Rhizosphere Bacterium Rhodococcus sp. P1Y Metabolizes Abscisic Acid to Form Dehydrovomifoliol
Abstract
1. Introduction
2. Materials and Methods
2.1. Labeling ABA with Tritium
2.2. Bacteria Cultivation
2.3. HPLC-MS Analysis Conditions
2.4. Isolation of Metabolite I
2.5. Spectrometric Analyses
2.6. Molecular Geometry Calculations
2.7. Statistical Analysis
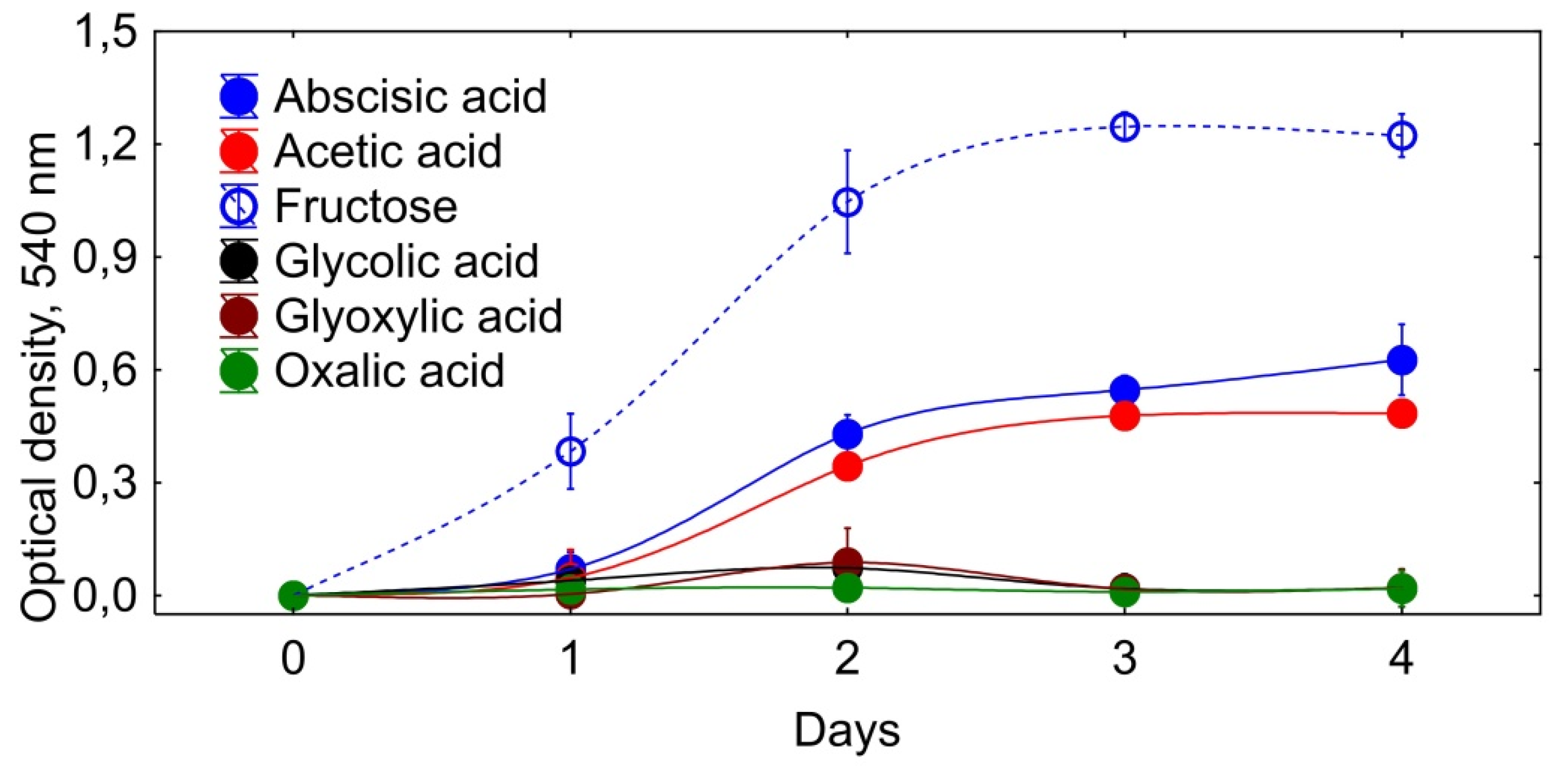
3. Results
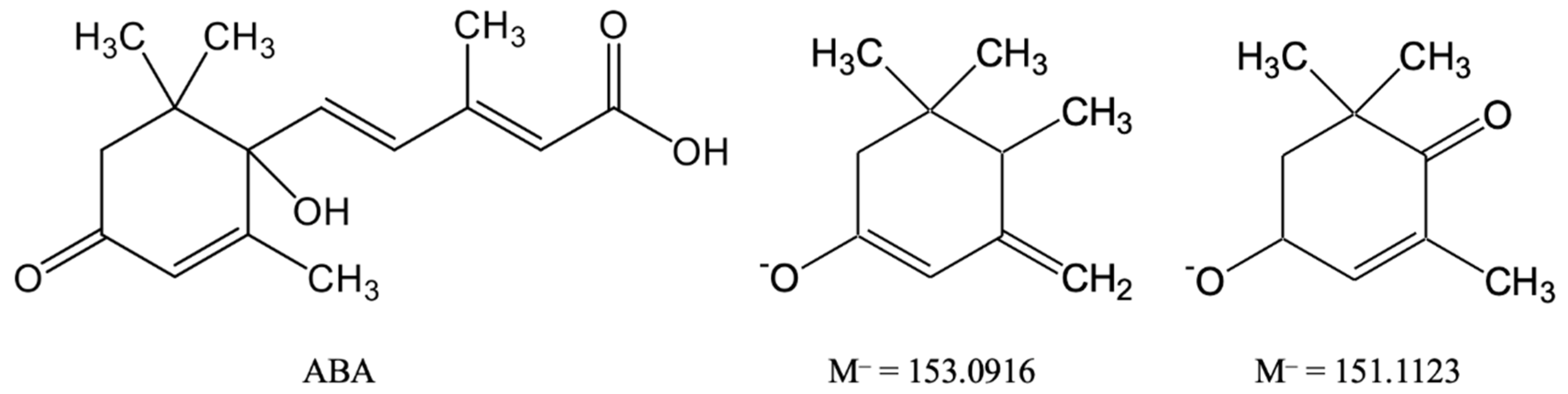
3.1. Identification and Isolation of Metabolite I
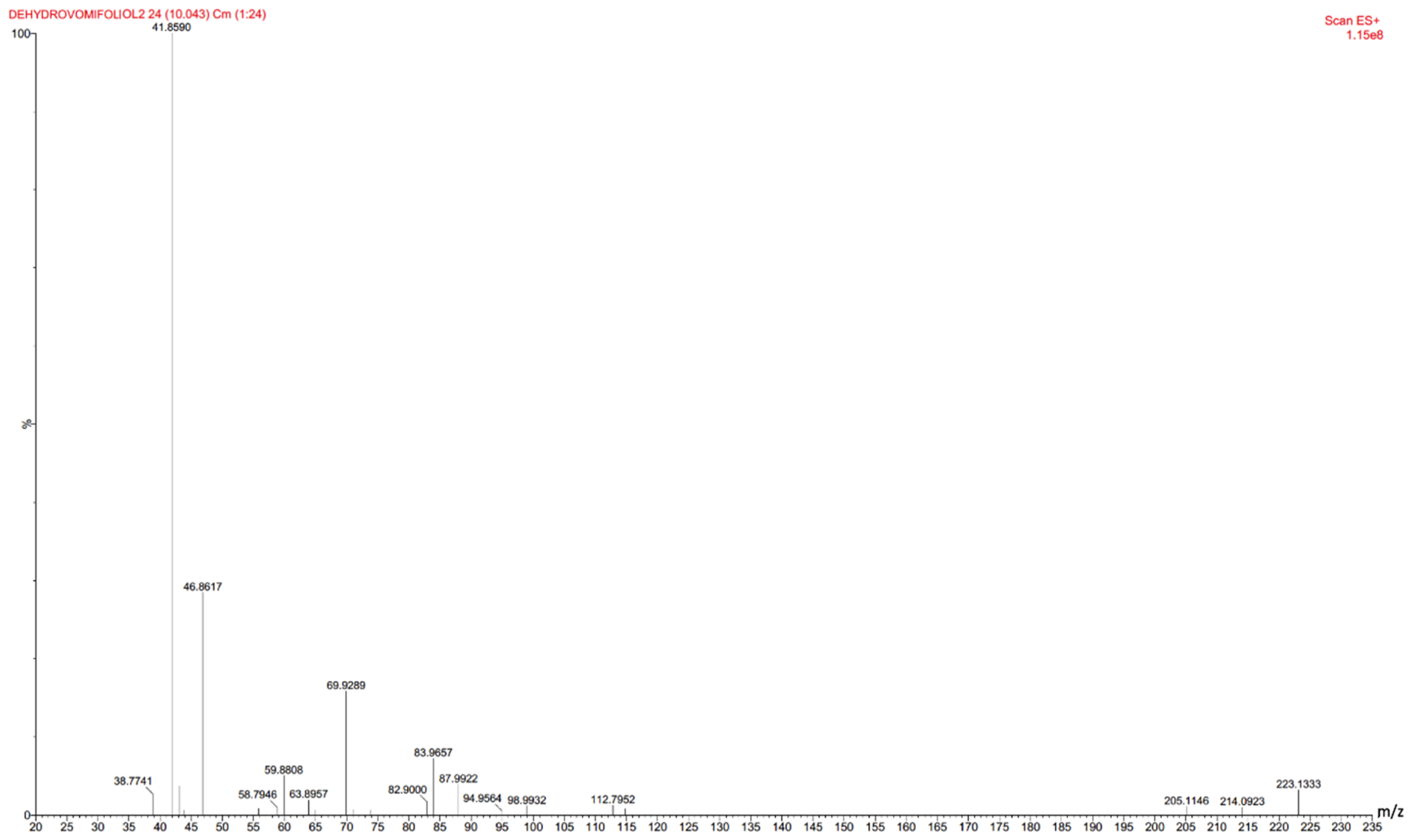
3.2. Chromatography-Mass Spectrometric Measurements
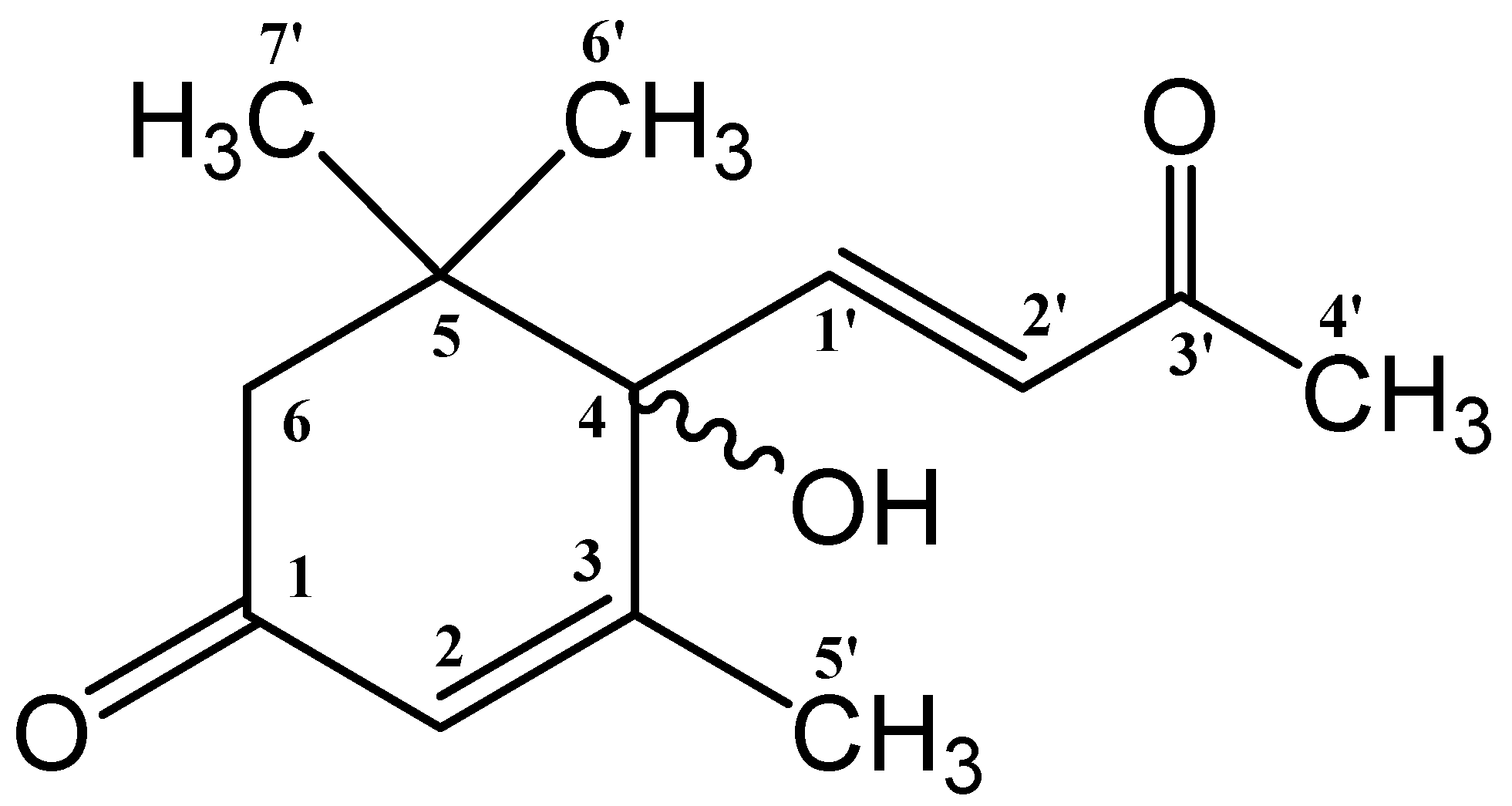
3.3. NMR Spectroscopy
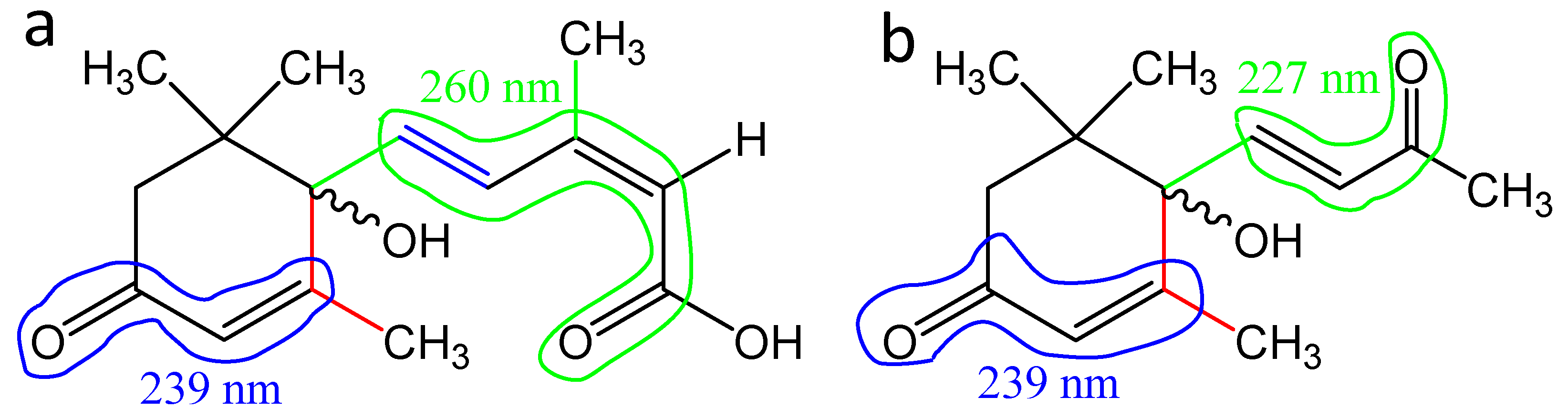
3.4. Optical Spectroscopy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeevaart, J.A.D.; Creelman, R.A. Metabolism and Physiology of Abscisic Acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 439–473. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic Acid Biosynthesis and Catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling and functions in plants. J. Integr. Plant. Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef]
- Cutler, A.J.; Krochko, J.E. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478. [Google Scholar] [CrossRef]
- Hartung, W.; Sauter, A.; Turner, N.; Fillery, I.; Heilmeier, H. Abscisic acid in soils: What is its function and which factors and mechanisms influence its concentration? Plant Soil 1996, 184, 105–110. [Google Scholar] [CrossRef]
- Hartung, W. The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct. Plant Biol. 2010, 37, 806–812. [Google Scholar] [CrossRef]
- Olds, C.L.; Glennon, E.K.K.; Luckhart, S. Abscisic acid: New perspectives on an ancient universal stress signaling molecule. Microbes Infect. 2018, 20, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Negri, P.; Maggi, M.D.; Ramirez, L.; De Feudis, L.; Szwarski, N.; Quintana, S.; Equaras, M.J.; Lamarttina, L. Abscisic acid enhances the immune response in Apis mellifera and contributes to the colony fitness. Apidologie 2015, 46, 542–557. [Google Scholar] [CrossRef]
- Magnone, M.; Ameri, P.; Salis, A.; Andraghetti, G.; Emionite, L.; Murialdo, G.; De Flora, A.; Zocchi, E. Microgram amounts of abscisic acid in fruit extracts improve glucose tolerance and reduce insulinemia in rats and in humans. FASEB J. 2015, 29, 4783–4893. [Google Scholar] [CrossRef] [PubMed]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvares, D.; Abdala, G. Endophytic bacteria in sunflower (Helianthus annuus L.): Isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 2007, 76, 1145–1152. [Google Scholar] [CrossRef]
- Cohen, C.; Travaglia, N.; Bottini, R.; Piccoli, N. Participation of abscisic acid and gibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botany 2009, 87, 455–462. [Google Scholar] [CrossRef]
- Piccoli, P.; Lucangeli, D.; Schneider, G.; Bottini, R. Hydrolysis of [17,17-2H2] Gibberellin A20-Glucoside and [17,17-2H2] Gibberellin A20-glucosyl ester by Azospirillum lipoferum cultured in a nitrogen-free biotin-based chemically-defined medium. Plant Growth Regul. 1997, 23, 179–182. [Google Scholar] [CrossRef]
- Sgroy, V.; Cassán, F.; Masciarelli, O.; Del Papa, M.; Lagares, A.; Luna, V. Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 2009, 85, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Crocoll, C.; Kettner, J.; Dorffling, K. Abscisic acid in saprophytic and parasitic species of fungi. Phytochemistry 1991, 30, 1059–1060. [Google Scholar] [CrossRef]
- Schmidt, K.; Pflugmacher, M.; Klages, S.; Mäser, A.; Mock, A.; Stahl, D. Accumulation of the hormone abscisic acid (ABA) at the infection site of the fungus Cercospora beticola supports the role of ABA as a repressor of plant defence in sugar beet. Mol. Plant Pathol. 2009, 9, 661–673. [Google Scholar] [CrossRef]
- Spence, C.; Bias, H. Role of plant growth regulators as chemical signals in plant-microbe interactions: A double-edged sword. Curr. Opin. Plant Biol. 2015, 27, 52–58. [Google Scholar] [CrossRef]
- Davies, W.J.; Zhang, J. Root signals and the regulation of growth and development of plants in drying soil. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 55–76. [Google Scholar] [CrossRef]
- Dodd, I.C.; Puértolas, J.; Huber, K.; Pérez-Pérez, J.G.; Wright, H.R.; Blackwell, M.S. The importance of soil drying and re-wetting in crop phytohormonal and nutritional responses to deficit irrigation. J. Exp. Bot. 2015, 66, 2239–2252. [Google Scholar] [CrossRef]
- Mulkey, T.J.; Evans, M.L.; Kuzmanoff, K.M. The kinetics of abscisic acid action on root growth and gravitropism. Planta 1983, 157, 150–157. [Google Scholar] [CrossRef]
- Pilet, P.E.; Rebeaud, J.E. Effect of abscisic acid on growth and indolyl-3- acetic acid levels in maize roots. Plant Sci. Lett. 1983, 31, 117–122. [Google Scholar] [CrossRef]
- Sharp, R.E.; LeNoble, M.E. ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 2002, 53, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Kutschera, U.; Schopfer, P. Effect of auxin and abscisic acid on cell wall extensibility in maize coleoptiles. Planta 1986, 167, 527–535. [Google Scholar] [CrossRef]
- Gaciarrubio, A.; Legaria, J.P.; Covarrubias, A.A. Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 1997, 203, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Henfling, J.W.D.M.; Bostock, R.; Kuc, J. Effect of abscisic acid on rishtin and lubimin accumulation and resistance to Phytophtora binfestans and Cladosporium cucumerinum in potato tuber tissue slices. Phytopathology 1980, 70, 1074–1078. [Google Scholar] [CrossRef]
- Mohr, P.G.; Cahill, D.M. Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct. Plant Biol. 2003, 30, 461–469. [Google Scholar] [CrossRef]
- Ward, E.W.B.; Cahill, D.M.; Bhattacharyya, M.K. Abscisic acid suppression of phenylalanine ammonia-lyase activity and mRNA and resistance of soybeans to Phytophthora megasperma f.sp. glycinea. Plant Physiol. 1989, 91, 23–27. [Google Scholar] [CrossRef]
- Asselbergh, B.; Achuo, A.E.; Höfte, M.; Van Gijsegem, F. Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi. Mol. Plant Pathol. 2008, 9, 11–24. [Google Scholar] [CrossRef]
- Dodd, I.; Zinovkina, N.; Safronova, V.; Belimov, A. Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 2010, 157, 361–379. [Google Scholar] [CrossRef]
- Belimov, A.A.; Dodd, I.C.; Safronova, V.I.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Davies, W.J.; Tikhonovich, I.A. Rhizobacteria that produce auxins and contain ACC deaminase decrease amino acid concentrations in the rhizosphere and improve growth and yield of well-watered and water-limited potato (Solanum tuberosum). Ann. Appl. Biol. 2015, 167, 11–25. [Google Scholar] [CrossRef]
- Zúñiga, A.; Poupin, M.J.; Donoso, R.; Ledger, T.; Guiliani, N.; Gutiérrez, R.A.; González, B. Quorum Sensing and Indole-3-Acetic Acid Degradation Play a Role in Colonization and Plant Growth Promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol. Plant. Microbe Interact. 2013, 26, 546–553. [Google Scholar] [CrossRef]
- Leveau, J.H.J.; Gerards, S. Discovery of a bacterial gene cluster for catabolism of the plant hormone indole 3-acetic acid. FEMS Microbiol. Ecol. 2008, 65, 238–250. [Google Scholar] [CrossRef]
- Olesen, M.R.; Jochimsen, B.U. Identification of enzymes involved in indole-3-acetic acid degradation. Plant Soil 1996, 186, 143–149. [Google Scholar] [CrossRef]
- Taylor, J.; Zaharia, L.; Chen, H.; Anderson, E.; Abrams, S. Biotransformation of adenine and cytokinins by the rhizobacterium Serratia proteamaculans. Phytochemistry 2006, 67, 1887–1894. [Google Scholar] [CrossRef]
- Cassán, F.; Bottini, R.; Schneider, G.; Piccoli, P. Azospirillum brasilense and Azospirillum lipoferum hydrolyze conjugates of GA20 and metabolize the resultant aglycones to GA1 in seedlings of rice dwarf mutants. Plant Physiol. 2001, 125, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Yen, K.M.; Serdar, C.M. Genetics of naphthalene catabolism in pseudomonads. Crit. Rev. Microbiol. 1988, 15, 247–268. [Google Scholar] [CrossRef] [PubMed]
- Sazonova, O.I.; Izmalkova, T.Y.; Kosheleva, I.A.; Boronin, A.M. Salicylate degradation by Pseudomonas putida strains not involving the “Classical” nah2 operon. Microbiology 2008, 77, 710–716. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Glick, B.R.; Rossi, M.J. Isolation and characterization of novel soil- and plant-associated bacteria with multiple phytohormone-degrading activities using a targeted methodology. Access Microbiol. 2019, 1, e000053. [Google Scholar] [CrossRef]
- Lievens, L.; Pollier, J.; Goossens, A.; Beyaert, R.; Staal, J. Abscisic Acid as Pathogen Effector and Immune Regulator. Front. Plant Sci. 2017, 8, 587. [Google Scholar] [CrossRef]
- Hasegawa, S.; Poling, S.; Mayer, V.; Bennett, R. Metabolism of abscisic acid: Bacterial conversion to dehydrovomifoliol and vomifoliol dehydrogenase activity. Phytochemistry 1984, 23, 2769–2771. [Google Scholar] [CrossRef]
- Belimov, A.A.; Dodd, I.C.; Safronova, V.I.; Dumova, V.A.; Shaposhnikov, A.I.; Ladatko, A.G.; Davies, W.J. Abscisic acid metabolizing rhizobacteria decrease ABA concentrations in planta and alter plant growth. Plant Physiol. Biochem. 2014, 7, 84–91. [Google Scholar] [CrossRef]
- Shevchenko, V.P.; Nagaev, I.Y.; Shaposhnikov, A.I.; Shevchenko, K.V.; Belimov, A.A.; Batasheva, S.N.; Gogoleva, N.E.; Myasoedov, N.F. Synthesis and Testing of Abscisic Acid with Predominant Replacement of Protium Atoms by Tritium in the Cyclohexene Moiety. Dokl. Chem. 2018, 483, 268–271. [Google Scholar] [CrossRef]
- Zaman, F.; Iwasaki, A.; Suenaga, K.; Kato-Noguchi, H. Two allelopathic substances from Paspalum commersonii Lam. Acta Agric. Scand. Soil Plant Sci. 2018, 68, 342–348. [Google Scholar]
- Schievano, E.; Morelato, E.; Facchin, C.; Mammi, C. Characterization of Markers of Botanical Origin and Other Compounds Extracted from Unifloral Honeys. J. Agric. Food Chem. 2013, 61, 1747–1755. [Google Scholar] [CrossRef] [PubMed]
- Lasch, P.; Naumann, D. Infrared spectroscopy in microbiology. In Encyclopedia of Analytical Chemistry; Meyers, R.A., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Kamnev, A.A.; Tugarova, A.V.; Dyatlova, Y.A.; Tarantilis, P.A.; Grigoryeva, O.P.; Fainleib, A.M.; De Luca, S. Methodological effects in Fourier transform infrared (FTIR) spectroscopy: Implications for structural analyses of biomacromolecular samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 193, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Tugarova, A.V.; Dyatlova, Y.A.; Kenzhegulov, O.A.; Kamnev, A.A. Poly-3-hydroxybutyrate synthesis by different Azospirillum brasilense strains under varying nitrogen deficiency: A comparative in-situ FTIR spectroscopic analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 252, 119458. [Google Scholar] [CrossRef]
- Takasugi, M.; Anetai, M.; Katsui, N.; Masamune, T. The occurrence of vomifoliol, dehydrovomifoliol and dehydrophaseic acid in the roots of “kidney bean” (Phaseolus vulgaris L.). Chem. Lett. 1973, 2, 245–248. [Google Scholar] [CrossRef]
- Häusler, M.; Montag, A. Isolation, identification and quantitative determination of the norisoprenoid (S)-(+)-dehydrovomifoliol in honey. Z. Lebensm. Unters. Forsch. 1989, 189, 113–115. [Google Scholar] [CrossRef]
- Bellamy, L.J. The Infra-Red Spectra of Complex Molecules; John Wiley & Sons: Hoboken, NJ, USA, 1957. [Google Scholar]
- Brown, D.W.; Floyd, A.J.; Sainsbury, M. Organic Spectroscopy; Wiley: Chichester, UK, 1988; 258p. [Google Scholar]
- Nhi-Cong, L.T.; Mikolasch, A.; Awe, S.; Sheikhany, H.; Klenk, H.-P.; Schauer, F. Oxidation of aliphatic, branched chain, and aromatic hydrocarbons by Nocardia cyriacigeorgica isolated from oil-polluted sand samples collected in the Saudi Arabian Desert. J. Basic Microbiol. 2010, 50, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Minerdi, D.; Zgrablic, I.; Sadeghi, S.J.; Gilardi, G. Identification of a novel Baeyer-Villiger monooxygenase fromAcinetobacter radioresistens: Close relationship to the Mycobacterium tuberculosis prodrug activator EtaA. Microb. Biotechnol. 2012, 5, 700–716. [Google Scholar] [CrossRef]
- Gogoleva, N.E.; Nikolaichik, Y.A.; Ismailov, T.T.; Khlopko, Y.A.; Dmitrieva, S.A.; Konnova, T.A.; Ermekkaliev, T.S.; Safronova, V.I.; Belimov, A.A.; Gogolev, Y.V. Complete Genome Sequence of Abscisic Acid-Metabolizing Rhizobacterium Rhodococcus sp. Strain P1Y. Microbiol. Resour. Announc. 2019, 8, e01591-18. [Google Scholar] [CrossRef] [PubMed]
- Gogoleva, N.E.; Nikolaichik, Y.A.; Ismailov, T.T.; Khlopko, Y.A.; Dmitrieva, S.A.; Konnova, T.A.; Ermekkaliev, T.S.; Safronova, V.I.; Belimov, A.A.; Gogolev, Y.V. Complete genome sequence of the abscisic acid-utilizing strain Novosphingobium sp. P6W. 3 Biotech 2019, 9, 94. [Google Scholar] [CrossRef] [PubMed]


| No. of Fraction | Starting Solvents Ratio, Vol % | Final Solvents Ratio, Vol % | Radioactivity, cpm µL−1 |
|---|---|---|---|
| 1 | A 100 | A-B 90:10 | 10.1 1 |
| 2 | A-B 90:10 | A-B 80:20 | 7.0 |
| 3 | A-B 80:20 | A-B 70:30 | 2.2 |
| 4 | A-B 70:30 | A-B 60:40 | 2.5 |
| 5 | A-B 60:40 | A-B 50:50 | 1.0 |
| 6 | A-B 50:50 | A-B 40:60 | - |
| 7 | A-B 40:60 | A-B 30:70 | - |
| 8 | A-B 30:70 | A-B 20:80 | - |
| 9 | A-B 20:80 | A-B 10:90 | 3.5 |
| 10 | A-B 10:90 | B 100 | 34.5 |
| 11 | B 100 | B-C 50:50 | 168.2 |
| 12 | B-C 50:50 | C 100 | 164.3 |
| 13 | C 100 | C 100 | 5.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuzikhin, O.S.; Gogoleva, N.E.; Shaposhnikov, A.I.; Konnova, T.A.; Osipova, E.V.; Syrova, D.S.; Ermakova, E.A.; Shevchenko, V.P.; Nagaev, I.Y.; Shevchenko, K.V.; et al. Rhizosphere Bacterium Rhodococcus sp. P1Y Metabolizes Abscisic Acid to Form Dehydrovomifoliol. Biomolecules 2021, 11, 345. https://doi.org/10.3390/biom11030345
Yuzikhin OS, Gogoleva NE, Shaposhnikov AI, Konnova TA, Osipova EV, Syrova DS, Ermakova EA, Shevchenko VP, Nagaev IY, Shevchenko KV, et al. Rhizosphere Bacterium Rhodococcus sp. P1Y Metabolizes Abscisic Acid to Form Dehydrovomifoliol. Biomolecules. 2021; 11(3):345. https://doi.org/10.3390/biom11030345
Chicago/Turabian StyleYuzikhin, Oleg S., Natalia E. Gogoleva, Alexander I. Shaposhnikov, Tatyana A. Konnova, Elena V. Osipova, Darya S. Syrova, Elena A. Ermakova, Valerii P. Shevchenko, Igor Yu. Nagaev, Konstantin V. Shevchenko, and et al. 2021. "Rhizosphere Bacterium Rhodococcus sp. P1Y Metabolizes Abscisic Acid to Form Dehydrovomifoliol" Biomolecules 11, no. 3: 345. https://doi.org/10.3390/biom11030345
APA StyleYuzikhin, O. S., Gogoleva, N. E., Shaposhnikov, A. I., Konnova, T. A., Osipova, E. V., Syrova, D. S., Ermakova, E. A., Shevchenko, V. P., Nagaev, I. Y., Shevchenko, K. V., Myasoedov, N. F., Safronova, V. I., Shavarda, A. L., Nizhnikov, A. A., Belimov, A. A., & Gogolev, Y. V. (2021). Rhizosphere Bacterium Rhodococcus sp. P1Y Metabolizes Abscisic Acid to Form Dehydrovomifoliol. Biomolecules, 11(3), 345. https://doi.org/10.3390/biom11030345







