Convolutional Neural Network-Based Artificial Intelligence for Classification of Protein Localization Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Deep Learning
2.3. Training
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| CNN | Convolutional neural network |
| DL | Deep learning |
| ELU | Exponential linear unit |
| FCN | Fully convolutional neural network |
| ReLU | Rectified linear unit |
| SGD | Stochastic gradient descent |
References
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356. [Google Scholar] [CrossRef]
- Neumann, B.; Held, M.; Liebel, U.; Erfle, H.; Rogers, P.; Pepperkok, R.; Ellenberg, J. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods 2006, 3, 385. [Google Scholar] [CrossRef] [PubMed]
- Loo, L.H.; Wu, L.F.; Altschuler, S.J. Image-based multivariate profiling of drug responses from single cells. Nat. Methods 2007, 4, 445. [Google Scholar] [CrossRef] [PubMed]
- Snijder, B.; Sacher, R.; Rämö, P.; Damm, E.M.; Liberali, P.; Pelkmans, L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 2009, 461, 520. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Jones, T.R.; Kang, I.H.; Wheeler, D.B.; Lindquist, R.A.; Papallo, A.; Sabatini, D.M.; Golland, P.; Carpenter, A.E. CellProfiler Analyst: Data exploration and analysis software for complex image-based screens. BMC Bioinform. 2008, 9, 482. [Google Scholar] [CrossRef]
- Rämö, P.; Sacher, R.; Snijder, B.; Begemann, B.; Pelkmans, L. CellClassifier: Supervised learning of cellular phenotypes. Bioinformatics 2009, 25, 3028–3030. [Google Scholar] [CrossRef] [PubMed]
- Held, M.; Schmitz, M.H.; Fischer, B.; Walter, T.; Neumann, B.; Olma, M.H.; Peter, M.; Ellenberg, J.; Gerlich, D.W. CellCognition: Time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods 2010, 7, 747. [Google Scholar] [CrossRef]
- Piccinini, F.; Balassa, T.; Szkalisity, A.; Molnar, C.; Paavolainen, L.; Kujala, K.; Buzas, K.; Sarazova, M.; Pietiainen, V.; Kutay, U.; et al. Advanced cell classifier: User-friendly machine-learning-based software for discovering phenotypes in high-content imaging data. Cell Syst. 2017, 4, 651–655. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef]
- Wainberg, M.; Merico, D.; Delong, A.; Frey, B.J. Deep learning in biomedicine. Nat. Biotechnol. 2018, 36, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Goecks, J.; Jalili, V.; Heiser, L.M.; Gray, J.W. How machine learning will transform biomedicine. Cell 2020, 181, 92–101. [Google Scholar] [CrossRef]
- Fioravanti, D.; Giarratano, Y.; Maggio, V.; Agostinelli, C.; Chierici, M.; Jurman, G.; Furlanello, C. Phylogenetic convolutional neural networks in metagenomics. BMC Bioinform. 2018, 19, 49. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.R.; Snoek, J.; Rinn, J.L. Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016, 26, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Quang, D.; Xie, X. DanQ: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016, 44, e107. [Google Scholar] [CrossRef]
- Xuan, P.; Pan, S.; Zhang, T.; Liu, Y.; Sun, H. Graph convolutional network and convolutional neural network based method for predicting lncRNA-disease associations. Cells 2019, 8, 1012. [Google Scholar] [CrossRef]
- Le, N.Q.K. Fertility-GRU: Identifying fertility-related proteins by incorporating deep-gated recurrent units and original position-specific scoring matrix profiles. J. Proteome Res. 2019, 18, 3503–3511. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Stringer, C.; Wang, T.; Michaelos, M.; Pachitariu, M. Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 2021, 18, 100–106. [Google Scholar] [CrossRef]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Ouyang, W.; Winsnes, C.F.; Hjelmare, M.; Cesnik, A.J.; Åkesson, L.; Xu, H.; Sullivan, D.P.; Dai, S.; Lan, J.; Jinmo, P.; et al. Analysis of the Human Protein Atlas Image Classification competition. Nat. Methods 2019, 16, 1254–1261. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- Xie, W.; Noble, J.A.; Zisserman, A. Microscopy cell counting and detection with fully convolutional regression networks. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2018, 6, 283–292. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Valkonen, M.; Kartasalo, K.; Liimatainen, K.; Nykter, M.; Latonen, L.; Ruusuvuori, P. Dual structured convolutional neural network with feature augmentation for quantitative characterization of tissue histology. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Venice, Italy, 22–29 October 2017; pp. 27–35. [Google Scholar]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Sullivan, D.P.; Winsnes, C.F.; Ãkesson, L.; Hjelmare, M.; Wiking, M.; Schutten, R.; Campbell, L.; Leifsson, H.; Rhodes, S.; Nordgren, A.; et al. Deep learning is combined with massive-scale citizen science to improve large-scale image classification. Nat. Biotechnol. 2018, 36, 820–828. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics; Teh, Y.W., Titterington, M., Eds.; Chia Laguna Resort, PMLR: Sardinia, Italy, 2010; Volume 9, pp. 249–256. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Chollet, F.; Rahman, F.; Lee, T.; de Marmiesse, G.; Zabluda, O.; chenta, M.S.; Santana, E.; McColgan, T.; Snelgrove, X.; Zhu, Q.S.; et al. Keras. 2015. Available online: https://github.com/fchollet/keras (accessed on 10 February 2021).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2016. Available online: https://arxiv.org/abs/1603.04467 (accessed on 10 February 2021).
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Sorensen, T.A. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skar. 1948, 5, 1–34. [Google Scholar]
- Xu, Y.Y.; Zhou, H.; Murphy, R.F.; Shen, H.B. Consistency and variation of protein subcellular location annotations. Proteins Struct. Funct. Bioinform. 2021, 89, 242–250. [Google Scholar] [CrossRef] [PubMed]
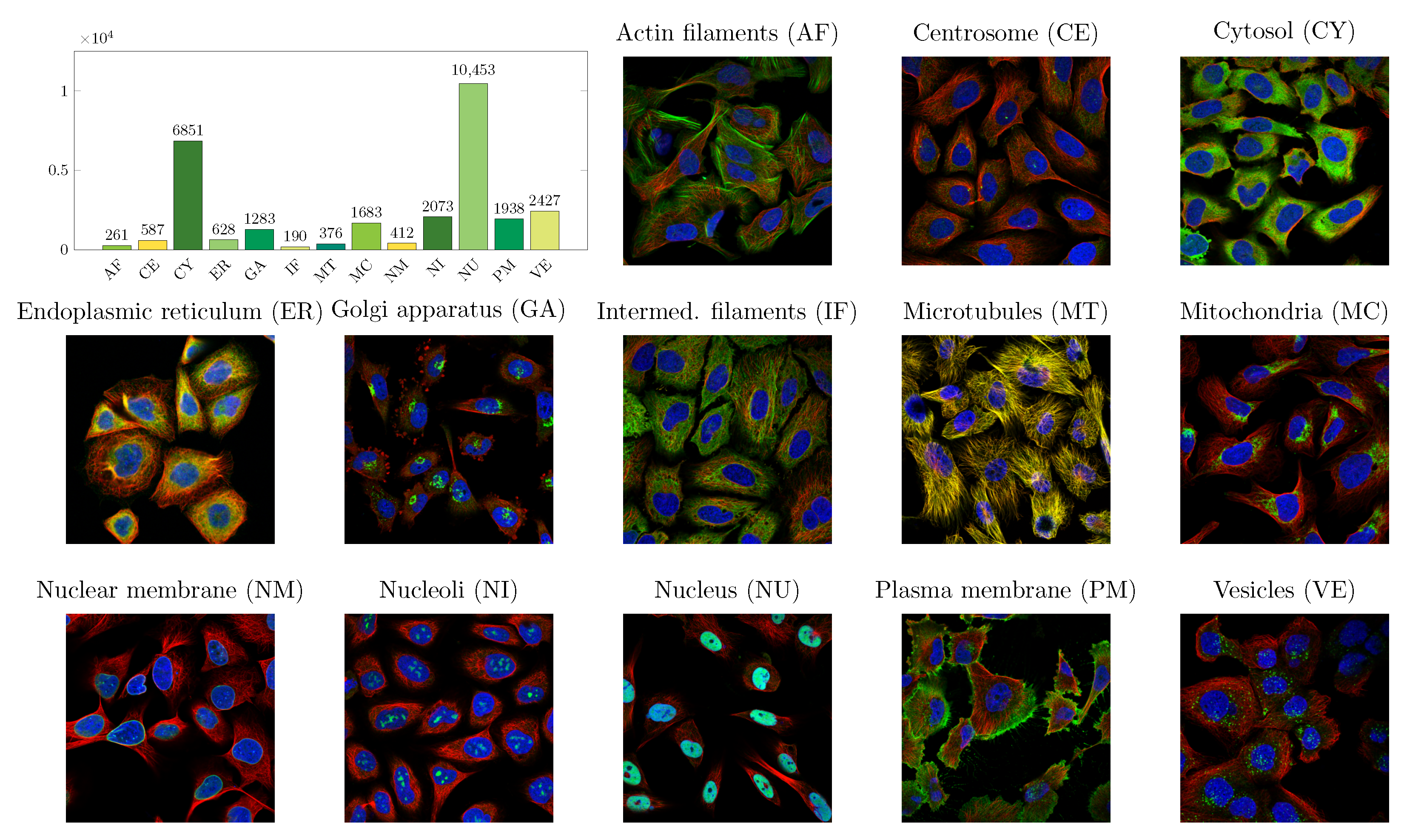
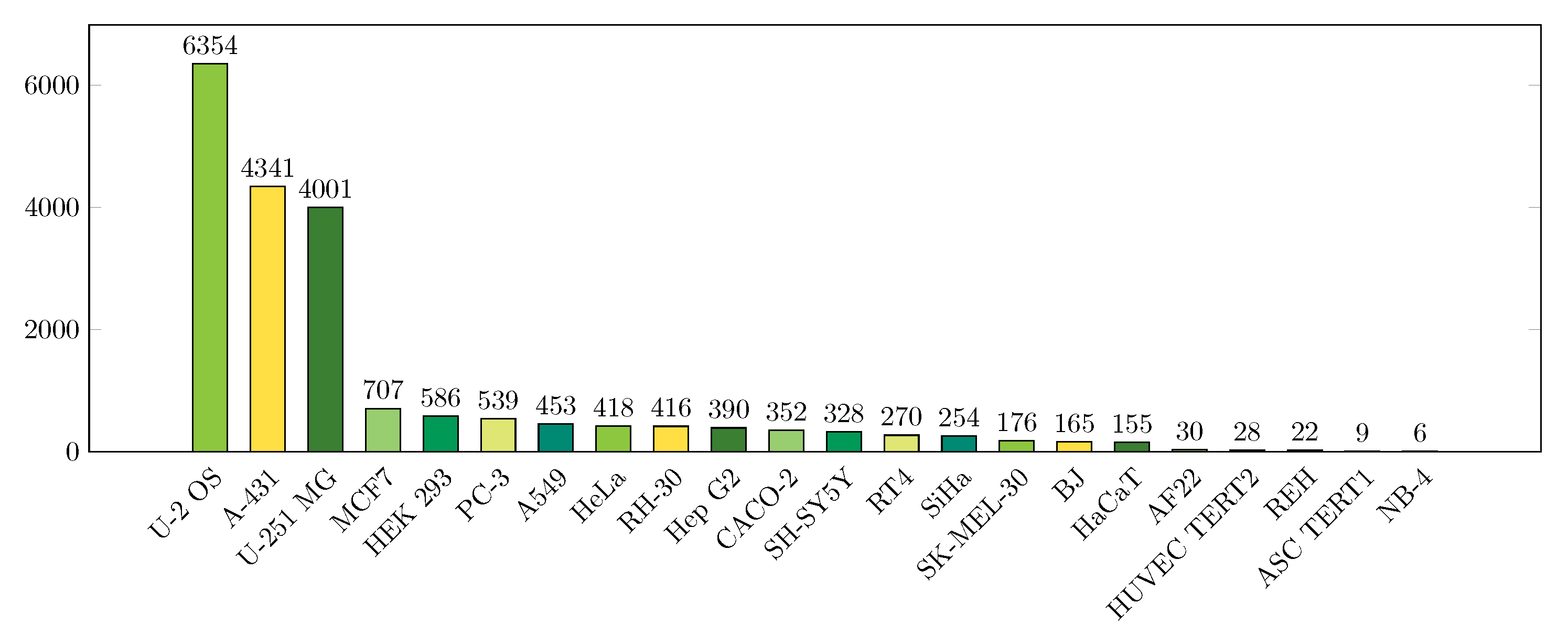
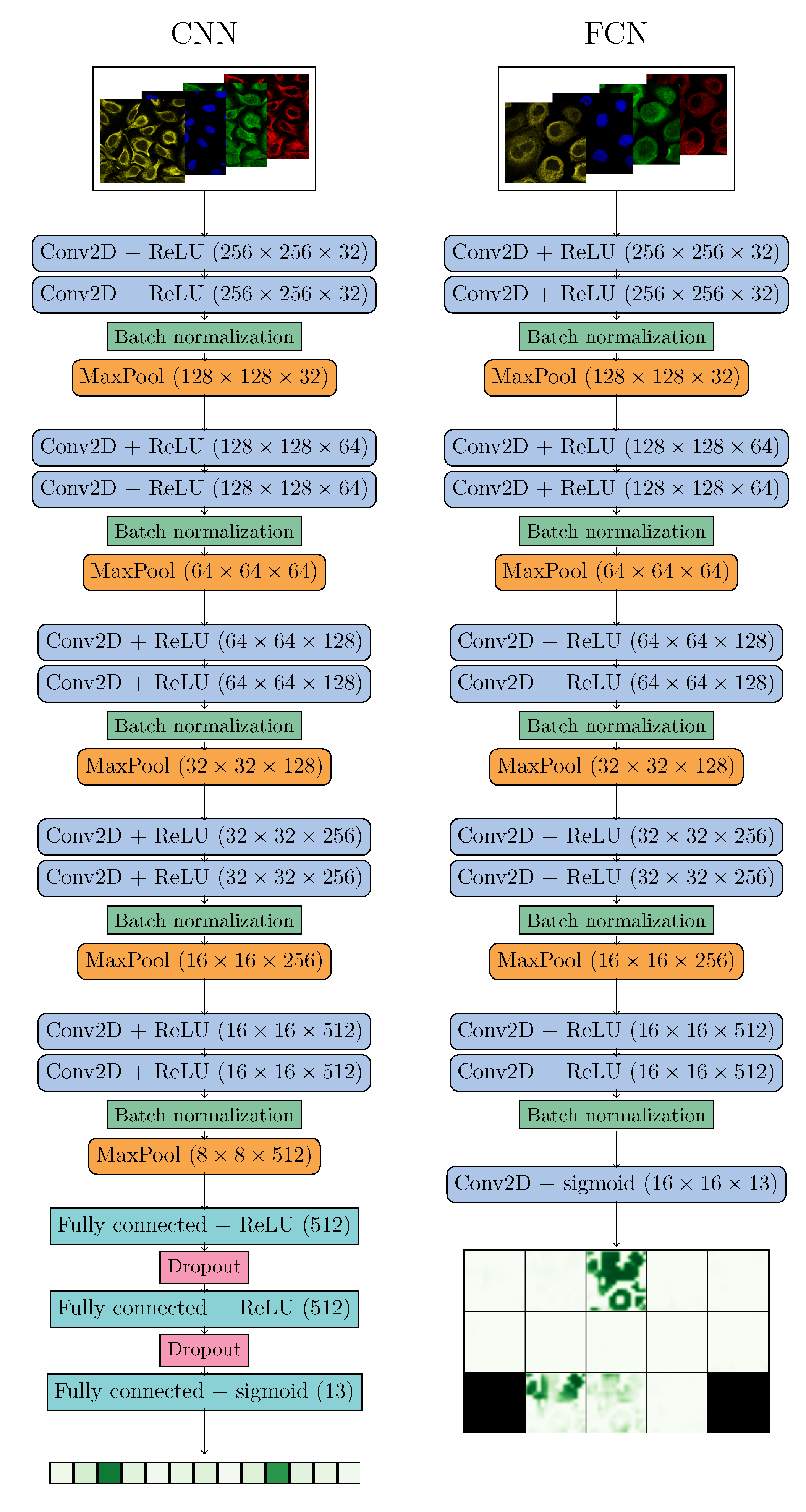
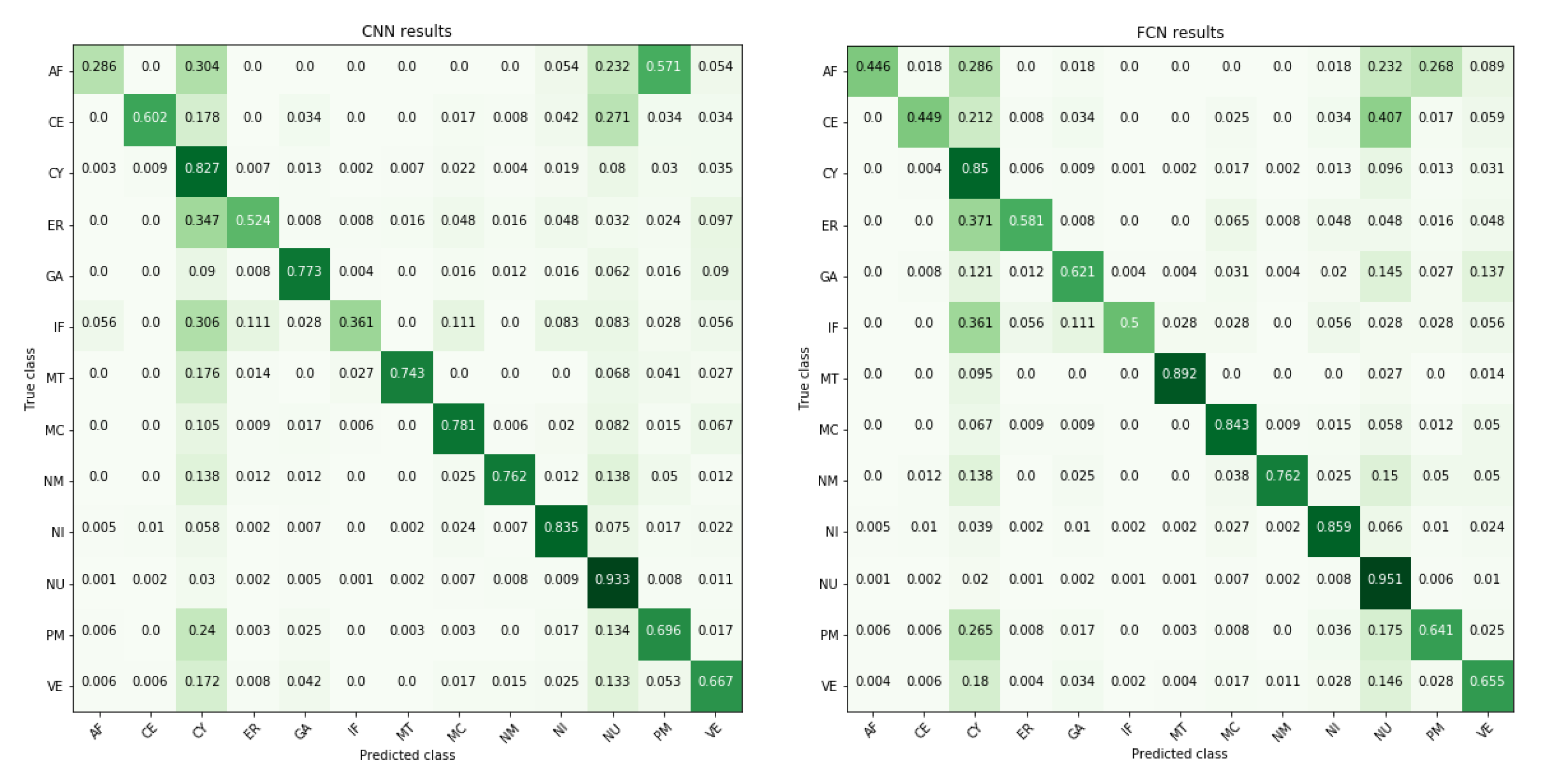
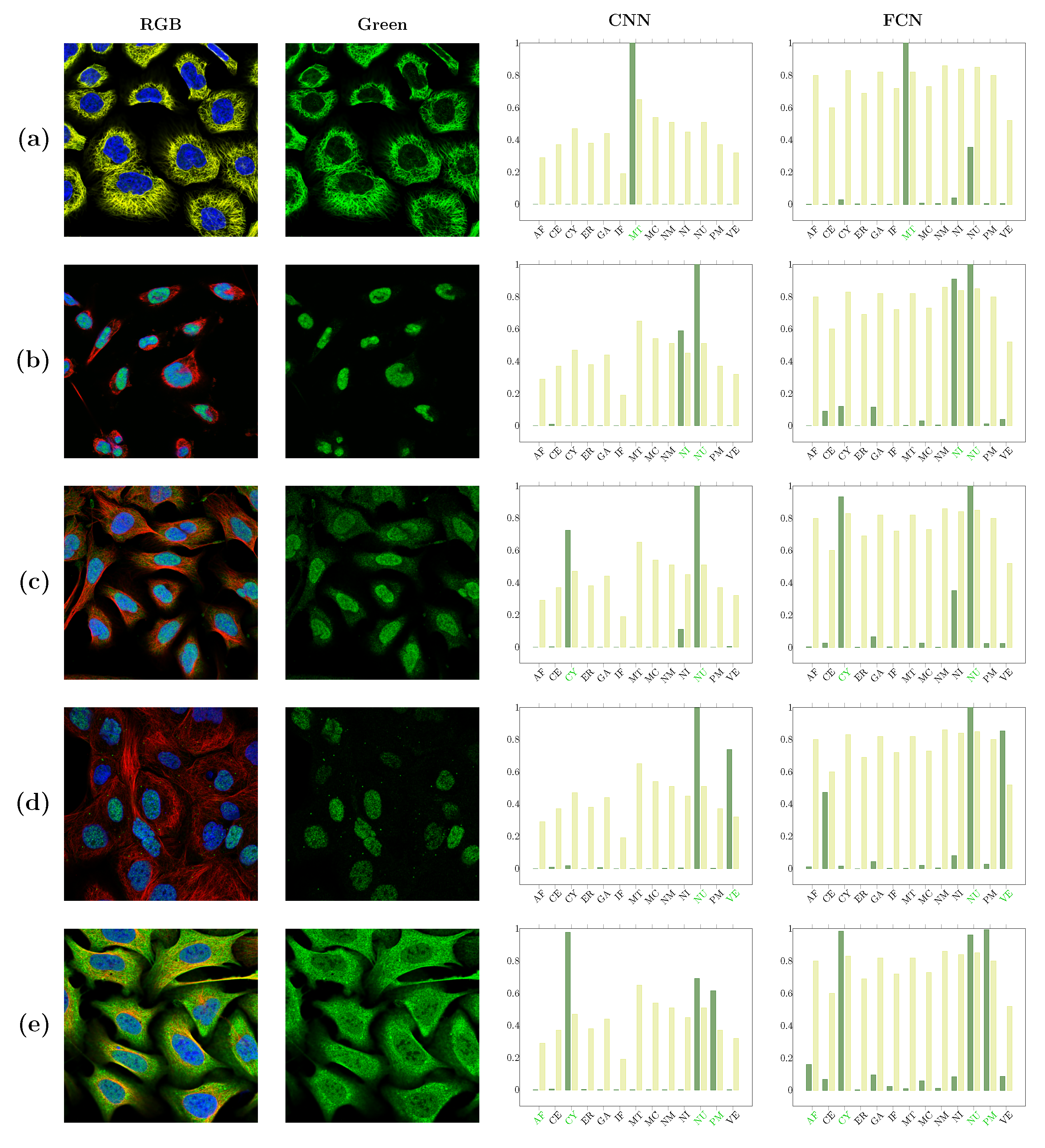
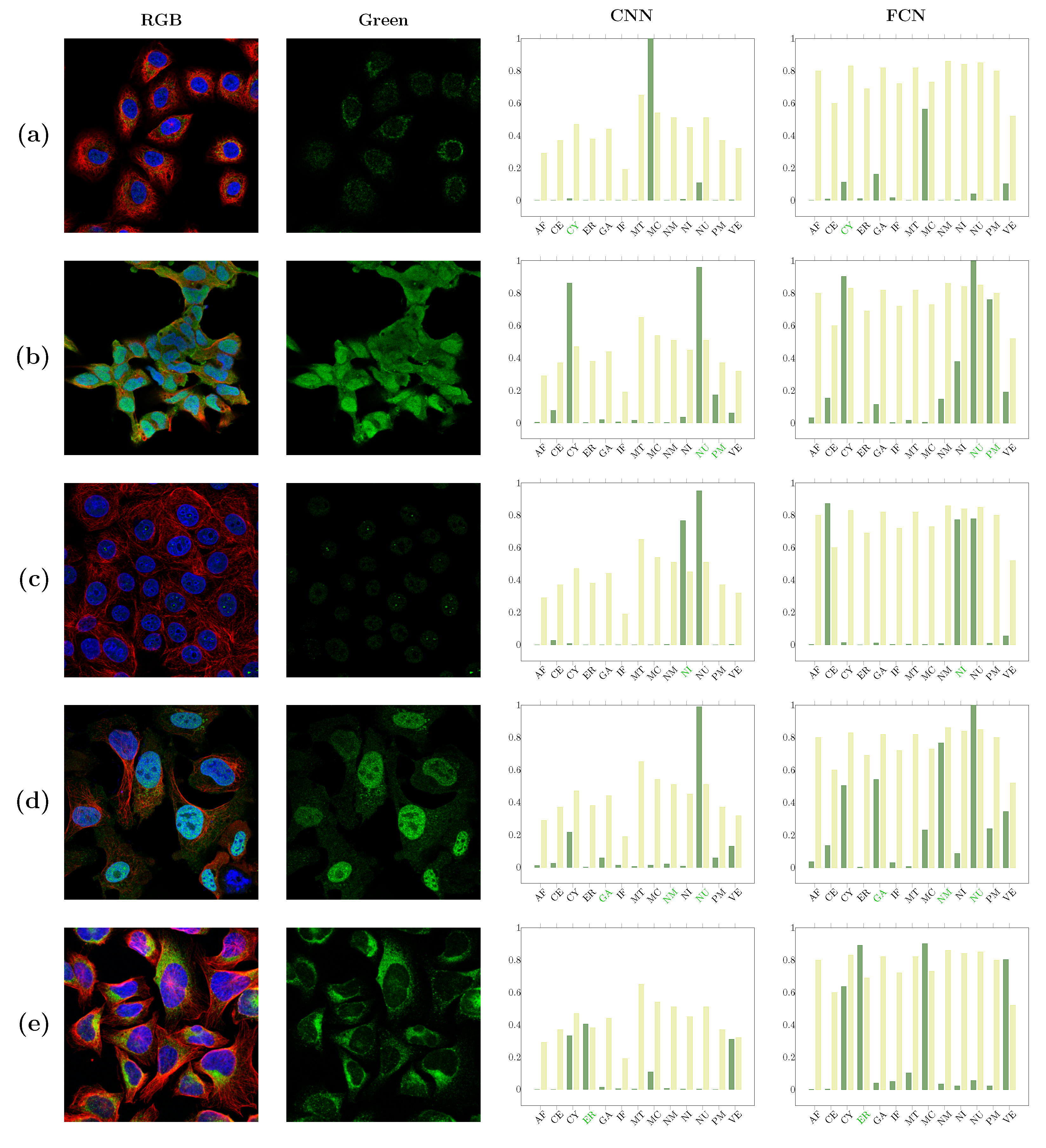
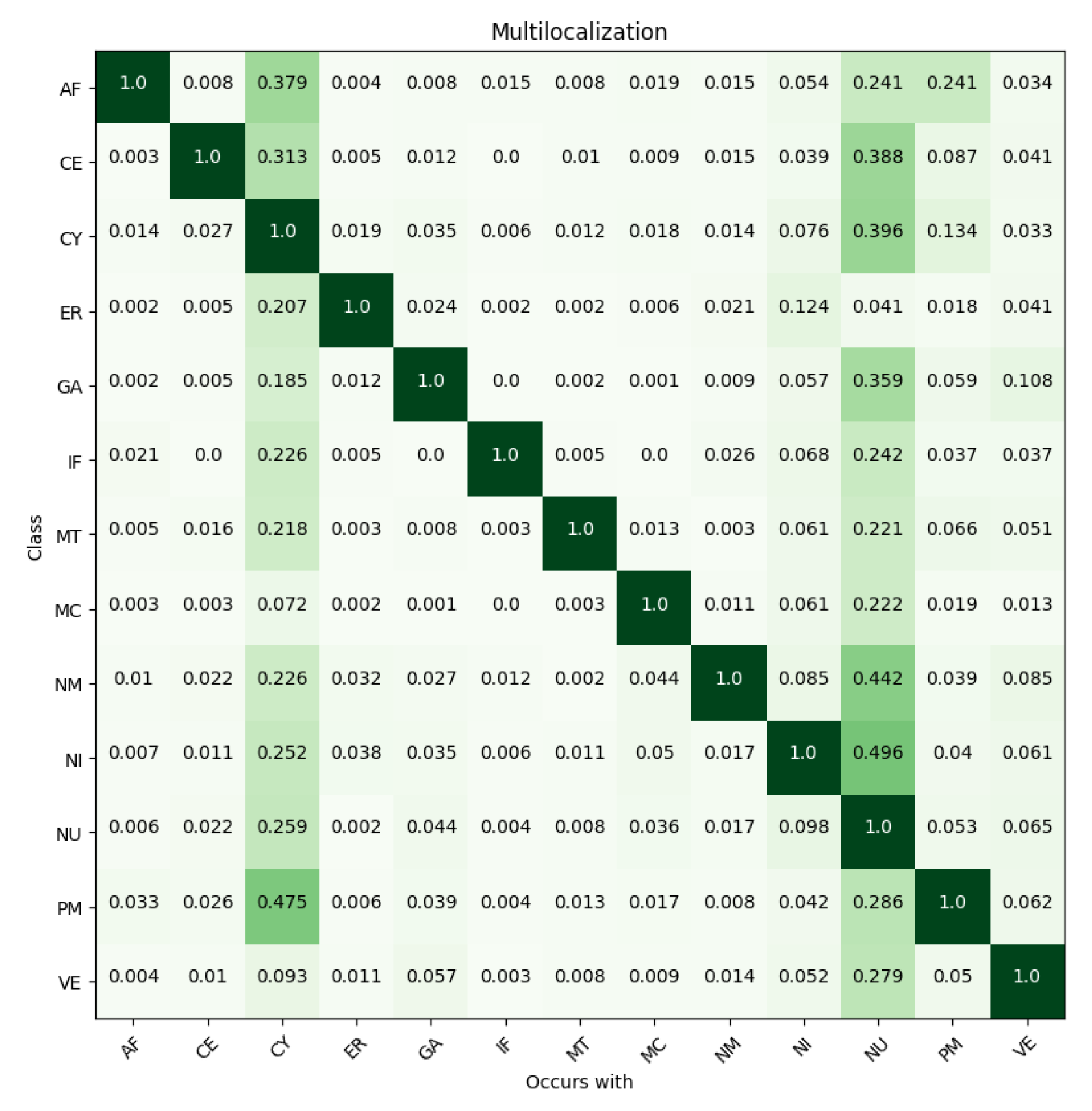


| Localizations | All Data (%) | Test Data (%) | Test Data (Amount) | Correct % (CNN) | Correct % (FCN) |
|---|---|---|---|---|---|
| 1 | 60.6 | 60.6 | 2424 | 92 | 90 |
| 2 | 33.3 | 32.8 | 1313 | 57 | 59 |
| 3 | 5.8 | 6.2 | 249 | 25 | 41 |
| 4 | 0.28 | 0.35 | 14 | 0 | 29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liimatainen, K.; Huttunen, R.; Latonen, L.; Ruusuvuori, P. Convolutional Neural Network-Based Artificial Intelligence for Classification of Protein Localization Patterns. Biomolecules 2021, 11, 264. https://doi.org/10.3390/biom11020264
Liimatainen K, Huttunen R, Latonen L, Ruusuvuori P. Convolutional Neural Network-Based Artificial Intelligence for Classification of Protein Localization Patterns. Biomolecules. 2021; 11(2):264. https://doi.org/10.3390/biom11020264
Chicago/Turabian StyleLiimatainen, Kaisa, Riku Huttunen, Leena Latonen, and Pekka Ruusuvuori. 2021. "Convolutional Neural Network-Based Artificial Intelligence for Classification of Protein Localization Patterns" Biomolecules 11, no. 2: 264. https://doi.org/10.3390/biom11020264
APA StyleLiimatainen, K., Huttunen, R., Latonen, L., & Ruusuvuori, P. (2021). Convolutional Neural Network-Based Artificial Intelligence for Classification of Protein Localization Patterns. Biomolecules, 11(2), 264. https://doi.org/10.3390/biom11020264







