increasing from Component 1 to 3, up to 0.7 (Figure S1a, Supplementary Materials). These data indicated very good validation and predictability for this model.
3.1.2. Euclidean Dendrogram and Correlation Heatmaps
Figure 2 shows the hierarchical clustering of different samples, displayed as a tree diagram called a dendrogram. The hierarchical clustering dendrogram was chosen, with an Euclidian distance measure and the Ward clustering algorithm, one of the options found in Metaboanalyst 5.0. Using the Euclidian algorithm, in the scale of distances from 1–60, one can see good similarities (distances of <35) between the individual samples from the CRC group. The pathologic CRC group (marked in green) showed a good similarity while the group C (marked red) was split into two subgroups.
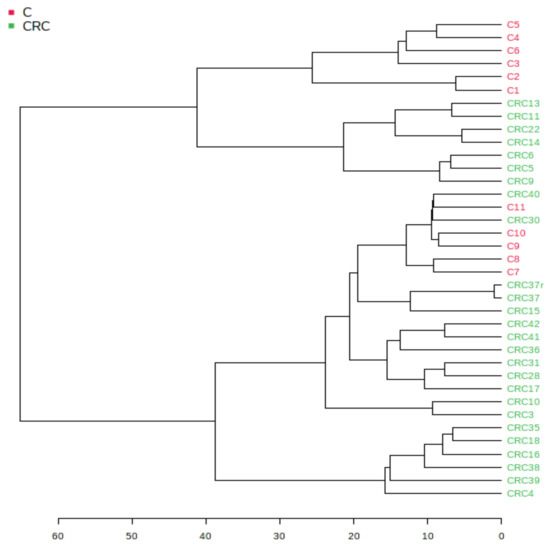
Figure 2.
Hierarchical clustering dendrogram of samples using the Euclidian distance measure and the Ward clustering algorithm.
Figure S2a–c (Supplementary Materials) represents the maps of correlation between variables (m/z values) and between samples, as well the heatmap showing the correlations between samples and variables. These maps were built considering the t-test and p-values obtained by applying the Metaboanalyst 5.0. algorithm. The colors represent positive (red) or negative (blue) correlations as determined by the t-test. One can discriminate different patterns for samples correlated with variables (blue zones differentiated from red zones). In the heatmap (Figure S2c), the red spots show the molecules which have increased levels in certain samples, while blue spots reflect decreases in certain molecules for specific samples.
3.1.3. The Random Forest Algorithm and Its Predictive Value
This algorithm was able to indicate the predictive value (as potential biomarkers) for some molecules which differentiated the CRC and C groups. Table 2 presents the m/z values of the first 30 molecules to be considered as predictive by the random forest algorithm. The MDA values from 0.012 to 0.002 were considered and the decrease (D) or increase (I) in the level of these molecules in the CRC vs. C groups was seen.

Table 2.
The m/z values of the first 30 molecules to be considered a predictive by the random forest algorithm. The MDA values from 0.012 to 0.002 were considered and a decrease (D) or increase (I) in the level of each molecule in the CRC vs. C groups.
3.1.4. Biomarker Analysis
According to Metaboanalyst software, biomarker analysis includes the receiver operating characteristic (ROC) curve as a useful tool to evaluate the diagnostic accuracy. Many biomarker combination methods rely on maximization of the area under the ROC curve (AUC). This parameter allowed the evaluation of the sensitivity versus specificity of each molecule to be considered a relevant biomarker. Higher values of AUC close to 1 for a certain molecule mean higher prediction to be considered as a biomarker. Table 3 shows the m/z value and putative identification, AUC value, p-values and log2FC values for each molecule identified, as well its variation in the CRC group vs. C group.

Table 3.
The m/z, area under the curve (AUC), p-values and log2FC values, and identification of molecules, based on HMDB and Lipidmaps. Variation in the PS group versus the CS group: I, increase; D, decrease. Identified only blood.
Significantly high AUC values above 0.750 showed that 25 molecules might be considered as putative biomarkers; these molecules belong to different lipid classes.
These data confirm that lipid molecules, mainly choline-dependent phospholipids, ceramides and different esters (of fatty acids or cholesterol), can be considered as predictive molecules with high prognostic values for CRC diagnosis.
3.2. Univariate Analysis ANOVA: Discrimination of CRC Stages
3.2.1. One-Way ANOVA to Identify Biomarkers for CRC Progression (Stages I to IV)
Applying the ANOVA univariate analysis, included in the Metaboanalyst software, Figure 3a,b presents the PCA and PLSDA plots. The PCA plot showed a covariance of 38.9%, while PLSDA showed a covariance of 28% that was able to discriminate between the C and CRCI–IV groups. In the PCA plot, the subgroups CRCIV and CRCII are well discriminated as well, in the opposite direction compared with the C group and the CRC I and III subgroups. Interestingly, in the PLSDA plot, the CRCIII subgroup had a significant difference from the other subgroups. Finally, considering the inputs from these plots, we consider that CRCIII and CRCIV are the CRC stages to show significant differences to be considered in chemo-statistic evaluations. The cross-validation graphic (Figure S1b) shows the high accuracy and significance of the PLSDA model: an accuracy value close to 1 indicates a very good description of the data by the model, whereas the R2 and high Q2 values confirm the model’s performance, increasing from Component 1 to 5, up to 0.7.
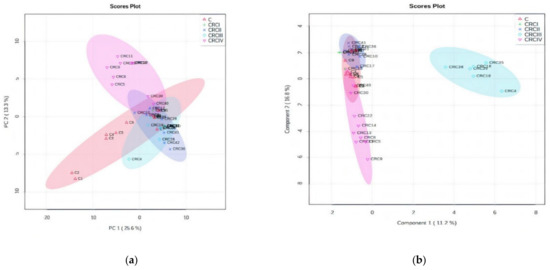
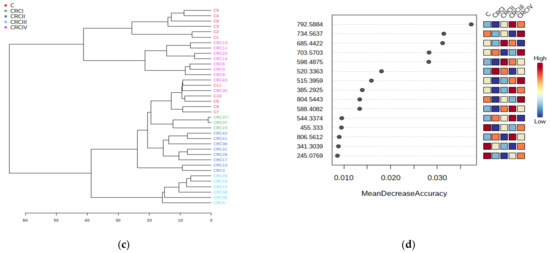
Figure 3.
ANOVA of CRC stages. (a) Principal component analysis (PCA) score plot (b) PLSDA score plot. (c) Dendrogram showing the clustering of CRC I–IV and C subgroups. (d) The graph of random forest analysis: MDA values for the first 15 predictive molecules.
The dendrogram (Figure 3c) shows the clustering of subgroups CRC I–IV and Figure 3d shows the MDA values of the model, as determined by the random forest algorithm, including the first 15 predictive molecules.
As can be seen in Figure 3a,b, clear delimitation of Stages III and IV were observed either by the PCA or PLSDA score plots. The loading analysis and MDA values showed which are the molecules considered to be responsible for this strong discrimination. Table 4 includes the first 30 molecules and their MDA values (up to 0.002). The increase (I) or decrease (D) of these molecules in the CRC vs. C group the included.

Table 4.
The m/z values of the first 30 molecules to be considered predictive by the random forest algorithm. The MDA values from 0.037 to 0.004 were considered, and the decrease (D) or increase (I) in the level of each molecule in the CRC vs. C groups is shown.
3.2.2. Statistical Analysis Based on MS Peak Intensity Values for CRC Subgroups
In order to compare the data obtained by multivariate analysis, we considered the initial matrices (peak intensity tables), considering the mean values for the CRCI-IV and C groups, and calculated the statistical differences between these groups (values from p < 0.1 to p < 0.01 indicate significance). From a total of 93 biomolecules (included in Table S1), significant deviations of these ratios (p < 0.01) were selected. The data released from LC-MS analysis were presented in a matrix representing the average values of MS peak intensity values for each molecule separated and selected according to the protocol presented above (n = 45). Table S1 includes the list of common molecules identified in blood serum from C and CRC groups. The ratios between the mean values of CRCIV and C, CRCIV and I, CRCIV and III, and CRCIII and C are presented in Table 5, as well the tentative identification of the molecules and their codification in the PubChem database.

Table 5.
M/z values and tentative identification of molecules which show different ratios between the mean values of CRCIV and C, CRCIV and I, CRCIV and III, and CRCIII and C. The codification in the PubChem database is included for each molecule. Significant increases (p < 0.01) in these ratios are marked with * symbol.
These data suggest general increases in different lipid subclasses such as choline-dependent glycerophspholipids, cholesterol and fatty acid esters, as well ceramides, especially in Stages III and IV compared with controls. Stearic and palmitic acids are mostly involved in such esters. These data are in good agreement with the multivariate analysis.
3.2.3. Enrichment and Pathway Analysis
Using enrichment analysis and pathway analysis by the GSEA algorithm for the matrix including the m/z values presented in Table 5 and decreasing p-values and t-scores, the possible pathways affected by CRC in different stages were obtained.
Figure 4 presents a general and detailed overview of the enriched metabolite classes sets and their significance.
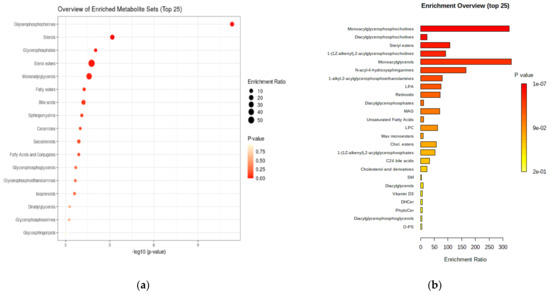
Figure 4.
(a) General overview of the enriched metabolite class sets (top 17) expressed as log 10 (p-value). (b) Detailed enrichment overview for molecule subsets and their enrichment ratios.
The results plotted above confirm the metabolite sets which can be considered as significant in CRC diagnosis and prognosis, including their classifications. The main class of metabolites is represented by the glycerophosphocholines (mono- and diacyl derivatives), followed by sterols and their esters, ceramides and sphingomyelins.
4. Discussion
The results of this study may contribute to the actual knowledge directed towards identification of the most relevant biomarkers of CRC and progression stage subtypes, compared with controls. These data are mostly in good agreement with previous findings which reflect the key role of lipid-mediated pathways in CRC diagnosis and prognosis.
As reported before, choline-related phospholipids can be considered good biomarkers for CRC [39,40]. Increases in PCs may be followed by decreases in LPCs, associated with body weight loss and activated inflammatory processes in CRC patients [41] but also by accumulation of some LPCs (16:0, 16:1 and 18:0) [42] oran increased degradation rate of some LPCs (20:4 and 22:6) as a result of the accelerated cell proliferation in CRC patients [36,43]. Another lipid metabolic signature represented by palmitic amide, oleamide, octadecanoic, hexadecanedioic, myristic and eicosatrienoic acids, LPCs(16:0, 18:2, 20:4, 22:6) was statistically significant and these lipid metabolites were considered potential biomarkers to discriminate early-stage patients from healthy controls, superior to the prediction made by carcinoembryonic antigen [40]. Endogenous synthesis of arachidonic and oleic acids was also reported to have an impact on CRC development, as well as the arachidonic acid metabolites (eicosanoids and their oxidized forms), which generate prostaglandin E2 which stimulates tumorigenesis [44]. Meanwhile, no significant differences between normal, polyp and cancer mucosa were noticed for oxidized lipids 12-hydroxyeicosatetraenoic acid (HETE), 15-HETE or leukotriene B4 levels, or decreased 13-hydroxyoctadecadienoic acid (HODE) and HETE levels in cancer and colorectal polyp mucosa [15,27,45]. The upregulated and downregulated metabolites through the various stages of CRC were found to be also benzoic, octanoic and decanoic acids, proportional to CRC stage [35,46,47]. Glyceraldehyde, hippuric and linolenic acids, glycochenodeoxycholate and glycocholate may also discriminate CRC from polyps [12], while β-hydroxibutyrate increased and tryptophan and indoleacrylic acid decreased from Stage I to Stage IV CRC [48,49,50]. By comparison, the healthy, polyp adenomas and CRC patients had different glycerolipid metabolism, reflected by higher levels of lipids and polyunsaturated fatty acids (PUFAs) and lower levels of glycerol [18].
In plasma, previous studies reported the upregulation of fatty acid synthesis, increased LPCs (26:0 and 28:0) [47,50] and other types of LPC [19,27,39], increased Monounsaturated fatty acid (MUFA)/PUFA ratios [51], and increased ethanolamine plasmalogens, polyunsaturated fatty acids and polar lipids [26,27,51,52]. In serum, there were noticeable increases in triglycerides containing C15:0,18:0, 18:1,18:2 and 18:3 [53,54] or decreases in lipids with C22:0, 24:0, 26:0, 30:0 and 18:1 [16]; and decreases in ultralong chain fatty acids [34]; increases in endocannabinoids and ceramides, sphingomyelins, eicosanoids [8,31,43,46], succinate, dimetihylguanosine, adenine, citraconic acid and methylguanosine [40]. Cerotic acid may also be a novel serum metabolic marker of colorectal malignancies [55], as well as plasma triacylglycerols [56,57].
To summarize the findings presented above, in relation to our results we can assume that in CRC, several classes of lipids including phosphatidylglycerols (PCs, phosphatidylethanolamines (PEs) and phosphatidic acids (PAs)), fatty acids and sterol esters, as well as ceramides, confirm the “lipogenic phenotype” for CRC development, dependent on lipogenesis and lipolysis, upregulated and associated with tumor progression. Both multivariate and univariate bioinformatics confirm these findings and the specificity of these metabolic pathways activated in CRC patients [58].
Further studies are under development using larger cohorts of patients in different CRC stages, with improved characterization and data processing.
5. Conclusions
Metabolomics has already proven a great potential as a high-value technology to realize proper metabolic signatures to discriminate significantly between healthy controls with benign polyps versus malignant CRC tumors. Specific classes of lipids involved in cellular signaling and energy provision proved to be good biomarkers for CRC in different stages and can be relevant prognosis factors. The lipid profile alterations presented in this study, many of them also confirmed by similar investigations, showed statistically significant differences and can be considered reliable biomarkers, differentiating between early and advanced stages of this malignancy, or serving as survival predictors. Complementary studies on larger cohorts of patients are needed for the development of clinically useful biomarkers, especially related to the signaling lipids.
Metabolomics have the potential to become a standard technology for future applications in translational cancer research, but further, large-scale studies and prospective validation are still needed. Moreover, bioinformatics tools offered by the online Metaboanalyst 5.0 software significantly helped refining of the key biomolecules which may be considered as putative biomarkers for CRC diagnosis and staging. These biomarkers are not only useful for diagnostics and patient stratification but can be mapped on a biochemical chart to identify the altered metabolic pathways involved in the initiation and progression of this invasive cancer.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/11/3/417/s1, Table S1. Matrix representing the m/z values of molecules separated by LC-MS (column 1) and the mean values of mass spectra peak intensity for the C group (column 2) and CRC subgroups representing Stages I, II, III and IV (Columns 3–6). Figure S1. (a) Cross-validation of PLSDA analysis for the CRC and C groups. For interpretation, see the main text. (b) Cross-validation of PLSDA analysis for CRC I to IV subgroups. For interpretation, see the manuscript. Figure S2. Correlation maps between variables (m/z values) (a) and samples (b). The heatmap (c) represents the correlation between samples and variables. All maps were built using the Metaboanalyst 5.0 algorithm (multivariate statistics and t-test).
Author Contributions
Conceptualization, C.R., D.T.E. and C.S., methodology; C.S. and M.A.S.; software, C.S. and M.A.S.; validation D.T.E. and N.A.H.; investigation, E.M., F.G. and N.A.H.; resources, C.R., D.T.E., E.M. and F.G.; data curation, C.R. and D.T.E.; writing—original draft preparation, C.R. and C.S.; writing—review and editing, C.R., C.S. and D.T.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Committee) of the University of Medicine and Pharmacy “Iuliu Hatieganu” Cluj-Napoca, Romania (protocols approved in May 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
The authors are grateful for the technical and logistic support offered by the Research center BIODIATECH (biodiatech.ro) and the Medical Center Medisyn from Cluj-Napoca, Romania.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Cer | ceramide |
| CRC | colorectal cancer |
| DG | diacylglycerol |
| FFA | free fatty acid |
| GlcCer | glucosylceramide |
| HETE | hydroxyeicosatetraenoic acid |
| HODE | 13-hydroxyoctadecadienoic acid |
| LA | linoleic acid |
| LPA | lysophosphatidic acid |
| LPC | lysophosphatidylcholine |
| MG | monoacylglycerol |
| MUFA | monounsaturated fatty acid |
| PA | phosphatidic acid |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| PL | phospholipid |
| PUFA | polyunsaturated fatty acid |
| SM | sphingomyelin |
| SPL | sphingolipid |
| TG | triacylglycerol. |
References
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A. Colorectal cancer statistics. CA Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Cancer Facts & Figures. 2019. Available online: https://www.cancer.org/research/cancer-facts-statistics.html (accessed on 24 January 2021).
- American Cancer Society. Colorectal Cancer Facts & Figures 2020–2022; American Cancer Society: Atlanta, GA, USA, 2020. [Google Scholar]
- Beggs, A.D.; Dilworth, M.P. Surgery in the era of the ’omics revolution. Br. J. Surg. 2015, 102, e29–e40. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98. [Google Scholar] [CrossRef]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Abu Zaid, M.; Chiorean, E.G.; Raftery, D. Targeted serum metabolite profiling and sequential metabolite ratio analysis for colorectal cancer progression monitoring. Anal. Bioanal. Chem. 2015, 407, 7857–7863. [Google Scholar] [CrossRef]
- Johnson, C.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell. Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, D.J.; Hoeferlin, L.A.; Chalfant, C.E. Lipidomics in translational research and the clinical significance of lipid-based biomarkers. Transl. Res. 2017, 189, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Muc-Wierzgoń, M.; Nowakowska-Zajdel, E.; Dzięgielewska-Gęsiak, S.; Kokot, T.; Klakla, K.; Fatyga, E.; Grochowska-Niedworok, E.; Waniczek, D.; Wierzgoń, J. Specific metabolic biomarkers as risk and prognostic factors in colorectal cancer. World Gastroenterol. 2014, 20, 9759–9774. [Google Scholar] [CrossRef]
- Djukovic, D.; Zhang, J.; Raftery, D. Colorectal cancer detection using targeted LC-MS metabolic profiling. In Colorectal Cancer. Methods in Molecular Biology; Beaulieu, J.F., Ed.; Humana Press: New York, NY, USA, 2018; Volume 1765, pp. 229–240. [Google Scholar] [CrossRef]
- Triebl, A.; Trötzmüller, M.; Hartler, J.; Stojakovic, T.; Köfeler, H.C. Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J. Chromatogr. B 2017, 1053, 72–80. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, H.; Liu, R.; Gao, R.; Yang, X.; Jin, W.; Zhang, Y.; Bi, K.; Li, Q. A novel strategy for targeted lipidomics based on LC-tandem-MS parameters prediction, quantification, and multiple statistical data mining: Evaluation of lysophosphatidylcholines as potential cancer biomarker. Anal. Chem. 2019, 91, 3389–3396. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, C.; Gui, Q.; Chen, Y.; Yang, Y. Ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry-based metabolic profiling of human serum prior to and following radical resection of colorectal carcinoma. Mol. Med. Rep. 2015, 12, 6879–6886. [Google Scholar] [CrossRef][Green Version]
- Kondo, Y.; Nishiumi, S.; Shinohara, M.; Hatano, N.; Ikeda, A.; Yoshie, T.; Kobayashi, T.; Shiomi, Y.; Irino, Y.; Takenawa, T.; et al. Serum fatty acid profiling of colorectal cancer by gas chromatography/mass spectrometry. Biomark. Med. 2011, 5, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Kawana, S.; Unno, Y.; Sakai, T.; Okamoto, K.; Yoshida, M. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget 2017, 8, 17115–17126. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics analysis in serum from patients with colorectal polyp and colorectal cancer by 1H-NMR spectrometry. Dis. Markers 2019. [Google Scholar] [CrossRef]
- Li, F.; Qin, X.; Chen, H.; Qiu, L.; Guo, Y.; Liu, H.; Li, Z.L. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 24–34. [Google Scholar] [CrossRef]
- Xu, X.D.; Shao, S.X.; Jiang, H.P.; Cao, Y.W.; Wang, Y.H.; Yang, X.C.; Wang, Y.L.; Wang, X.S.; Ni, H.T. Warburg effect or reverse Warburg effect? A review of cancer metabolism. Oncol. Res. Treat. 2015, 38, 117–122. [Google Scholar] [CrossRef]
- Chekulayev, V.; Mado, K.; Shevchuk, I.; Koit, A.; Kaldma, A.; Klepinin, A.; Ounpuua, L.; Heckc, K.; Truud, L.; Plankene, A.; et al. Metabolic remodeling in human colorectal cancer and surrounding tissues: Alterations in regulation of mitochondrial respiration and metabolic fluxes. Biochem. Biophys. Rep. 2015, 4, 111–125. [Google Scholar] [CrossRef]
- Hao, Y.; Samuels, Y.; Li, Q.; Krokowski, D.; Guan, B.-J.; Wang, C.; Wang, Z. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat. Commun. 2016, 7, 11971. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, Y.Y.; Harris, J.W.; Mitov, M.I.; Kim, J.T.; Butterfield, D.A.; Lee, E.Y.; Weiss, H.L.; Evers, B.M. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget 2015, 6, 18891–18904. [Google Scholar] [CrossRef] [PubMed]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and Cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29–38. [Google Scholar] [CrossRef]
- Shen, S.; Yang, L.; Li, L.; Bai, Y.; Cai, C.; Liu, H.C. A plasma lipidomics strategy reveals perturbed lipid metabolic pathways and potential lipid biomarkers of human colorectal cancer. J. Chromatogr. B 2017, 1068–1069, 41–48. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, B.; Zhang, J.; Li, J.; Yang, Q.; Zhong, Q.; Zhan, S.; Liu, H.; Cai, C. Serum polyunsaturated fatty acid metabolites as useful tool for screening potential biomarker of colorectal cancer. Prostaglandins Leukot. Essent. Fat Acids 2017, 120, 25–31. [Google Scholar] [CrossRef]
- Yan, F.; Zhao, H.; Zeng, Y. Lipidomics: A promising cancer biomarker. Clin. Trans. Med. 2018, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Charkoftaki, G.; Thompson, D.C.; Golla, J.P.; Garcia-Milian, R.; Lam, T.K.T.; Engel, J.; Vasiliou, V. Integrated multi-omics approach reveals a role of ALDH1A1 in lipid metabolism in human colon cancer cells. Chem. Biol. Interact. 2019, 304, 88–96. [Google Scholar] [CrossRef]
- Wang, Y.; Hinz, S.; Uckermann, O.; Hönscheid, P.; Von Schönfels, W.; Burmeister, G.; Hendricks, A. Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. BBA Mol. Cell Biol. Lipids 2020, 1865, 158579. [Google Scholar] [CrossRef]
- Jung, J.H.; Taniguchi, K.; Lee, H.M.; Lee, M.Y.; Bandu, R.; Komura, K.; Lee, K.Y.; Akao, Y.; Kim, K.P. Comparative lipidomics of 5-Fluorouracil–sensitive and –resistant colorectal cancer cells reveals altered sphingomyelin and ceramide controlled by acid sphingomyelinase (SMPD1). Sci. Rep. 2020, 10, 6124. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Cai, G.; Zhou, B.; Li, D.; Zhao, A.; Xie, G.; Li, H.A.; Cai, S.; Xie, D.; Huang, C.; et al. Distinct metabolic signature of human colorectal cancer with prognostic potential. Clin. Cancer Res. 2014, 20, 2136–2147. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Yoshida, M. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef] [PubMed]
- Perttula, K.; Schiffman, C.; Edmands, W.M.B.; Petrick, L.; Grigoryan, H.; Cai, X.; Rappaport, S.M. Untargeted lipidomic features associated with colorectal cancer in a prospective cohort. BMC Cancer 2018, 18, 996. [Google Scholar] [CrossRef] [PubMed]
- Kühn, T.; Floegel, A.; Sookthai, D.; Johnson, T.; Rolle-Kampczyk, U.; Otto, W.; Kaaks, R. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Xiang, Y.B.; Rothman, N.; Yu, D.; Li, H.L.; Yang, G.; Zheng, W. Prospective study of blood metabolites associated with colorectal cancer risk. Int. J. Cancer 2018, 143, 527–534. [Google Scholar] [CrossRef]
- Răchieriu, C.; Eniu, D.T.; Moiş, E.; Graur, F.; Socaciu, C.; Socaciu, M.A.; Alhajjar, N. Lipidomics: Advanced analytical technology to identify biomarkers of colorectal cancer. Studia Univ. Babes Bolyai Chem. 2020, 65, 203–215. [Google Scholar] [CrossRef]
- Răchieriu, C.; Graur, F.; Moiş, E.; Socaciu, C.; Eniu, D.T.; Alhajjar, N. Metabolomic profile of colorectal cancer patients and its clinical implications. Rom. Biotechnol. Lett. 2020, 25, 2045–2054. [Google Scholar] [CrossRef]
- Zhao, Z.; Xiao, Y.; Elson, P.; Tan, H.; Plummer, S.J.; Berk, M.; Aung, P.P.; Lavery, I.C.; Achkar, J.P.; Li, L.; et al. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J. Clin. Oncol. 2007, 25, 2696–2701. [Google Scholar] [CrossRef]
- Li, S.; Guo, B.; Song, J.W.; Deng, X.L.; Cong, Y.; Li, P.; Zhao, K.; Liu, L.; Xiao, G.; Xu, F.; et al. Plasma choline-containing phospholipids: Potential biomarkers for colorectal cancer progression. Metabolomics 2013, 9, 202–212. [Google Scholar] [CrossRef]
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Jia, W. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef] [PubMed]
- Kurabe, N.; Hayasaka, T.; Ogawa, M.; Masaki, N.; Ide, Y.; Waki, M.; Nakamura, T.; Kurachi, K.; Kahyo, T.; Shinmura, K.; et al. Accumulated phosphatidylcholine (16:0/16:1) in human colorectal cancer; possible involvement of LPCAT4. Cancer Sci. 2013, 104, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Geijsen, J.M.R.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.J.B.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer 2019, 145, 1221–1231. [Google Scholar] [CrossRef]
- Zuo, X.; Shureiqi, I. Eicosanoid profiling in colon cancer: Emergence of a pattern. Prostaglandins Other Lipid Mediat. 2013, 104–105, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Shureiqi, I.; Chen, D.; Day, R.S.; Zuo, X.; Hochman, F.L.; Ross, W.A.; Lippman, S.M. Profiling lipoxygenase metabolism in specific steps of colorectal tumorigenesis. Cancer Prev. Res. 2010, 3, 829–838. [Google Scholar] [CrossRef]
- Crotti, S.; Agnoletto, E.; Cancemi, G.; Di Marco, V.; Traldi, P.; Pucciarelli, S.; Agostini, M. Altered plasma levels of decanoic acid in colorectal cancer as a new diagnostic biomarker. Anal. Bioanal. Chem. 2016, 408, 6321–6328. [Google Scholar] [CrossRef]
- Uchiyama, K.; Yagi, N.; Mizushima, K.; Higashimura, Y.; Hirai, Y.; Okayama, T.; Naito, Y. Serum metabolomics analysis for early detection of colorectal cancer. J. Gastroenterol. 2017, 52, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Farshidfar, F.; Kopciuk, K.A.; Hilsden, R.; Mcgregor, S.E.; Mazurak, V.C.; Buie, W.D. A quantitative multimodal metabolomic assay for colorectal cancer. BMC Cancer 2018, 18, 26. [Google Scholar] [CrossRef]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.; Hilsden, S.E.; Mcgregor, F.; Buie, W.D.; Bathe, O.F. A validated metabolomic signature for colorectal cancer: Exploration of the clinical value of metabolomics. Br. J. Cancer 2016, 115, 848–857. [Google Scholar] [CrossRef]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.; Buie, W.D.; Maclean, A.; Dixon, E.; Sutherland, F.R.; Molckovsky, A.; Vogel, H.J.; Bathe, O.F. Serum metabolomic profile as a means to distinguish stage of colorectal cancer. Genome Med. 2012, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Butler, L.M.; Yuan, J.M.; Huang, J.; Su, J.J.; Wang, R.; Koh, W.-P.; Ong, C.-N. Plasma fatty acids and risk of colon and rectal cancers in the Singapore Chinese Health Study. Precis. Oncol. 2017, 1, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, L.; Ye, X.; Chen, L.; Zhang, L.; Gao, Y.; Kang, J.X.; Cai, C. Characteristics of fatty acid distribution is associated with colorectal cancer prognosis. Prostaglandins Leukot. Essent. Fat Acids 2013, 88, 355–360. [Google Scholar] [CrossRef]
- Liu, T.; Tan, Z.; Yu, J.; Peng, F.; Guo, J.; Meng, W.; Chen, Y.; Rao, T.; Liu, Z.; Peng, J. A conjunctive lipidomic approach reveals plasma ethanolamine plasmalogens and fatty acids as early diagnostic biomarkers for colorectal cancer patient. Expert Rev. Proteom. 2020, 17, 233–242. [Google Scholar] [CrossRef]
- Guo, S.; Wang, Y.; Zhou, D.; Li, Z. Significantly increased monounsaturated lipids relative to polyunsaturated lipids in six types of cancer microenvironment are observed by mass spectrometry imaging. Sci. Rep. 2014, 4, 5959. [Google Scholar] [CrossRef]
- Mika, A.; Kobiela, J.; Czumaj, A.; Chmielewski, M.; Stepnowski, P.; Sledzinski, T. Hyper-elongation in colorectal cancer tissue-cerotic acid is a potential novel serum metabolic marker of colorectal malignancies. Cell. Physiol. Biochem. 2017, 41, 722–730. [Google Scholar] [CrossRef]
- Mika, A.; Pakiet, A.; Czumaj, A.; Kaczynski, Z.; Liakh, I.; Kobiela, J.; Perdyan, A.; Adrych, K.; Makarewicz, W.; Sledzinski, T. Decreased triacylglycerol content and elevated contents of cell membrane lipids in colorectal cancer tissue: A lipidomic study. J. Clin. Med. 2020, 9, 1095. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Peng, F.; Yu, J. LC-MS-based lipid profile in colorectal cancer patients: TAGs are the main disturbed lipid markers of colorectal cancer progression. Anal. Bioanal. Chem. 2019, 411, 5079–5088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, Y.; Zhao, W.; Deng, K.; Wang, Z.; Yang, C.; Ma, L.; Openkova, M.S.; Hou, Y.; Li, K. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: A systematic review. Oncotarget 2017, 8, 35460–35472. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).